Key Points
Cells transformed by activating JAK1 mutations become resistant to JAK inhibitor by acquiring activating mutations in JAK3 and vice versa.
JAK1 and JAK3 mutants cooperatively activate STAT transcription factors.
Abstract
The acquisition of growth signal self-sufficiency is 1 of the hallmarks of cancer. We previously reported that the murine interleukin-9–dependent TS1 cell line gives rise to growth factor–independent clones with constitutive activation of the Janus kinase (JAK)- signal transducer and activator of transcription (STAT) pathway. Here, we show that this transforming event results from activating mutations either in JAK1, JAK3, or in both kinases. Transient and stable expression of JAK1 and/or JAK3 mutants showed that each mutant induces STAT activation and that their coexpression further increases this activation. The proliferation of growth factor–independent TS1 clones can be efficiently blocked by JAK inhibitors such as ruxolitinib or CMP6 in short-term assays. However, resistant clones occur upon long-term culture in the presence of inhibitors. Surprisingly, resistance to CMP6 was not caused by the acquisition of secondary mutations in the adenosine triphosphate–binding pocket of the JAK mutant. Indeed, cells that originally showed a JAK1-activating mutation became resistant to inhibitors by acquiring another activating mutation in JAK3, whereas cells that originally showed a JAK3-activating mutation became resistant to inhibitors by acquiring another activating mutation in JAK1. These observations underline the cooperation between JAK1 and JAK3 mutants in T-cell transformation and represent a new mechanism of acquisition of resistance against JAK inhibitors.
Introduction
Hematopoiesis is a tightly regulated process in which cytokines dictate fate (apoptosis, proliferation, or differentiation) of the various progenitors through binding to specific receptors at the surface of target cells. Most hematopoietic cytokine receptor chains associate with a member of the Janus kinase (JAK) family to form a competent cytokine–receptor complex. The JAK family comprises 4 tyrosine kinases (JAK1, JAK2, JAK3, and TYK2) that bind to a proper repertory of preassociated homo- or heterodimeric receptor chains. Structurally, JAK kinases are characterized by the presence of a pseudo-kinase domain directly adjacent to the kinase domain and are supposed to prevent inappropriate catalytic activity of the kinase domain in the absence of stimulus.1 The major substrates of activated JAKs are the cytoplasmic signal transducer and activator of transcription (STAT) molecules.
STAT factors play an active role not only in physiological hematopoiesis, but also in tumoral transformation because uncontrolled STAT activation leads to growth signal self-sufficiency.2 In hematological malignancies, a common mechanism for constitutive STAT activation is the deregulation of tyrosine kinase activity of the cytokine–receptor complex by gain-of-function alterations affecting the JAK itself or its associated receptor chain. A classical illustration is the activating JAK2V617F mutation identified in a substantial proportion of patients with chronic myeloproliferative neoplasms (MPNs).3-6 Among the JAK2V617F-negative MPNs, 5% of essential thrombocythemia patients harbor a mutation in the thrombopoietin receptor (commonly MPLW515L or W515K).7 In acute leukemias from lymphoid or myeloid origin, various JAK1, JAK2, and JAK3 mutations and translocations have been reported.8-18 In childhood acute lymphoblastic leukemia (ALL), gain-of-function mutations of the interleukin-7 receptor chain (IL-7Rα) were described in 10% of cases.19,20 JAK inhibitors therefore represent an appealing therapy for leukemia patients because a significant proportion (20% to 30% in childhood ALL) seems to acquire growth signal self-sufficiency through constitutive activation of the JAK-STAT pathway. Nevertheless, their efficacy has first to be experimentally assessed using preclinical leukemia models.
TS1 is an IL-9–dependent cell line derived from a murine CD4+ T-helper clone. Although normal T cells require antigen presentation for in vitro growth, repeated biweekly antigen stimulation led to gradual acquisition of IL-9 responsiveness, loss of the need for antigen presentation, and emergence of permanent IL-9–dependent cell lines such as TS1.21 The progressive deregulation undergone by T cells during acquisition of IL-9 responsiveness is somewhat reminiscent of the multistep process leading to cell transformation.22,23 Furthermore, the IL-9–responsive TS1 cell line could spontaneously give rise to growth factor (GF)-independent clones that were tumorigenic in vivo.24 Even though there was no autocrine cytokine production in the supernatants of these GF-independent clones, they showed constitutive phosphorylation of JAK1, JAK3, STAT3, and STAT5 comparable to those induced by IL-9 stimulation in parental TS1 cells.25
Because of its T-cell origin and its ability to spontaneously recapitulate the transformation process in vitro, the TS1 model provides us with a unique opportunity to identify T-cell–specific oncogenic events potentially implicated in ALL. The aims of our study were first to unravel the genetic mechanisms underlying constitutive JAK-STAT activation in this model and, second, to test the efficacy of short- and long-term treatment with JAK inhibitors.
Methods
Cell cultures
TS1 and BW5147 cells were cultured in Iscove-Dulbecco’s medium supplemented with 10% fetal bovine serum, 50 µM β-mercaptoethanol, 0.55 mM l-arginine, 0.24 mM l-asparagine, 1.25 mM l-glutamine, and, for TS1 cells, murine IL-9 (100 U/mL) was added. CMK cells were cultured in RPMI medium supplemented with 10% fetal bovine serum.
Stable DNA transfections and selection of clones
Wild-type (WT) JAK3 and human IL-9Rα complementary DNA (cDNA) were subcloned into the pMX-IRES-CD4 biscistronic retroviral vector upstream of the IRES. WT JAK1 and human γ chain (γc) cDNA were subcloned into the pMX-IRES-GFP biscistronic retroviral vector upstream of the IRES. The JAK3 (R657H, V674A) and JAK1 (Y652H and A634D) mutant cDNA were generated using QuickChange XL II site-directed mutagenesis kit (Stratagene). For stable transduction, retroviral supernatants were generated by transient transfection of the BOSC packaging cell line and used for infection of 0.5 × 106 cells as described.26 Populations of cells expressing markers were isolated by FACS sorting, using a phycoerythrin-coupled antibody against hCD4 (#555347, BD Pharmingen) diluted 1/30.
For the selection of GF-independent TS1 clones, IL-9–dependent TS1 cells were washed 3 times in phosphate-buffered saline and seeded in 96-well plates at a concentration of 5000 cells/well in the absence of cytokines. After 1 to 2 weeks, GF-independent clones were picked up from plates in which less than 20% of wells were positive for proliferation, corresponding to a probability of clonality superior to 0.9 according to the Poisson distribution. For the selection of JAK inhibitor–resistant subclones, GF-independent TS1 clones were seeded in 96-well plates at a density of 30 000 cells/well in the presence of 300 to 600 nM of either pan-JAK inhibitor CMP6 (Calbiochem, cat#420097) or ruxolitinib (Haoyuan Chemexpress Co., INCB018424). After 2 to 3 weeks, wells containing proliferating cells were counted. Resistant cells from plates meeting clonality conditions were picked up and amplified in the presence of the respective JAK inhibitor.
Luciferase assays
STAT5 transcriptional activity was assessed by measurements of luciferase expression in HEK293 cells upon transient transfection of appropriate cDNA and pLHRE-luc vector harboring tandem copies of the STAT5-inducible lactogenic hormone response element of the rat β-casein gene promoter, inserted upstream a luciferase gene.27 Another reporter plasmid, Renilla luciferase (pRLTk, Promega), was cotransfected as an internal transfection control. Transient transfection of HEK293 cells by LipofectAMINE (Invitrogen) was previously described.28 Twenty-four hours after transfection, luciferase assays were performed using the Dual Luciferase Reporter Assay kit (Promega).
Western blots
For western blot analysis, 2 × 106 BW5147 or CMK cells were lysed in 200 µL of lysis buffer containing 50 mM tris(hydroxymethyl)aminomethane-HCl, 1% NP40, 150 mM NaCl, 1 mM EDTA, 15% glycerol, Halt Protease Inhibitor Cocktail 1/100, phenylmethylsulfonyl fluoride 0.1 mM, and sodium orthovanadate 0.1 mM. A total of 35 µL of lysates was loaded on Bolt 4% to 12% Bis-tris precast gels (Novex) and electrophoretically transferred to nitrocellulose membranes (i-blot Gel Transfer Stacks, Novex). Phosphorylation of signaling proteins was investigated with the following phospho-specific antibodies from Cell Signaling Technology: anti-pY705 STAT3 (#9131) and anti-pY694 STAT5 (#9351). Blots were reprobed with anti-JAK1 (#3332), anti-JAK3 (#3775), anti-STAT3 (#9132) (Cell Signaling Technology), anti-STAT5 (#835, Santa Cruz), or anti-β-actin (#A5441, Sigma) antibodies as a control.
RNA extraction, cDNA synthesis, PCR, and sequencing
Total RNA was isolated from 106 TS1 cells using TriPure Isolation Reagent (Roche). Reverse transcription was performed on 1 µg of total RNA using an oligo-dT primer and M-MULV Reverse transcriptase (RevertAid Reverse Transcriptase, Thermo Scientific). Taq DNA polymerase-mediated polymerase chain reaction (PCR) amplification (TaKaRa) was performed and PCR products were sequenced using the DYEnamic Big Dye Terminator Kit (Amersham).
Results
GF-independent TS1 clones acquired point mutations in the pseudo-kinase and kinase domains of JAK1 and/or JAK3
TS1, a T-lymphocyte cell line, is dependent on IL-9 for proliferation. Removal of IL-9 from the culture medium results in rapid cell death, but few cells occasionally succeed to proliferate independently of GF (at a frequency of approximately 1/20 000). We selected 50 GF-independent TS1 clones. Constitutive STAT5, STAT3, AKT, JAK1, and JAK3 phosphorylation could be observed in all tested IL-9–independent clones, but not in IL-9–starved parental TS1 cells25 (supplemental Figure 1, available on the Blood Web site). GF-independent TS1 clones remained sensitive to JAK inhibitors, indicating that their proliferation still relies on JAK activity (data not shown).
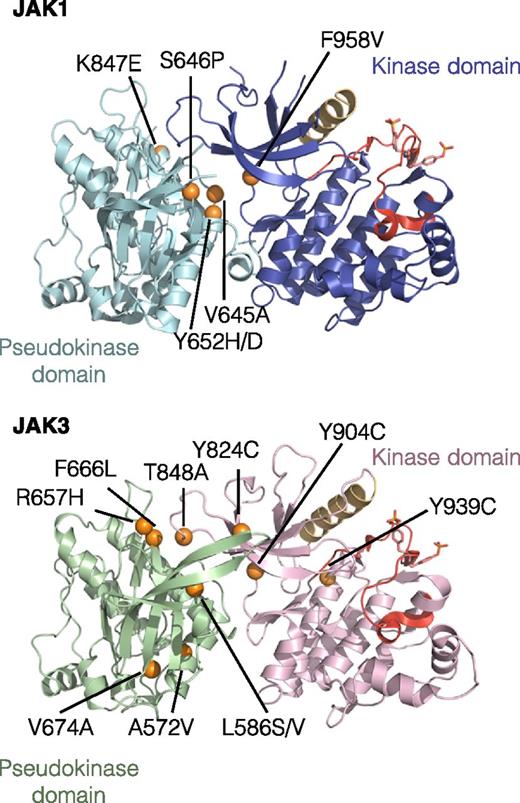
Because both JAK1 and JAK3 were constitutively phosphorylated, we sequenced the JAK1 and JAK3 cDNA in our collection of 50 GF-independent TS1 clones and found that all of them had acquired a missense point mutation affecting a residue of the pseudo-kinase or kinase domains of either JAK1, JAK3, or both (Table 1). A majority of clones harbored a single mutation in 1 JAK (23 with JAK1 mutation vs 22 with JAK3 mutation), but some clones (5 of 50) acquired a mutation in both kinases of the IL-9 receptor complex. Altogether, we identified 16 different mutations (6 in JAK1 and 10 in JAK3) affecting 14 residues conserved in mouse and human JAK. As shown in Figure 1, the great majority of mutations map to the interface between pseudo-kinase and kinase domains of JAK1 and JAK3 that was recently shown to mediate autoinhibitory mechanisms.29 For 7 of the 14 residues mutated in our clones, mutations have been also described in human cancers (supplemental Table 1). Although the total number of mutations of JAK1 and JAK3 was equal (28 JAK1 mutations vs 27 JAK3 mutations), statistics confirm that they were not randomly distributed among experiments suggesting that the relative expansion of the mutated cells occurred before cytokine withdrawal (chi-squared test, P < .05) (supplemental Table 2).
Localization of the 16 identified missense point mutations in JAK1 and JAK3. The figure represents the predicted 3-dimensional structure of kinase and pseudo-kinase domains of JAK1 and JAK3 based on the recent TYK2 crystal structure.29 Molecular images were obtained with the PyMOL molecular visualization system. The pseudo-kinase domain is shown in light blue for JAK1 and green for JAK3. The adjacent kinase domain is shown in violet for JAK1 and pink for JAK3. The αC helix is shown in beige and activation loop in red. Mutated residues are indicated with orange balls.
Localization of the 16 identified missense point mutations in JAK1 and JAK3. The figure represents the predicted 3-dimensional structure of kinase and pseudo-kinase domains of JAK1 and JAK3 based on the recent TYK2 crystal structure.29 Molecular images were obtained with the PyMOL molecular visualization system. The pseudo-kinase domain is shown in light blue for JAK1 and green for JAK3. The adjacent kinase domain is shown in violet for JAK1 and pink for JAK3. The αC helix is shown in beige and activation loop in red. Mutated residues are indicated with orange balls.
Activated JAK1 and JAK3 mutants cooperate for downstream signal transduction
The observation that some GF-independent clones had acquired mutations both in JAK1 and JAK3 raised the hypothesis that these mutants might cooperatively contribute to the activation of the signaling pathways necessary for survival and proliferation. To address this hypothesis, we compared the activation of the JAK-STAT pathway induced by expression of a single JAK mutant vs coexpression of the JAK1Y652H together with JAK3V674A or JAK3R657H mutants found in ALL patients. To avoid any bias during selection of stable transfectants, these mutants were expressed in the murine lymphoma T cells BW5147, which do not rely on the JAK-STAT pathway for proliferation.
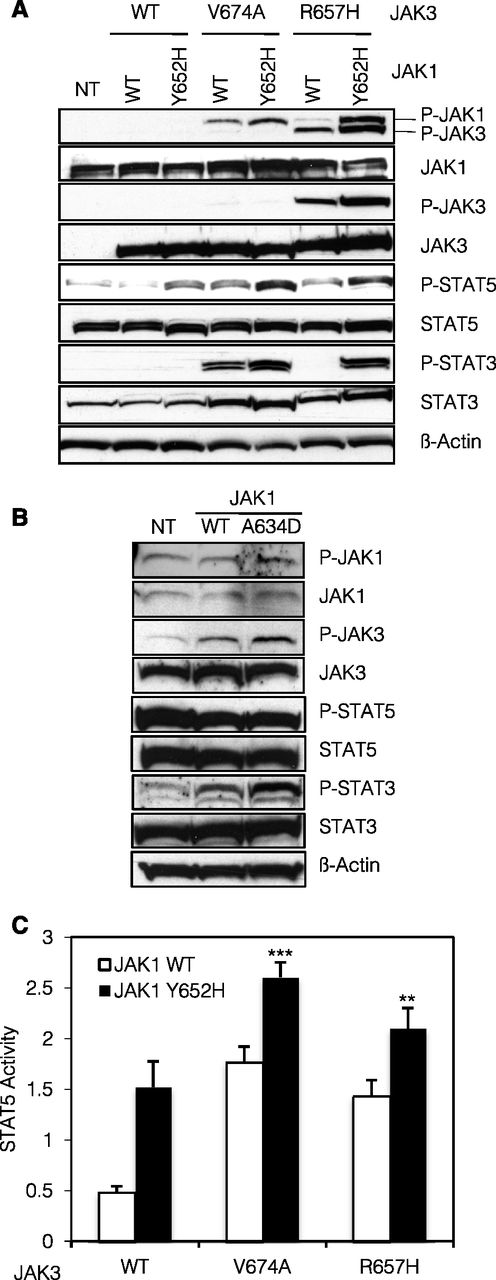
As shown in Figure 2A, stable expression of JAK1Y652H, JAK3V674A, or JAK3R657H alone resulted in constitutive phosphorylation of STAT5 (Figure 2A, lanes 3, 4, and 6). This phosphorylation was further increased when JAK1 and JAK3 mutants were coexpressed (Figure 2A, lanes 5 and 7). For JAK1, JAK3, and STAT3, phosphorylation was either exclusively observed in the presence of a combination of 2 mutants or increased in comparison with each mutant separately. This indicates that, in BW5147 cells, JAK1 and JAK3 mutants cooperate in their transphosphorylation and subsequent activation of downstream signaling molecules STAT3 and STAT5. These observations were corroborated in the human acute megakaryoblastic leukemia CMK cell line harboring a heterozygous activating mutation in JAK3 (A572V).13 As displayed in Figure 2B, stable expression of JAK1A634D enhanced the phosphorylation of JAK3 and STAT3 providing evidence that a JAK1 activated mutant can collaborate with an endogenous JAK3 mutant.
JAK1 and JAK3 mutants cooperatively activate the JAK-STAT pathway. (A) A total of 2 × 106 BW5147 cells nontransduced (NT) or stably transduced with JAK1 (WT or Y652H mutant) together with JAK3 (WT, V674A, or R657H mutants) were lysed and subjected to western blot analysis. The basal phosphorylation level of STAT and JAK proteins was detected using anti-pY694 STAT5, anti-pY705 STAT3, anti-pY1034/1035 JAK1, and anti-pY980/981 JAK3 antibodies. Cells expressed equivalent levels of JAK1 and JAK3, as shown with anti-JAK1 and anti-JAK3 antibodies. Membranes were reprobed with anti-STAT3, anti-STAT5, and anti–β-actin antibodies as loading control. (B) A total of 2 × 106 CMK cells NT or stably transduced with JAK1 (WT or A634D mutant) were lysed and subjected to western blot analysis. Basal phosphorylation level of STAT and JAK proteins was detected using anti-pY694 STAT5, anti-pY705 STAT3, anti-pY1034/1035 JAK1, and anti-pY980/981 JAK3 antibodies. Membranes were reprobed with anti-STAT3, anti-STAT5, anti-JAK1, anti-JAK3, and anti–β-actin antibodies as loading control. (C) HEK293 cells were transiently cotransfected with JAK1 (WT or Y652H mutant), JAK3 (WT, V674A, or R657H mutants), the IL-9Ra chain, and common γ-chain, in addition to the STAT5-responsive luciferase reporter construct pLHRE and the pRLTK plasmid as transfection control. Twenty-four hours posttranfection, cells were lysed and subjected to dual luciferase activity assay. Histograms are means ± standard error of the mean of 5 independent experiments. A 1-way analysis of variance test was performed to compare the condition with the 2 mutants vs each condition with 1 mutant only (**P < .01).
JAK1 and JAK3 mutants cooperatively activate the JAK-STAT pathway. (A) A total of 2 × 106 BW5147 cells nontransduced (NT) or stably transduced with JAK1 (WT or Y652H mutant) together with JAK3 (WT, V674A, or R657H mutants) were lysed and subjected to western blot analysis. The basal phosphorylation level of STAT and JAK proteins was detected using anti-pY694 STAT5, anti-pY705 STAT3, anti-pY1034/1035 JAK1, and anti-pY980/981 JAK3 antibodies. Cells expressed equivalent levels of JAK1 and JAK3, as shown with anti-JAK1 and anti-JAK3 antibodies. Membranes were reprobed with anti-STAT3, anti-STAT5, and anti–β-actin antibodies as loading control. (B) A total of 2 × 106 CMK cells NT or stably transduced with JAK1 (WT or A634D mutant) were lysed and subjected to western blot analysis. Basal phosphorylation level of STAT and JAK proteins was detected using anti-pY694 STAT5, anti-pY705 STAT3, anti-pY1034/1035 JAK1, and anti-pY980/981 JAK3 antibodies. Membranes were reprobed with anti-STAT3, anti-STAT5, anti-JAK1, anti-JAK3, and anti–β-actin antibodies as loading control. (C) HEK293 cells were transiently cotransfected with JAK1 (WT or Y652H mutant), JAK3 (WT, V674A, or R657H mutants), the IL-9Ra chain, and common γ-chain, in addition to the STAT5-responsive luciferase reporter construct pLHRE and the pRLTK plasmid as transfection control. Twenty-four hours posttranfection, cells were lysed and subjected to dual luciferase activity assay. Histograms are means ± standard error of the mean of 5 independent experiments. A 1-way analysis of variance test was performed to compare the condition with the 2 mutants vs each condition with 1 mutant only (**P < .01).
Furthermore, in HEK293 cells transiently transfected with a STAT5-responsive luciferase reporter construct, single expression of JAK1Y652H, JAK3V674A, or JAK3R657H induced STAT5 activation that was further exacerbated by coexpression of JAK1 and JAK3 mutants (Figure 3). Altogether, these 2 different experimental models show the cooperation of JAK1 and JAK3 mutants in the activation of the JAK-STAT pathway in 3 cell lines and support the hypothesis that JAK1 and JAK3 collaborate toward the transformation of IL-9–dependent TS1 cells into GF-independent clones.
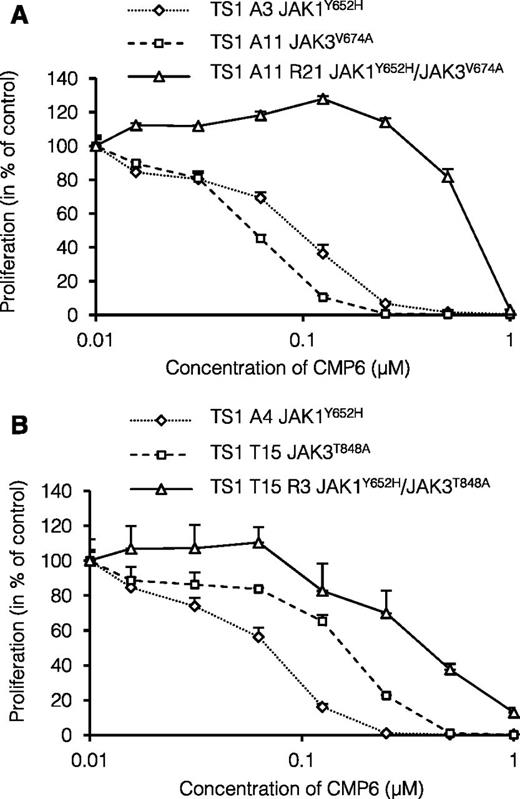
Sensitivity to pan-JAK inhibitor CMP6 of GF-independent and CMP6-selected TS1 clones. (A-B) GF-independent TS1 clones with endogenous JAK1 or JAK3 mutants and CMP6-selected TS1 clones with endogenous JAK1 and JAK3 mutants were seeded in 96-well plates at a density of 1000 cells/well with increasing concentrations of pan-JAK inhibitor CMP6 (0 to 1 μM). After 48 hours, methyl-3H thymidine was added to the cells for 4 hours and thymidine incorporation was measured. Results are means ± standard deviation of triplicate cultures represented in % of control untreated cells.
Sensitivity to pan-JAK inhibitor CMP6 of GF-independent and CMP6-selected TS1 clones. (A-B) GF-independent TS1 clones with endogenous JAK1 or JAK3 mutants and CMP6-selected TS1 clones with endogenous JAK1 and JAK3 mutants were seeded in 96-well plates at a density of 1000 cells/well with increasing concentrations of pan-JAK inhibitor CMP6 (0 to 1 μM). After 48 hours, methyl-3H thymidine was added to the cells for 4 hours and thymidine incorporation was measured. Results are means ± standard deviation of triplicate cultures represented in % of control untreated cells.
Secondary mutations in the other JAK of the cytokine receptor complex confer increased resistance to JAK inhibitors
CMP6 is an adenosine triphosphate (ATP)-competitive tyrosine kinase inhibitor that effectively inhibits all JAK. In short-term assay, GF-independent proliferation of TS1 clones could be efficiently blocked by CMP6 with a half maximal inhibitory concentration (IC50) close to 0.1 µM (data not shown). GF-independent TS1 clones harboring primary mutations in either JAK1 (A3, A4) or JAK3 (A6, A11, T4, T15) were seeded in 96-well plates in the presence of CMP6 concentrations inhibiting 100% of cell proliferation within 2 days (300 to 600 nM). Upon long-term culture, CMP6-resistant clones arose at a low frequency (1 in 2 million cells). Using this experimental setup, 20 CMP6-selected subclones were obtained from 2 independent TS1 clones with the JAK1Y652H mutation, and 56 CMP6-selected subclones were obtained from 4 independent clones with 3 different JAK3 mutations (L586S, V674A, and T848A). Contrary to our expectations, none of these subclones had acquired any secondary mutations in the originally mutated JAK. By contrast, 16 of the 20 subclones (80%) with a primary JAK1 mutation had acquired a mutation in JAK3, whereas 32 of the 56 subclones (57%) with a primary JAK3 mutation had acquired a mutation in JAK1 (Table 2).
Most of the secondary mutations affect the homologous residues F958 in JAK1 and Y904 in JAK3 located in the ATP-binding pocket of the kinase domain. We previously showed that mutations targeting the F958 residue of JAK1 not only confer constitutive kinase activity but also resistance to ATP-competitive JAK inhibitors.30 However, other secondary mutations were located in the pseudo-kinase domain of JAK1 (Y652H and F804L) or in the kinase domain of JAK3 outside the ATP-binding pocket (R840C) and are not expected to directly confer resistance to JAK inhibitor. For instance, the JAK1Y652H mutation was previously shown to transform BaF3 cells but did not change their sensitivity to JAK inhibitors.30 As illustrated in Figure 3A-B, the proliferation of TS1 clones harboring the primary JAK1Y652H mutation (clones A3 and A4) was blocked by CMP6 with an IC50 ≤0.1 µM in the same range as clones with the JAK3V674A or JAK3T848A mutations (clones A11 and T15, respectively). However, the CMP6-selected subclones A11R21 and T15R3, which both had acquired the JAK1Y652H as a secondary mutation, showed a 4 to 7 times higher IC50 of 0.4 to 0.7 µM. This observation indicated that increased resistance to a kinase inhibitor such as CMP6 can be conferred by cooperativity between 2 active mutants of JAK1 and JAK3, which are both intrinsically CMP6-sensitive.
To test this hypothesis in another model, we took again advantage of the human CMK cell line carrying the JAK3A572V mutation. Proliferation of CMK cells can be efficiently blocked by the JAK1/2 inhibitor ruxolitinib. However, CMK cells transduced with the JAK1A634D mutant exhibited a 2-fold shift in IC50 compared with cells transduced with JAK1 WT or nontransduced (nontransduced: 88 nM; JAK1 WT: 75 nM; JAK1A634D: 163 nM) (data not shown).
These experiments show that acquiring a secondary activating mutation in another JAK allows for increased resistance to JAK inhibitors that block both JAKs. However, a striking observation from the CMP6-selected subclones described in Table 2, is that such secondary mutations did not occur in the other allele of the originally mutated JAK. This contrasts with JAK2V617F-MPNs patients, where homozygosity is often observed because of homologous recombination. JAK2 associates with homodimeric receptors such as the erythropoietin-receptor, whereas JAK1 and JAK3 are involved in signaling by heterodimeric cytokine receptor complexes using the common γc. In this respect, our data suggest that signaling by heterodimeric receptor complexes is further boosted by acquiring another heterozygous activating mutation in the JAK partner rather than by having both alleles of a single JAK mutated.
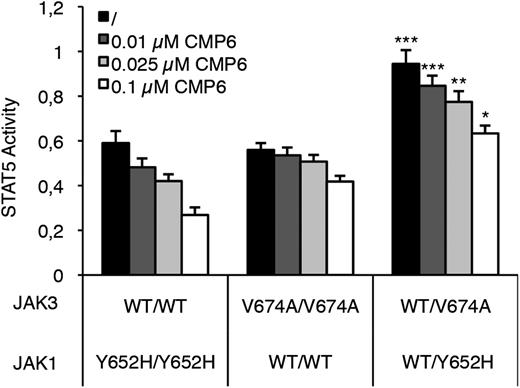
To address this question, we reconstituted homozygous and heterozygous JAK configurations in HEK293 cells by transiently transfecting JAK1 (WT, Y652H, or an equimolar mix of both) and JAK3 (WT, V674A, or both) together with the IL-9Rα and γc cDNA. Cells were then treated or not with increasing concentrations of CMP6 (0.01 to 0.1 µM) and STAT5 activity was assessed 24 hours posttransfection. As shown in Figure 4, the basal level of STAT5 activity was higher in cells having 50% from each activated JAK mutant compared with cells having 100% of mutated JAK1 or JAK3 (black histograms). This boost in JAK-STAT pathway activation could be explained by the fact that both JAK1 and JAK3 mutants interact within the same heterodimeric receptor complex, whereas such an association is not possible in a homozygous situation. CMP6 treatment decreased STAT5 activity in all transfected cells but for each inhibitor concentration, STAT5 activity remained significantly higher in the double heterozygous condition compared with the homozygous ones (Figure 4). Complete titration allowing to assess IC50 concentrations confirmed that the intrinsic ability of CMP6 to inhibit STAT5 phosphorylation is not altered in the double heterozygous condition (JAK1 homozygous: 61 nM; JAK3 homozygous: 365 nM; double heterozygous: 250 nM) (data not shown). These results support the hypothesis that acquiring a secondary mutation in the JAK partner from heterodimeric cytokine receptor complex activates the pathway further than becoming homozygous. In addition, when basal STAT5 activity is boosted to strong levels by the 2 mutated JAKs, CMP6 still interferes with STAT5 activation, but higher concentrations would be required to reach a critical threshold.
Inhibition by CMP6 of STAT5 activity in cells expressing JAK1 and/or JAK3 mutants. HEK293 cells were transiently cotransfected with JAK1 (WT, Y652H mutant, or both in equimolar concentration to mimic heterozygous vs homozygous configuration), JAK3 (WT, V674A mutant, or both), IL-9Rα, and γc, in addition to the STAT5-responsive luciferase reporter construct pLHRE and the pRLTK plasmid as transfection control. Cells were maintained in the presence of increasing concentrations of pan-JAK inhibitor CMP6 or left untreated. Twenty-four hours posttransfection, cells were lysed and subjected to dual luciferase activity assay. Raw values were standardized with BCR-ABL–induced STAT5 activity. Histograms are means ± standard error of the mean of 5 independent experiments performed in triplicate. A 1-way analysis of variance test was used to determine P values between the heterozygous and the 2 homozygous configurations for each CMP6 concentration (*P < .05, ** P < .01, ***P < .001).
Inhibition by CMP6 of STAT5 activity in cells expressing JAK1 and/or JAK3 mutants. HEK293 cells were transiently cotransfected with JAK1 (WT, Y652H mutant, or both in equimolar concentration to mimic heterozygous vs homozygous configuration), JAK3 (WT, V674A mutant, or both), IL-9Rα, and γc, in addition to the STAT5-responsive luciferase reporter construct pLHRE and the pRLTK plasmid as transfection control. Cells were maintained in the presence of increasing concentrations of pan-JAK inhibitor CMP6 or left untreated. Twenty-four hours posttransfection, cells were lysed and subjected to dual luciferase activity assay. Raw values were standardized with BCR-ABL–induced STAT5 activity. Histograms are means ± standard error of the mean of 5 independent experiments performed in triplicate. A 1-way analysis of variance test was used to determine P values between the heterozygous and the 2 homozygous configurations for each CMP6 concentration (*P < .05, ** P < .01, ***P < .001).
Secondary mutations in JAK3 confer increased resistance to a JAK1-specific inhibitor
It is well known that the transforming effect of an active JAK mutant depends on its association with cytokine receptor chains allowing homo- or heterodimerization with another JAK and subsequent cross-phosphorylation.31,32 For instance, an active JAK3 mutant requires a functional JAK1 partner for constitutive signaling.33 When CMP6, a pan-JAK inhibitor, was used to select resistant TS1 subclones, selective pressure was exerted on both the mutated JAK and its partner, and secondary mutations in the JAK partner provided a selective advantage. We thus wondered whether the same process of selection would occur if only JAK1 activity is blocked with a more selective kinase inhibitor such as ruxolitinib, which is at least 100 times less potent on JAK3.34
To address this question, we seeded 6 different GF-independent TS1 clones harboring the primary JAK1Y652H mutation in the presence of ruxolitinib. Ruxolitinib-resistant subclones arose at a frequency comparable to what was observed during selection with CMP6. JAK1 and JAK3 cDNA sequencing revealed that 44 of 53 ruxolitinib-selected TS1 subclones had acquired secondary mutations in the JAKs. Twenty-five of them (57%) had a secondary mutation in JAK3, similar to the selection with CMP6, although ruxolitinib is not supposed to block JAK3. However, contrasting with CMP6 selection, 18 of the ruxolitinib-selected subclones had a secondary mutation in JAK1, either within or outside the ATP-binding pocket, including 4 subclones that became homozygous for the JAK1Y652H mutation (Table 3). Finally, 1 of the ruxolitinib-selected subclones had acquired 2 secondary mutations, JAK1F958S and JAK3R840C. As for the primary mutations, the observation of a single predominant mutation within an experiment (eg, JAK3Y824D in A3) is consistent with the hypothesis of preexisting secondary mutations before selective pressure with the inhibitor. Altogether, our results indicate that when 1 JAK from a heterodimeric cytokine receptor complex is specifically targeted by a tyrosine kinase inhibitor, resistance can be conferred either by a secondary mutation in the other JAK partner or by a secondary activating mutation in the same JAK. In both cases, and in line with CMP6-selected clones, the secondary mutations are not restricted to the ATP-binding pocket.
Discussion
In the present study, we describe a new mechanism of resistance to JAK kinase inhibitors. We show that 2 concomitant spontaneous activating mutations in endogenous JAK1 and JAK3 cooperatively activate the JAK-STAT pathway leading to increased resistance to CMP6 or Ruxolitinib. Interestingly, such resistance could be conferred by combining mutations that do not affect the sensitivity of cells to JAK inhibitors when present individually.
In our series of subclones selected in the presence of tyrosine kinase inhibitors (TKI), TKI resistance was not primarily conferred by impairing drug binding to the oncogenic kinase, but rather by increasing the activity of the heterodimeric JAK–receptor complex. Indeed, secondary mutations associated with CMP6 resistance did not affect the originally mutated JAK. In addition, the secondary mutations in the JAK partner observed here were previously described as activating mutations per se, associated with oncogenesis in vitro and in vivo (supplemental Table 1; Hornakova et al30 ). However, these activating mutations can be further divided into 2 types. First, a series of mutations can either lead to GF independence with normal TKI sensitivity when occurring as primary mutations or increase TKI resistance when occurring as secondary mutations. This first set of mutants is illustrated in Figure 4 with JAK1Y652H. On the other hand, another series of secondary mutations target the JAK1 F958 residue or the JAK3 Y904 residue, which correspond to homologous amino acids that were previously found to confer both constitutive activation and lower sensitivity to JAK inhibitors.30 Thus, selective pressure with the JAK inhibitor selects essentially for activating mutations, but with a further advantage for gain-of-function mutations that also affect drug binding.
In patients relapsing after treatment with TKI targeting oncogenic proteins such as BCR-ABL, c-Kit and epidermal growth factor receptor (EGFR) secondary mutations frequently occur in the kinase domain of the respective target. A great part of those substitutions affect the conserved threonine gatekeeper residue regulating access of TKIs to the ATP binding pocket, pointing to impaired drug binding as the main cause of resistance. Classic examples include the BCR-ABLT315I mutation in chronic myeloid leukemia (CML),35 the c-KitT670I mutation in gastrointestinal stromal tumors36 and EGFRT790M in lung adenocarcinomas.37 There are other substitutions described in imatinib-resistant CML patients that are not located near the ATP-binding site of BCR-ABL, but are postulated to destabilize the conformation of the kinase domain required for inhibitor binding.38
Interestingly, some of the BCR-ABL imatinib-resistant mutations (Y235F/E255K/T315I) have also been reported to enhance the oncogenic potency or kinase activity of the fusion protein independently of their effect on drug sensitivity and are therefore considered as gain-of-function mutations.39-41 Using high-sensitivity detection methods, BCR-ABL imatinib-resistant mutations could be detected at low levels in a considerable fraction (12.5% to 22%) of untreated CML patients, indicating that resistant mutations can preexist to the onset of treatment.42-44 Noticeably, the majority of pretreatment secondary mutations are gain-of-function substitutions, suggesting that they might confer a slight proliferation advantage over parental BCR-ABL in the absence of imatinib, and expand during treatment under selective pressure.43,44 In our TS1 model, a similar selection process of preexisting activating JAK1 and JAK3 mutations might take place. During culture, spontaneous mutations occur randomly throughout the TS1 cell genome and some of them activate JAK1 or JAK3. GF-independent cells with a single activating JAK mutation that acquired by chance a secondary activating mutation in the JAK partner activate further the JAK-STAT pathway, which provides a subtle growth advantage allowing for their persistence and slow relative expansion in the culture. JAK inhibitor selection pressure will foster the emergence of these double-mutated cells because their higher basal level of JAK-STAT activation will raise the threshold inhibitor concentration needed to induce apoptosis. The hypothesis that the secondary mutations preexisted before selection with JAK inhibitors is supported by the fact that, in several experiments, a majority of TKI-resistant subclones showed the same secondary mutation (Tables 2 and 3).
In ALL patients, activating mutations in either JAK1, JAK3, and IL-7Rα have been described in approximately 20% of the patients,18,45 raising a potential indication for JAK inhibitors. Our data suggest that such treatment could favor the selection of tumor clones with preexisting mutations in both JAK1 and JAK3. This possibility is supported by the fact that several cases of leukemia patients with concomitant mutations in the pseudo-kinase or kinase domains of JAK1 and JAK3 have already been reported (supplemental Table 3). 18,46-48
Secondary mutations are not the exclusive way for transformed cells to survive in the presence of TKIs, and some of our TKI-resistant subclones did not show any secondary mutation in JAK1 or JAK3 (Tables 2 and 3). The 2 types of JAK inhibitor–selected subclones (mutated and nonmutated) cannot be distinguished based on their sensitivity to inhibitors. All JAK inhibitor–selected subclones do not exhibit an absolute resistance, but rather a relative shift in sensitivity (Figure 3A-B). This gives us a rationale to speculate that the unknown mechanism allowing the nonmutated subclones to proliferate in the presence of inhibitor still relies on JAK activity. However, no mutations in JAK2 and TYK2 were found in 20 nonmutated subclones analyzed so far. Alternatively, a shift in oncogene addiction from the JAK-STAT toward phosphatidylinositol 3-kinase or mitogen-activated protein kinase pathways is not excluded.
In JAK2V617F-transformed cell lines treated with gradually increasing doses of ruxolitinib in vitro, JAK2 overexpression was associated with a reversible stage of so-called JAK2 inhibitor persistence.49 In our laboratory, we observed a similar increase in the expression of JAK1 messenger RNA in BAF3 cells expressing the endogenous JAK1F958V mutant when cells were cultured in the presence of CMP6.30 However, we did not observe any significant increase in JAK1 and JAK3 messenger RNA expression levels for our TKI-resistant subclones without secondary mutations in JAK (data not shown). An alternative resistance mechanism might result from loss of negative regulators of the pathway, such as PTPN2, a phosphatase active on phospho-tyrosine residues located in the activation loop of JAK1 and JAK3 and necessary for full kinase activity.50 Deletions of PTPN2 were identified in 6% of T-cell ALL cases51 and knock-down of PTPN2 reduced sensitivity of cells to JAK inhibition.52
Because of its high versatility, the TS1 model described here represents an invaluable tool for further studies aimed at unraveling new secondary mutation in such genes that might cause increased TKI resistance.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the Belgian Program on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister’s Office, Science Policy Programming (IAP-P7/39) (J.C.R.) and (IAP-P7/43) (S.N.C.); the Actions de Recherche Concertées of the Communauté Française de Belgique (ARC10/15-027) (S.N.C.) and (ARC09/14-021) (J.C.R.); the Fondation Contre le Cancer, the Foundation Salus Sanguinis, Belgium; Fonds pour la formation a la recherche dans l'industrie et l'agriculture (FRIA); Opération Télévie, Belgium; and Associazione Italiana per la Ricerca sul Cancro (AIRC) (IG8803) (M.T.). L.K. is a fellow and S.N.C. is a senior research associate of the Fonds National de la Recherche Scientifique Belgium.
Authorship
Contribution: L.S., T.H., E.L., and F.L. carried out experiments, analyzed the data, and performed the statistical analysis; S.N.C., E.F., and M.T. generated and provided mutated JAK constructs; L.S., T.H., S.N.C., M.T., L.K., and J.C.R. conceived the study and participated in the design and coordination of the experiments; E.L. performed the modeling work; L.S., J.C.R., and L.K. wrote the paper; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jean-Christophe Renauld, Ludwig Institute for Cancer Research, Ave Hippocrate 74, B-1200 Brussels, Belgium; jean-christophe.renauld@bru.licr.org.
References
Author notes
L.S. and T.H. contributed equally to this manuscript.
L.K. and J.-C.R. shared senior coauthorship of this manuscript.