Key Points
A recurrent 4-bp deletion in the NFKBIE gene is a common event during lymphomagenesis in various lymphoid malignancies.
The deletion occurs in 22.7% of PMBL cases and is associated with a particularly aggressive clinical disease course.
Abstract
We recently reported a truncating deletion in the NFKBIE gene, which encodes IκBε, a negative feedback regulator of NF-κB, in clinically aggressive chronic lymphocytic leukemia (CLL). Because preliminary data indicate enrichment of NFKBIE aberrations in other lymphoid malignancies, we screened a large patient cohort (n = 1460) diagnosed with different lymphoid neoplasms. While NFKBIE deletions were infrequent in follicular lymphoma, splenic marginal zone lymphoma, and T-cell acute lymphoblastic leukemia (<2%), slightly higher frequencies were seen in diffuse large B-cell lymphoma, mantle cell lymphoma, and primary central nervous system lymphoma (3% to 4%). In contrast, a remarkably high frequency of NFKBIE aberrations (46/203 cases [22.7%]) was observed in primary mediastinal B-cell lymphoma (PMBL) and Hodgkin lymphoma (3/11 cases [27.3%]). NFKBIE-deleted PMBL patients were more often therapy refractory (P = .022) and displayed inferior outcome compared with wild-type patients (5-year survival, 59% vs 78%; P = .034); however, they appeared to benefit from radiotherapy (P = .022) and rituximab-containing regimens (P = .074). NFKBIE aberrations remained an independent factor in multivariate analysis (P = .003) and when restricting the analysis to immunochemotherapy-treated patients (P = .008). Whole-exome sequencing and gene expression profiling verified the importance of NF-κB deregulation in PMBL. In summary, we identify NFKBIE aberrations as a common genetic event across B-cell malignancies and highlight NFKBIE deletions as a novel poor-prognostic marker in PMBL.
Introduction
Deregulated NF-κB signaling is a hallmark of most lymphoid malignancies, and recurrent mutations in NF-κB transcription factors or upstream signaling components, such as CD79B, CARD11, MYD88, or TNFAIP3, are common findings in B-cell neoplasms.1-3 Genetic aberrations in both the canonical and noncanonical NF-κB pathway are known to lead to NF-κB activation.4 However, the full compendium of NF-κB pathway genes affected by recurrent mutations in lymphoid malignancies remains to be elucidated.
Recently, we reported a 4-bp truncating mutation in the NFKBIE gene, which encodes IκBε, a negative regulator of NF-κB in normal B cells, in chronic lymphocytic leukemia (CLL).5-8 NFKBIE deletions were enriched in clinically aggressive CLL patients (7% to 8%) and associated with shorter time to first treatment. At the functional level, NFKBIE-deleted cases showed reduced IκBε levels and decreased p65 inhibition, along with increased phosphorylation and nuclear translocation of p65, compared with wild-type patients. Similarly, loss of IκBε was reported to lead to constitutive NF-κB transcriptional activation in C57B16 mice, which in turn resulted in increased B-cell proliferation and survival.9
Preliminary data from limited patient series indicate an increased frequency of NFKBIE aberrations in other lymphoid malignancies, such as relapsed/refractory diffuse large B-cell lymphomas (DLBCL) and primary mediastinal large B-cell lymphomas (PMBL).10,11 By screening a large cohort of 1460 patients with different lymphoid neoplasms, we provide further evidence that NFKBIE deletions are a common event during lymphomagenesis and highlight an enrichment in PMBL linked to a particularly poor outcome.
Study design
Patients
In total, 1460 patients diagnosed with different lymphoid malignancies were included from collaborating institutions in Denmark, Germany, Greece, France, Spain, Sweden, and the United Kingdom. The study cohort comprised DLBCL (n = 520), follicular lymphoma (n = 225), PMBL (n = 203),12,13 mantle cell lymphoma (n = 189), splenic marginal zone lymphoma (n = 175),14 T-cell acute lymphoblastic leukemia (n = 94), primary central nervous system lymphoma (n = 34), classical Hodgkin lymphoma (cHL; n = 11), and small lymphocytic lymphoma (n = 9). All samples were collected before start of therapy and diagnosed according to the WHO classification.15 Written consent was obtained in accordance with the Declaration of Helsinki and with ethical approval obtained from the local ethics committees.
Mutational analysis of NFKBIE
The entire coding region of NFKBIE was investigated by Sanger sequencing in a representative subset (n = 292) of the study cohort, while the 4-bp deletion hotspot located in exon 1 was analyzed either by Sanger sequencing (n = 350) or GeneScan analysis (n = 807) in the remaining cases (detailed in supplemental Methods, available on the Blood Web site). In addition, targeted deep sequencing was performed in 44 PMBL and 22 DLBCL samples using the Nextera XT kit (Illumina) for library preparation (supplemental Methods).6,7,16,17 For the cHL samples, Hodgkin and Reed/Sternberg (HRS) cells were microdissected and sequenced as detailed in supplemental Methods.
Whole-exome sequencing
Whole-exome sequencing (WES) was performed in 7 matched tumor or germline PMBL cases (detailed in supplemental Methods); in addition, previously reported WES data on 7 PMBLs were included.10,18 WES data have been deposited at the European Nucleotide Archive (http://www.ebi.ac.uk/ena), which is hosted at the European Bioinformatics Institute, under accession number PRJEB15361.
Gene expression profiling
To assess gene expression differences in NFKBIE-deleted (n = 8) and NFKBIE wild-type (n = 21) PMBL, we applied the NanoString PanCancer Pathways Panel to quantify transcript levels of 770 genes representing 13 canonical cancer pathways plus 30 selected genes reported to be important in PMBL and/or the NF-κB pathway8,19 (described in supplemental Methods). Gene expression data have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus database and are accessible through Gene Expression Omnibus Series accession number GSE86815.
Statistical analysis
Pairwise comparisons of variables for exploratory purposes were performed using the Mann-Whitney test or the χ2 test. Overall survival (OS) was calculated from time of diagnosis until date of death or last follow-up. Kaplan-Meier analysis was performed to construct survival curves, and the log-rank test was applied to evaluate differences between subgroups. Cox regression analysis was applied for multivariate analysis. All analyses were carried out using the software package SPSS Version 23.0 (IBM, Armonk, NY).
Results and discussion
Overall, 86 NFKBIE mutations were identified in 84 of 1460 patients (5.8%). All but 4 patients (L389P, P440L, and L495V missense mutations and a 4-bp splice-site deletion) exhibited a heterozygous 4-bp deletion (delTTAC), known to result in a truncated protein.6 Two PMBL cases with the recurrent 4-bp deletion showed additional NFKBIE frameshift deletions (a 7-bp and 38-bp deletion). The somatic nature of the 4-bp deletion has previously been confirmed.5-7,11,18 Using a cutoff of >5% for the mutant allele, GeneScan analysis and deep sequencing revealed a high concordance of allele frequencies between both techniques (n = 10, r = 0.80, P = .005), with variant allele frequencies ranging from 5% to 62% in NFKBIE-deleted cases. This finding indicates that NFKBIE mutations may occur at different time points of lymphomagenesis (supplemental Tables 1 and 2; Figure 1).
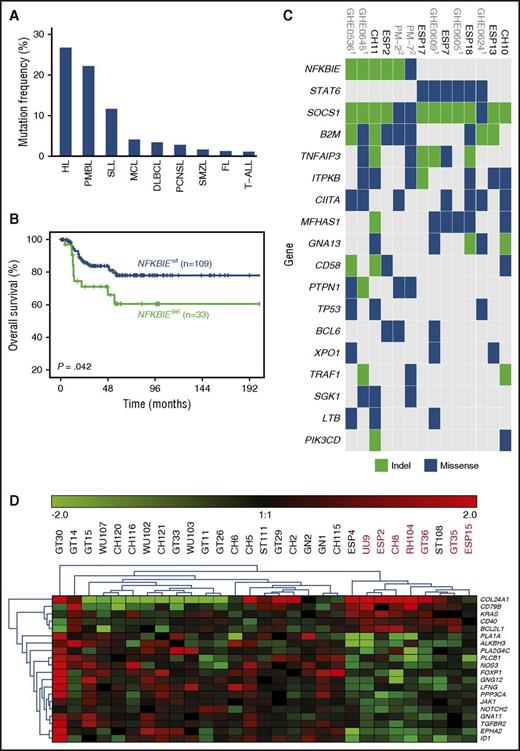
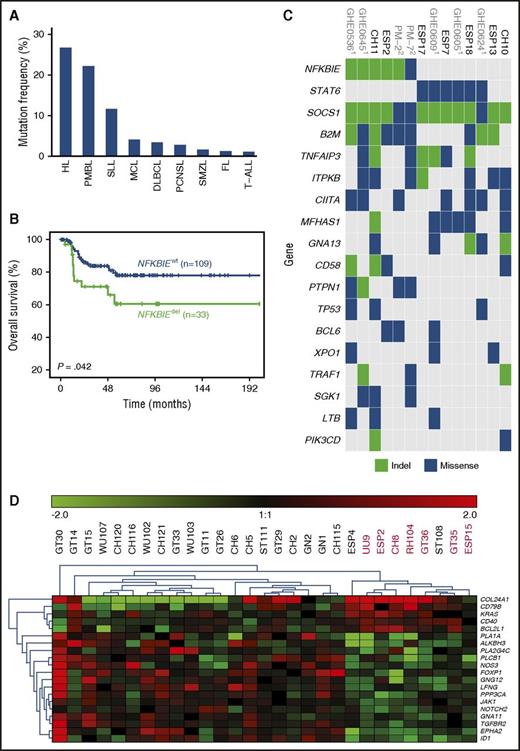
Assessment of NFKBIE mutations in 1460 patients diagnosed with different lymphoid malignancies. (A) NFKBIE mutation frequency per disease entity. (B) OS in 143 PMBL patients according to NFKBIE mutation status (P value refers to log-rank test). (C) Recurrently mutated genes in PMBL. Based on WES generated on 7 PMBL cases from this series and available exome data on 7 cases from Mareschal et al.10 and Gunawardana et al.18 (D) Differentially expressed genes in NFKBIE wild-type (n = 21) vs deleted (n = 8) PMBL cases based on the NanoString PanCancer Pathways Panel plus an additional 30 genes (detailed in supplemental Methods). FL, follicular lymphoma; HL, Hodgkin lymphoma; MCL, mantle cell lymphoma; PCNSL, primary central nervous system lymphoma; SMZL, splenic marginal zone lymphoma; SLL, small lymphocytic lymphoma; T-ALL, T-cell acute lymphoblastic leukemia.
Assessment of NFKBIE mutations in 1460 patients diagnosed with different lymphoid malignancies. (A) NFKBIE mutation frequency per disease entity. (B) OS in 143 PMBL patients according to NFKBIE mutation status (P value refers to log-rank test). (C) Recurrently mutated genes in PMBL. Based on WES generated on 7 PMBL cases from this series and available exome data on 7 cases from Mareschal et al.10 and Gunawardana et al.18 (D) Differentially expressed genes in NFKBIE wild-type (n = 21) vs deleted (n = 8) PMBL cases based on the NanoString PanCancer Pathways Panel plus an additional 30 genes (detailed in supplemental Methods). FL, follicular lymphoma; HL, Hodgkin lymphoma; MCL, mantle cell lymphoma; PCNSL, primary central nervous system lymphoma; SMZL, splenic marginal zone lymphoma; SLL, small lymphocytic lymphoma; T-ALL, T-cell acute lymphoblastic leukemia.
While NFKBIE deletions were relatively rare in patients diagnosed with follicular lymphoma (3/225 [1.3%]), splenic marginal zone lymphoma (3/175 [1.7%]), and T-cell acute lymphoblastic leukemia (1/94 [1.1%]), slightly higher frequencies were detected among DLBCL (18/520 [3.5%]), mantle cell lymphoma (8/189 [4.2%]), primary central nervous system lymphoma (1/34 [2.9%]), and small lymphocytic lymphoma (1/9 [11.1%]). In contrast, PMBL patients showed a marked enrichment, with 46 of 203 cases harboring a NFKBIE deletion (22.7% vs 2.9% [38/1257 in other entities]; P < .001; Figure 1A). Notably, the prevalence of NFKBIE-deleted PMBL cases was independent of contributing center (supplemental Table 3).
In an ongoing exome sequencing analysis of microdissected HRS cells in cHL, we obtained indication for NFKBIE mutations in 4 out of 11 cases. From these 4 cases, we isolated HRS cells and confirmed somatic NFKBIE aberrations (1 deletion and 2 missense mutations) in 3 out of 4 cases by Sanger sequencing (supplemental Table 4). Hence, NFKBIE aberrations are also a recurrent event in cHL (Figure 1A) and further explain biologic similarities reported between both lymphoma entities.19-21
For 142 out of 203 investigated PMBL cases, clinical follow-up data were available; the median follow-up time for patients alive was 61 months (range, 1-258 months). There were no significant differences between NFKBIE-deleted and wild-type PMBL patients with respect to clinical characteristics (Table 1). However, NFKBIE-deleted PMBL patients were more likely than wild-type patients to be refractory to primary chemotherapy (25% vs 6%; P = .022). Furthermore, these patients had a significantly shorter OS as compared with wild-type patients (5-year survival, 59% vs 78%; P = .034; Figure 1B). In multivariate analysis, NFKBIE deletion status (n = 111; 95% confidence interval, 1.65-12.04; hazard ratio, 4.46; P = .003) remained an independent factor for poor outcome (supplemental Table 5), even when restricting the analysis to patients treated with immunochemotherapy (n = 91; 95% confidence interval, 1.72-39.82; hazard ratio, 8.27; P = .008; supplemental Table 6). In contrast, no significant difference in treatment response and OS was seen between 16 NFKBIE-deleted and 428 wild-type DLBCL patients (median OS, 65 vs 52 months; P = .804; supplemental Table 7) or within immunohistochemistry-based germinal center B-cell–like and non–germinal center B-cell–like subtypes (supplemental Figure 2).
An improved patient outcome was recently demonstrated in PMBL with the addition of rituximab to dose-intense chemotherapy (ie, dose-adjusted etoposide, doxorubicin, cyclophosphamide, vincristine, and prednisone [CHOEP]), resulting in an OS rate of 97%.22 Patients in the present series received heterogeneous treatment regimens, and data interpretation warrants caution. Nevertheless, all patients received CHOP-based treatment; in ∼75% of cases, rituximab was added, and ∼25% were treated with dose-intensified schemes. For the latter, the vast majority received CHOEP, while individual cases were treated with mega-CHOEP; or a combination of doxorubicin, cyclophosphamide, vindesine, bleomycin, and prednisone (ACVBP). While dose escalation did not improve outcome for NFKBIE-deleted patients, these patients appeared to benefit from the addition of rituximab (n = 33, P = .074) or, particularly, radiotherapy (n = 30, P = .022). After restricting this analysis to nonprogressive patients, the benefit of radiation showed borderline significance (P = .083; supplemental Figure 3).
Several studies have reported high frequencies of TNFAIP3 aberrations (ranging from 30% to 60%10,18,23,24 ), another key regulator of NF-κB, in PMBL. Based on exome data on 14 cases,10,18 most PMBL cases demonstrated a very high number of somatic variants (average 218 mutations). While 3 NFKBIE-mutated cases showed concomitant TNFAIP3 aberrations (Figure 1C), other members of the NF-κB pathway or interrelated pathways, such as the JAK/STAT pathway,25 were also affected. In addition, we performed gene expression profiling in NFKBIE-deleted (n = 8) vs wild-type (n = 21) PMBL cases using NanoString technology and identified 79 differentially expressed genes, including several NF-κB target genes or upstream mediators, such as BCL2L1, NFKBIB, EGFR, CD79B, and CD40 (Figure 1D; supplemental Table 8). Together, these findings support deregulation of NF-κB signaling as a major factor in PMBL pathobiology.
In conclusion, our findings highlight NFKBIE deletions as a common, recurrent genetic event among different lymphoid malignancies. NFKBIE deletions emerged among the most frequent genetic aberrations in PMBL and were associated with chemorefractoriness and an inferior clinical outcome. Further validation studies are now warranted to confirm this novel finding.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors would like to thank Hedwig Lammert, Stefanie Mende, Philip Abstoß, and Alexandra Stege for technical assistance.
This study was supported by grants from the Berliner Krebsgesellschaft (#DAFF201503), the Else-Kröner-Fresenius-Stiftung (#2015_A09), the Deutsche Forschungsgemeinschaft (#DA1787/1-1), the Lady Tata Memorial Trust (all F.D.); the Swedish Cancer Society, the Swedish Research Council, Uppsala University, Uppsala University Hospital, the Lion’s Cancer Research Foundation (Uppsala), and Selander’s Foundation, Uppsala (all R.R.); and the H2020 “MEDGENET, Medical Genomics and Epigenomics Network” (grant 692298), funded by the European Union (K.S. and R.R.). R.K. and M.-L.H. were supported by the Wilhelm Sander Stiftung (2014.136.1). E.C. is an Academia Researcher of the “Institució Catalana de Recerca i Estudis Avançats” of the Generalitat de Catalunya and is supported by the Spanish Ministerio de Economía y Competitividad SAF2015-64885-R. F.D. is a fellow of the Charité Clinical Scientist Program funded by the Charité University Medical Center Berlin and the Berlin Institute of Health.
Authorship
Contribution: L.M., D.N., R.R., and F.D. designed the research; L.M., D.N., E.Y., E.M., M. Abdulla, M.F., F.A., V.L., M.S., K.Y., A.S., T.P., B.G., A.T., N.W., A.R.-D., M. Angelopoulou, M.Z., C.M.A., L.C., D.L., C.D.B., C.B., J.O., J.F., B.D., H.G.D., D.R.-W., C.A.S., H.D.M.-P., T.Z., M.-L.H., J.C.S., G.E., O.A.B., E.R., M.E., P.K., M. Hultdin, T.P., K.G., A.L.-G., R.K., S.O., K.S., N.S., G.K., A.R., E.C., R.-M.A., G.O., T.P.V., M. Hummel, R.R., and F.D. performed the research and/or contributed patient samples and clinical data; L.M., D.N., E.Y., R.R., and F.D. analyzed the data; and L.M, D.N., R.R., and F.D. wrote the paper. All authors read and agreed to the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Frederik Damm, Department of Hematology, Oncology, and Tumor Immunology, Charité, University Medical Center, Augustenburger Platz 1, 13353 Berlin, Germany; e-mail: frederik.damm@charite.de.
References
Author notes
L.M., D.N., and E.Y. contributed equally to this study as joint first authors.
R.R. and F.D. contributed equally as joint senior authors.