Abstract
Depsipeptide is in clinical trials for chronic lymphocytic leukemia (CLL) on the basis of earlier observations demonstrating selective in vitro activity in CLL. We sought to determine the relationship of histone H3 and H4 acetylation, inhibition of histone deacetylase, and apoptosis observed in CLL cells to justify a pharmacodynamic end point in these clinical trials. We demonstrate that in vitro depsipeptide induces histone H3 and H4 acetylation and histone deacetylase enzyme inhibition at concentrations corresponding to the LC50 (concentration producing 50% cell death) for cultured CLL cells (0.038 μM depsipeptide). The changes in histone acetylation are lysine specific, involving H4 K5, H4 K12, and H3 K9, and to a lesser extent H4 K8, but not H4 K16 or H3 K14. Depsipeptide-induced apoptosis is caspase dependent, selectively involving the tumor necrosis factor (TNF) receptor (extrinsic pathway) initiating caspase 8 and effector caspase 3. Activation of caspase 8 was accompanied by the down-regulation of cellular FLICE-inhibitory protein (c-FLIP, I-FLICE) without evidence of Fas (CD95) up-regulation. Changes in other apoptotic proteins, including Bcl-2, Bax, Mcl-1, and X-linked inhibitor of apoptosis (XIAP), were not observed. Our results demonstrate a relationship between target enzyme inhibition of histone deacetylase, histone H3 and H4 acetylation, and apoptosis involving the TNF-receptor pathway of apoptosis that is not used by other therapeutic agents in CLL. These data suggest use of histone H3 and H4 acetylation, inhibition of histone deacetylase, and down-regulation of FLIP as pharmacodynamic end points for further evaluation of this drug in patients.
Introduction
Chronic lymphocytic leukemia (CLL) is one of the most common types of leukemia diagnosed in the Western hemisphere, with 7000 projected new cases in the United States in the year 2002 and a nationwide prevalence of approximately 50 000 persons.1 CLL is characterized by disrupted apoptosis, as opposed to proliferation, leading to a gradual accumulation of leukemia cells, eventually producing symptoms related to cytopenias or organomegaly.2,3 A small proportion of CLL patients have indolent disease, but the majority of patients either present with advanced-stage disease or progress to the state of requiring treatment. The traditional therapeutic approach to CLL has been to use chlorambucil or fludarabine as initial therapy, although recently completed studies favor the latter with improved response rate and progression-free survival over alkylator-based regimens.4,5
Unfortunately, the majority of patients either fail to gain a complete response to fludarabine therapy or eventually experience a relapse. This emphasizes the importance of focusing upon identification of new and specific therapies for CLL. Ideally, such treatments would work through activation of an apoptotic pathway different from that used in standard regimens and would involve a specific drug target for which the minimally effective pharmacologic dose can be determined in vivo, thus avoiding unnecessary toxicity. Current treatments for CLL (eg, fludarabine, cladribine, and alkylator-based therapies) induce apoptosis via a mitochondria-dependent pathway involving activation of the protease caspase 9.6-9 An alternative pathway for apoptosis involves cell death protease caspase 8 (FLICE) and is triggered by Fas (CD95) and other tumor necrosis factor (TNF) receptor family members. This cytokine-mediated pathway is generally not functional in B-cell CLL.9-11 Attempts to increase expression of the Fas receptor on CLL cells with CD40 ligand (CD40L) or bryostatin have been successful,12 but these leukemic cells remain resistant to Fas-mediated ligation, implying a postreceptor block to apoptosis. Since defects in the mitochondrial pathway of apoptosis are likely to exist in chemotherapy-refractory CLL, identification of agents that exert their cytotoxic effect via the Fas/TNF receptor pathway of apoptosis would represent a major therapeutic advance for the treatment of patients with CLL and present a new opportunity for combination therapies.
Depsipeptide, currently in phase 1 clinical trials,13,14 is one such unique compound that has promise for CLL patient therapy. Our group recently reported15 that this agent exhibits selective cytotoxicity toward CLL B cells as compared with both normal mononuclear cells and bone marrow progenitor cells, with maximal cytotoxicity observed following a 4-hour drug exposure. Others have demonstrated that depsipeptide has selective cytotoxicity toward drug-resistant P388 leukemia cell lines as compared with nonresistant P388 cells, causing down-regulation of c-myc and morphologic normalization of Ras-transformed cells.16-19 In such dividing cell lines, it appears that depsipeptide mediates its effects through inhibition of the enzyme histone deacetylase.20 Indeed, histone acetylation and DNA methylation are two of the primary mechanisms that control gene transcription.21-25 Inhibition of histone deacetylase (HDAC) in tumor cell lines potentially activates differentiation-related or tumor suppressor genes by removing transcriptional repression.21,22
The relationship between alteration of histone acetylation and apoptosis by depsipeptide observed in nondividing CLL cells is presently uncertain. We therefore studied histone acetylation and HDAC inhibition at depsipeptide concentrations that result in increased apoptosis in cultured CLL cells. We also further dissected the pathway of apoptosis used by depsipeptide and examined its treatment effect on levels of select apoptotic regulatory proteins.
Patients, materials, and methods
Patients, cell separation, and culture conditions
Approval for patient blood collection was obtained from The Ohio State University Institutional Review Board for these studies. Informed consent was provided according to the Declaration of Helsinki. Cells were procured from patients who had previously received a diagnosis of CLL as defined by the modified National Cancer Institute (NCI) criteria.26 All of the CLL patients had been without prior therapy for a minimum of 2 months. Mononuclear cells were isolated from the peripheral blood by means of density gradient centrifugation (Ficoll-Paque Plus; Pharmacia Biotech, Piscataway, NJ). Isolation of mononuclear cells in this manner provides more than 90% positive coexpressing CD19 and CD5 clonal B lymphocytes. HeLa (CCL-2) and K562 (CCL-243) cell lines were obtained from American Type Culture Collection (ATCC; Manassas, VA). Cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin-G, 100 μg/mL streptomycin, and 2 mM l-glutamine (Life Technologies, Grand Island, NY). The broad caspase inhibitor carbobenzoxy-valyl-alanyl-aspartyl-fluoromethyl ketone (Z-VAD-fmk) was obtained from Kamiya Biochemical (Seattle, WA). Depsipeptide (FR901228 or NSC649890) and 2-fluoroadenine-9-β-D-arabinofuranoside (2-F-ara-A) were obtained from the Developmental Therapeutics Program, Division of Cancer Treatment, National Cancer Institute.
Apoptosis and flow cytometric studies
Apoptosis was assessed in permeabilized cells by means of a phycoerythrin (PE)–labeled antiactive caspase 3 polyclonal antibody (BD Pharmingen, San Diego, CA) that is specific for the active form of this enzyme. After drug treatment, the cells were washed twice in cold phosphate-buffered saline (PBS). Cells were fixed for 20 minutes with cold Cytofix/Cytoperm solution (BD Pharmingen). Cells were then washed twice with Perm/Wash buffer (BD Pharmingen) and then stained with the PE-labeled antiactive caspase 3 antibody for 30 minutes at room temperature. After a final wash, the cells were analyzed by flow cytometry.
Cationic dyes such as rhodamine-123 readily accumulate in actively respiring mitochondria to a degree dependent of the mitochondrial membrane potential. Thus, rhodamine-123 can be used to monitor the integrity of mitochondria following depsipeptide treatment. Media- and depsipeptide-treated cells were washed once in RPMI 1640 media and then incubated in RPMI 1640 media containing 50 ng/mL rhodamine-123 (Molecular Probes, Eugene, OR) for 30 minutes at 37°C. Stained cells were washed once in RPMI 1640 media, placed on ice, and then quickly analyzed by flow cytometry.
The surface expression of CD95 on CLL cells was assessed with anti-CD95 PE and anti-CD19 fluorescein isothiocyanate (FITC) antibodies with appropriate isotype controls (Becton Dickinson, San Jose, CA). Cells were washed with PBS and then analyzed by flow cytometry.
Histone extraction
For acetylated histone studies, approximately 5 × 107 human CLL cells were used. Nuclei were isolated after 10 minutes' incubation with nuclear isolation buffer NIB plus Triton X (10 mM Tris [tris(hydroxymethyl) aminomethane], pH 7.5; 1.5 mM MgCl2; 1.0 mM CaCl2; 2.0 mM ZnSO4; 0.25 M sucrose; 0.2 mM phenylmethyl sulfonyl fluoride [PMSF]; 0.5% Triton X-100). Cells were transferred to a teflon homogenizer and broken with 20 strokes. Nuclei were pelleted by centrifugation at 2000g for 5 minutes. Nuclear pellets were washed twice in NIB. The washed nuclear pellets were resuspended in 1 mL 0.4 N sulfuric acid and incubated on ice for 30 minutes, followed by a 20-minute centrifugation at maximum speed in a microfuge. Then, 100% trichloroacetic acid (TCA) was added to the supernatant to a final concentration of 20% TCA, and this was incubated at least 30 minutes on ice. After centrifuging for 10 minutes at maximum speed in a microfuge, cold acetone washes with shaking were used to remove salts from the pellet. Histones were then removed from the pellet by shaking with 0.5 mL distilled water and were stored at -20°C.
Western blot analysis
Whole cellular lysates were prepared as previously described.27 Total protein in each sample was quantified by the BCA method (Pierce, Rockford, IL). Lysates or extracted histones were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE)/immunoblotting with antibodies recognizing acetylated or methylated histones (Upstate Biotechnology, Lake Placid, NY) that are specific for various histone acetylation or histone methylation sites (Serotec, Raleigh, NC); pro-caspase 3 (Santa Cruz Biotechnology, Santa Cruz, CA); caspase 8 (BD Pharmingen); caspase 9 (Oncogene Research Products, San Diego, CA); X-linked inhibitor of apoptosis (XIAP) (Transduction Laboratories); Bcl-2 (Santa Cruz Biotechnology); Bax (Santa Cruz Biotechnology); BH3 interacting domain death agonist (BID; Cell Signaling Technology, Beverly MA); poly-adenosine 5′-diphosphate-ribose polymerase (PARP; Oncogene Research Products); and cellular FLICE-inhibitory protein (c-FLIP) (a kind gift from Dr Marcus Peter, University of Chicago, Chicago, IL). Protein samples were separated along with molecular weight markers (Bio-Rad, Hercules, CA) in 10% to 14% polyacrylamide gels. Gels were transferred onto 0.45-μm nitrocellulose membranes (Schleicher and Schuell, Keene, NH). Gel loading equivalence was confirmed by Coomassie blue stain (Sigma, St Louis, MO) of membranes or by probing with antibodies for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Chemicon International, Temecula, CA) or actin (I-19; Santa Cruz Biotechnology). Species-specific immunoglobulin G–horseradish peroxidase (IgG-HRP) secondary antibodies were purchased from Bio-Rad. Blots were developed with chemiluminescent substrate (Pierce Super-Signal; Pierce), and autoradiography was performed with the use of X-OMAT film (Kodak, Rochester, NY). Protein bands were quantified by computer densitometry (Image-Quant; Amersham Biosciences, Sunnyvale, CA).
Immunocytochemistry studies of acetylation on histone H3 and histone H4 proteins
Slides were prepared by depositing 1 × 105 cells isolated from CLL patient blood onto glass slides with the use of a Cytospin 3 centrifuge (Thermo Shandon, Pittsburgh, PA). Slides were Wright-Giemsa stained or fixed for 1 minute at room temperature in a solution of 95% ethanol and 5% glacial acetic acid, and then washed 2 times in PBS. Cells were permeabilized for 10 minutes at room temperature with 0.2% Triton X-100, and then blocked in 10% normal donkey serum (NDS) in PBS for 1 hour at room temperature before incubating overnight at 4°C with antiacetylated histone H3 (Upstate Biotechnology) diluted 1:150 or with antiacetylated histone H4 (Upstate Biotechnology) diluted 1:100 in 2% NDS in PBS. After washing 3 times in PBS, the slides were stained with donkey antirabbit cyanin 3 (Cy3)–conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1.5 hours, and mounted with Cytoseal 60 (Electron Microscopy Science, Fort Washington, PA). In a parallel control experiment, it was observed that omission of either primary antibody eliminated staining. Slides were analyzed by means of an Olympus BX51 microscope equipped with an Olympus PM30 camera (Melville, NY).
Caspase 3, caspase 8, and caspase 9 enzyme activity
A colorimetric reaction assay kit (R&D Systems, Minneapolis, MN) was used to determine the enzymatic activities of caspases 3, 8, and 9 according to the manufacturer's instructions. The colorimetric reaction products were measured by means of an Anthos 2001 microplate reader (Anthos Analytical, Durham, NC) at 405-nm wavelength light.
Histone deacetylase assay
Assays were performed with the use of cell lysates containing 50 μg protein, 40 μL 10 mM Tris-HCl pH 7.0, plus 1 mM benzamidine, Sigma protease inhibitor cocktail (1:100 dilution), 1 mM PMSF, and 2 mM NaVO4; 3H acetate–labeled histones extracted from K562 cells at approximately 1000 cpm; and distilled water to a total volume of 200 μL. Assay mixtures were centrifuged briefly to collect components in the bottom of the tube. Incubations were performed in an Eppendorf Thermomixer at 37°C at 750 rpm for 3 hours. Each reaction was stopped with the use of 50 μL Quenching Solution (3 M HCl plus 0.6 M glacial acetic acid), and tubes were vortexed. Ethyl acetate (0.6 mL) was added to each reaction; tubes were vortexed vigorously for 1 minute; and samples were centrifuged to separate the phases. Radioactivity was determined in two 200-μL aliquots of the ethyl acetate phase by means of a beta-counter (Packard Instrument, Downer's Grove, IL). Counts per minute were corrected for extraction efficiency (50%) and reported as a percentage of the total counts per minute of 3H histone added per assay. To provide a positive and a negative control, HeLa cell lysates incubated with and without 250 mM sodium butyrate were included in all experiments. Samples were assayed in duplicate.
Histone protein substrate preparation
Approximately 1 × 108 K562 cells were grown in RPMI media with additions as described above. Cells were preincubated in media with 200 μg/mL cycloheximide and 10 mM sodium butyrate for 60 minutes at 37°C in a 75-mL Corning (Corning, NY) tissue culture flask. Cells were then incubated in 10 mL media with 300 μCi (11.1 MBq) 3H acetate (DuPont NEN, Boston, MA) in a 50-mL centrifuge tube at 37°C. Cells were washed 3 times in 10 mL PBS plus 10 mM sodium butyrate. The histones were then isolated as described.
Results
Depsipeptide induces a dose-dependent, lysine-specific increase in histone acetylation
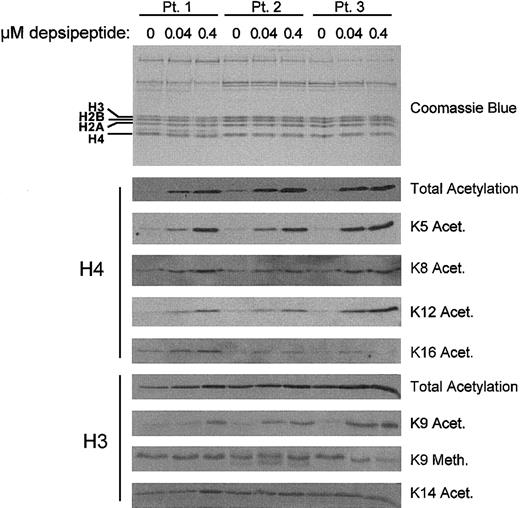
The majority of studies with HDAC inhibitors such as depsipeptide have been performed in dividing tumor cell lines rather than nondividing patient cells. Using nonproliferating cells from CLL patients (n = 5), we sought to determine if a short exposure (4 hours) to concentrations (0.04 to 0.4 μM) of depsipeptide, similar to those attained in phase 1 studies, could promote histone acetylation. Figure 1 shows the in vitro treatment of 3 representative CLL samples with depsipeptide. These studies show that depsipeptide induces a dose-dependent increase in acetylation of total histone H4. Baseline acetylation of histone H3 is greater than histone H4, but also increases modestly with depsipeptide treatment. No change in acetylation was noted at concentrations lower than those reported here (data not shown). By comparison, incubation with the active metabolite of fludarabine, 2-F-ara-A (1 μM), did not promote histone acetylation (data not shown).
Depsipeptide induction of lysine-specific changes in histone acetylation. The amount of global and lysine-specific acetylated H3 and H4 in CLL cells increases in a dose-dependent manner following a 4-hour incubation with depsipeptide. Patients' mononuclear cells were isolated and cultured in media or depsipeptide (0.04 and 0.4 μM) for 4 hours. Histone extractions were performed at 4 hours and analyzed by SDS-PAGE/immunoblotting with antihuman antibodies for acetylated H3 and H4 and specific lysine residues on these histones. Equivalent loading was verified by staining with Coomassie blue.
Depsipeptide induction of lysine-specific changes in histone acetylation. The amount of global and lysine-specific acetylated H3 and H4 in CLL cells increases in a dose-dependent manner following a 4-hour incubation with depsipeptide. Patients' mononuclear cells were isolated and cultured in media or depsipeptide (0.04 and 0.4 μM) for 4 hours. Histone extractions were performed at 4 hours and analyzed by SDS-PAGE/immunoblotting with antihuman antibodies for acetylated H3 and H4 and specific lysine residues on these histones. Equivalent loading was verified by staining with Coomassie blue.
Acetylation of specific lysine residues on histone H3 and histone H4 has been associated with transcriptional activation, differentiation, and deposition of synthesized histones onto newly replicated DNA. We analyzed the histone acetylation patterns of 3 separate CLL patient samples treated with depsipeptide to quantify increases in global histone acetylation and to determine the lysine specificity of the acetylation. The pattern of lysine residue acetylation demonstrated increases in H4 K5, H4 K12, and H3 K9 acetylation in all patient specimens examined. A small change in H4 K8 was observed, and no changes in H4 K16 acetylation, H3 K14 acetylation, or H3 K9 methylation were observed. These findings suggest that inhibition of HDAC by depsipeptide induces acetylation of specific lysine residues on H3 and H4.
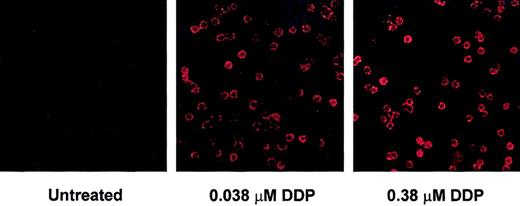
Analysis of changes in histone acetylation by immunoblot does not distinguish whether modifications are occurring in large increments in a small proportion of tumor cells versus equivalent increments among the majority of tumor cells. We therefore examined changes in global H4 and H3 acetylation by immunohistochemistry where individual cells can be evaluated. These experiments revealed changes in H3 (Figure 2) and H4 (data not shown) acetylation in virtually all the CLL cells examined.
Depsipeptide induction of histone H3 acetylation in CLL cells. Histone H3 acetylation increases in the majority of CLL cells following treatment with depsipeptide. CLL cells were incubated in media or depsipeptide (0.038 and 0.38 μM) for 4 hours. Acetylation of H3 was analyzed at 4 hours by immunofluorescence detection with Cy3-conjugated antiacetylated H3 and appropriate negative control antibodies. Magnification is × 100 in all panels.
Depsipeptide induction of histone H3 acetylation in CLL cells. Histone H3 acetylation increases in the majority of CLL cells following treatment with depsipeptide. CLL cells were incubated in media or depsipeptide (0.038 and 0.38 μM) for 4 hours. Acetylation of H3 was analyzed at 4 hours by immunofluorescence detection with Cy3-conjugated antiacetylated H3 and appropriate negative control antibodies. Magnification is × 100 in all panels.
Depsipeptide-induced histone acetylation in CLL cells is promoted by inhibition of histone deacetylase
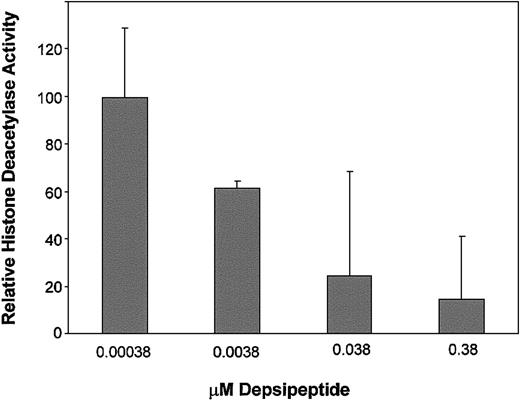
Several compounds, including arsenic trioxide,28 have been noted to induce histone acetylation even in the absence of enzyme inhibition of HDAC. To determine the specificity of the change in histone acetylation with inhibition of the target enzyme HDAC, we studied the kinetics of inhibition of depsipeptide following a 4-hour treatment with this agent in 3 patients. Figure 3 demonstrates that significant (P < .001) inhibition of HDAC activity in primary CLL tumor cells occurs following 4-hour exposure to 0.038 μM and 0.38 μM concentration of depsipeptide as compared with cells incubated in media alone. Specifically, 76% inhibition of global HDAC activity was observed at the depsipeptide concentration of 0.038 μM, which represents the LC50 concentration (concentration producing 50% cell death) at 4 days identified in our previous study. No HDAC inhibition was observed following incubation of cells with 2-Fara-A (data not shown). These data demonstrate that depsipeptide-mediated histone acetylation in nonproliferating cells occurs through direct inhibition of the HDAC enzyme.
Depsipeptide inhibition of histone deacetylase activity in CLL cells. Depsipeptide inhibits histone acetylase at concentrations that promote histone acetylation in vitro in CLL cells. CLL cells were incubated in media or depsipeptide (0.000 38, 0.0038, 0.038 and 0.38 μM) for 4 hours. Histone deacetylase activity was measured by conversion of a tritiated K562 histone substrate. Histone deacetylase activity in the media control is set at 100%, and the depsipeptide treatment data are expressed relative to this. Error bars represent 95% confidence intervals (CI).
Depsipeptide inhibition of histone deacetylase activity in CLL cells. Depsipeptide inhibits histone acetylase at concentrations that promote histone acetylation in vitro in CLL cells. CLL cells were incubated in media or depsipeptide (0.000 38, 0.0038, 0.038 and 0.38 μM) for 4 hours. Histone deacetylase activity was measured by conversion of a tritiated K562 histone substrate. Histone deacetylase activity in the media control is set at 100%, and the depsipeptide treatment data are expressed relative to this. Error bars represent 95% confidence intervals (CI).
Depsipeptide induces caspase-dependent apoptosis
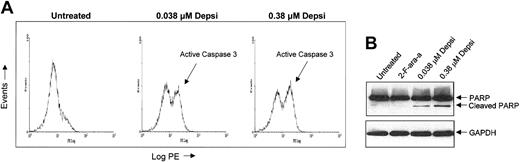
We previously documented that a 4-hour incubation of CLL cells with depsipeptide induces apoptosis in a dose-dependent fashion15 and have demonstrated (Figure 3) that greatest inhibition of HDAC activity occurs at concentrations corresponding to where apoptosis is noted. Apoptosis can occur through caspase-dependent and independent pathways. In the caspase-dependent pathway, caspase 3 serves as an effector molecule by cleaving cellular proteins, including PARP, that are key for cell survival. We detected caspase 3 activation in depsipeptide-treated cells, as assessed by means of flow cytometric analysis with a PE-directed antibody specific for the active cleavage product (Figure 4A). In addition, we demonstrated both cleavage of caspase 3 with appearance of the 17-kDa active protease (data not shown) and cleavage of the downstream substrate PARP by immunoblotting (Figure 4B). Furthermore, addition of the pan-caspase inhibitor Z-VAD-fmk inhibits this process (data not shown).
Effect of depsipeptide on caspase 3 activation and PARP cleavage. (A) Depsipeptide treatment results in activation of caspase 3 as assessed by flow cytometry with the use of a PE-directed antibody specific for the active cleavage product of caspase 3. Depsipeptide in human CLL cells activates caspase 3. CLL cells were treated with media or depsipeptide (0.038 and 0.38 μM) for 4 hours and subsequently incubated in media for 20 hours. Cells were washed, permeated, and stained with a PE-directed antibody specific for the active cleavage product of caspase 3. (B) Depsipeptide treatment results in cleavage of PARP. Depsipeptide-mediated apoptosis promotes processing of PARP. To determine if depsipeptide treatment caused alteration of a caspase 3 substrate, we examined both the unprocessed and the processed forms of PARP in fresh human CLL cells at 24 hours following a 4-hour incubation of CLL cells with media, 0.038 μM depsipeptide, or 0.38 μM depsipeptide. Protein lysates were prepared, and 50 μg protein per lane was separated on a 14% SDS-PAGE gel. Loading equivalence was confirmed by blotting with an antibody for constitutively expressed protein GAPDH. PARP and its cleaved product were detected by means of an anti-PARP polyclonal antibody.
Effect of depsipeptide on caspase 3 activation and PARP cleavage. (A) Depsipeptide treatment results in activation of caspase 3 as assessed by flow cytometry with the use of a PE-directed antibody specific for the active cleavage product of caspase 3. Depsipeptide in human CLL cells activates caspase 3. CLL cells were treated with media or depsipeptide (0.038 and 0.38 μM) for 4 hours and subsequently incubated in media for 20 hours. Cells were washed, permeated, and stained with a PE-directed antibody specific for the active cleavage product of caspase 3. (B) Depsipeptide treatment results in cleavage of PARP. Depsipeptide-mediated apoptosis promotes processing of PARP. To determine if depsipeptide treatment caused alteration of a caspase 3 substrate, we examined both the unprocessed and the processed forms of PARP in fresh human CLL cells at 24 hours following a 4-hour incubation of CLL cells with media, 0.038 μM depsipeptide, or 0.38 μM depsipeptide. Protein lysates were prepared, and 50 μg protein per lane was separated on a 14% SDS-PAGE gel. Loading equivalence was confirmed by blotting with an antibody for constitutively expressed protein GAPDH. PARP and its cleaved product were detected by means of an anti-PARP polyclonal antibody.
Depsipeptide-induced apoptosis occurs via the tumor necrosis factor–receptor pathway
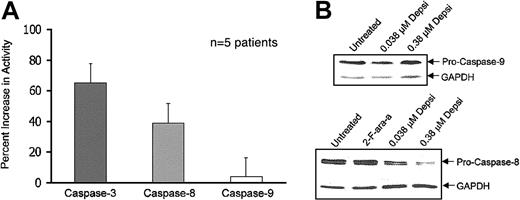
Caspase-dependent apoptosis can be initiated through a variety of signal cascades, including the TNF receptor (caspase 8) and mitochondria (caspase 9) pathway of apoptosis. These initiator caspases subsequently activate the effector caspase 3. To further characterize the apoptotic pathway used by depsipeptide to activate caspase 3, we examined whether the activity levels of either caspase 8 or caspase 9 increased following treatment, an indication of proteolytic processing and activation. Five patient samples were exposed to media or depsipeptide (0.38 μM) for 4 hours and subsequently analyzed for caspase 8 and caspase 9 activity. These data, depicted in Figure 5A, demonstrate that caspase 3 and caspase 8 activities increase over baseline at 1 day, but that caspase 9 activity changes only minimally following treatment.
Effect of depsipeptide on activation of caspases 3, 8, and 9 and on processing of caspases 8 and 9. (A) Depsipeptide induces activation of caspases 3 and 8 but not caspase 9. Human CLL cells were exposed to depsipeptide (0.38 μM) for 4 hours and compared with media control at 24 hours. Changes in caspase 3, caspase 8, and caspase 9 activities were determined by a colorimetric reaction assay. Error bars represent 95% confidence intervals (CI). (B) Depsipeptide induces processing of the caspase 8 but not the caspase 9 pro-form. To confirm the findings of selective caspase 8 activation in CLL cells, protein lysates were prepared, and 50 μg protein per lane was separated on a 14% SDS-PAGE gel. Loading equivalence was confirmed by blotting with an antibody for constitutively expressed protein GAPDH. The unprocessed form of caspase 8 and caspase 9 was detected with the use of appropriate antibodies.
Effect of depsipeptide on activation of caspases 3, 8, and 9 and on processing of caspases 8 and 9. (A) Depsipeptide induces activation of caspases 3 and 8 but not caspase 9. Human CLL cells were exposed to depsipeptide (0.38 μM) for 4 hours and compared with media control at 24 hours. Changes in caspase 3, caspase 8, and caspase 9 activities were determined by a colorimetric reaction assay. Error bars represent 95% confidence intervals (CI). (B) Depsipeptide induces processing of the caspase 8 but not the caspase 9 pro-form. To confirm the findings of selective caspase 8 activation in CLL cells, protein lysates were prepared, and 50 μg protein per lane was separated on a 14% SDS-PAGE gel. Loading equivalence was confirmed by blotting with an antibody for constitutively expressed protein GAPDH. The unprocessed form of caspase 8 and caspase 9 was detected with the use of appropriate antibodies.
These data were confirmed by 5 separate in vitro experiments that showed no decrease in the noncleaved caspase 9 but a dose-dependent decline in uncleaved caspase 8, generally corresponding to activation of this caspase following exposure to increasing concentrations of depsipeptide (Figure 5B). This suggests that depsipeptide uses the TNF-receptor pathway of apoptosis to activate caspase 8, which leads to recruitment of caspase 3 and subsequent cleavage of PARP. While caspase 8 can cross-activate caspase 9 through cleavage of BID in some systems, we were unable to demonstrate either baseline or induced expression of BID (data not shown) in CLL cells as one other group has also reported.29 Furthermore, we did not detect caspase 9 processing, as shown in Figure 5B.
FLIP expression decreases without CD95 induction following depsipeptide exposure
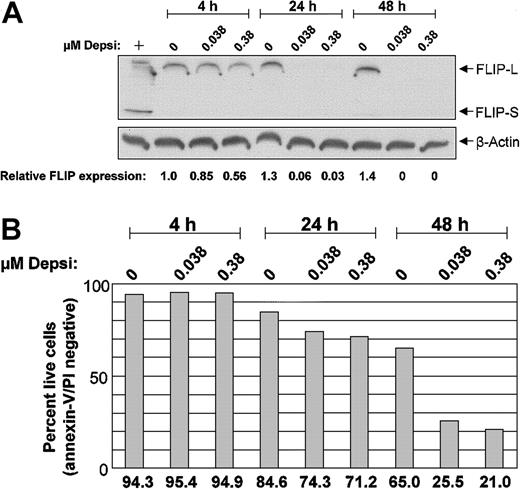
Caspase 8 activation can occur through a variety of TNF-receptor pathways, including signaling via Fas and Fas ligand. The FLIP acts downstream of Fas to inhibit TNF-receptor–mediated apoptosis. FLIP can bind to the adaptor protein Fas-associated death domain (FADD) and to caspase 8, interfering with caspase 8 activation. Given that Fas ligation promotes, and FLIP impedes, caspase 8 activation, we assessed expression of these proteins on days 1 and 2 following in vitro exposure to depsipeptide in cells from 6 patients with CLL. At baseline, no patient samples expressed Fas on more than 10% of the CLL cells, and we detected neither an increased population of Fas-expressing CLL cells nor an up-regulation of cell surface Fas expression following depsipeptide exposure. In contrast, FLIP was noted to decrease as early as 4 hours in 7 of the 9 CLL patient samples treated with depsipeptide. As decreased levels of c-FLIP following depsipeptide could represent degradation occurring as an event associated with apoptosis, we performed a parallel assessment of apoptosis by annexin/propidium iodide (PI), mitochondrial membrane potential, and c-FLIP expression analyses as demonstrated in Figure 6 in 4 patients. Similar results were noted for all 4 patients; representative data from 1 patient are shown in Figure 6. In each case, at the time apoptosis was noted by annexin-V/PI staining (24 hours and beyond; Figure 6B), loss of mitochondrial membrane potential occurred. Notably, depsipeptide treatment resulted in a substantial decrease in c-FLIP-L expression prior to detectable induction of apoptosis as early as 4 hours. The splice variant FLIP-S was not detected in these samples. These findings suggest that depsipeptide induces apoptosis via a caspase 8 pathway that does not require Fas expression and ligation, but instead involves FLIP down-modulation. Examination of other antiapoptotic proteins, including Bcl-2, Bax, Mcl-1, and XIAP, at these same time points demonstrated no change with depsipeptide treatment (data not shown).
Effect of depsipeptide on c-FLIP in CLL cells. Following in vitro exposure to depsipeptide, c-FLIP decreases in CLL cells. CLL patient cells were incubated with or without depsipeptide (DDP) for 4, 24, and 48 hours. (A) Protein lysates were prepared at each time point, separated on a 14% SDS-PAGE gel, and immunoblotted with polyclonal anti–c-FLIP. Gel loading equivalence was confirmed by blotting with an antibody for constitutively expressed protein beta-actin. FLIP expression in these samples was measured by laser densitometry, and is shown relative to the 4-hour untreated sample after equalizing to actin. The positive control (+) is lysate from CLL cells stimulated with CD40L for 12 hours. (B) Prior to lysing, an aliquot of cells from each condition was assessed for early apoptosis by flow cytometry with annexin-V FITC and propidium iodide.
Effect of depsipeptide on c-FLIP in CLL cells. Following in vitro exposure to depsipeptide, c-FLIP decreases in CLL cells. CLL patient cells were incubated with or without depsipeptide (DDP) for 4, 24, and 48 hours. (A) Protein lysates were prepared at each time point, separated on a 14% SDS-PAGE gel, and immunoblotted with polyclonal anti–c-FLIP. Gel loading equivalence was confirmed by blotting with an antibody for constitutively expressed protein beta-actin. FLIP expression in these samples was measured by laser densitometry, and is shown relative to the 4-hour untreated sample after equalizing to actin. The positive control (+) is lysate from CLL cells stimulated with CD40L for 12 hours. (B) Prior to lysing, an aliquot of cells from each condition was assessed for early apoptosis by flow cytometry with annexin-V FITC and propidium iodide.
Discussion
Previous preclinical studies of CLL cells derived from patients with this disease have demonstrated that depsipeptide selectively induces apoptosis of tumor cells, relative to normal mononuclear cells or bone marrow progenitor cells.15 In the study presented here, we have demonstrated that apoptosis induced by depsipeptide in CLL B cells corresponds to increases in histone H3 and H4 acetylation that is restricted to specific lysine residues. This increase in histone acetylation is noted early following depsipeptide exposure and occurs as a consequence of inhibition of the enzyme histone deacetylase, confirming the results of one prior study of this agent in proliferating cell lines.20 We further demonstrate that apoptosis induced by depsipeptide involves a caspase-dependent pathway, using the TNF-receptor pathway of apoptosis (caspase 8) followed by activation of the effector caspase 3 to promote apoptosis in human CLL cells. Activation of the caspase 8 pathway does not appear to involve induction of expression of Fas (CD95) or of its ligand CD95L. However, we observed that c-FLIP, a protein that inhibits caspase 8 activation, is down-modulated by depsipeptide treatment at a time before apoptosis is noted. The observation that depsipeptide operates via a caspase 8–mediated process in human CLL cells is quite significant, as this pathway is not activated by any other therapeutic agents currently used in the treatment of this disease.6-9
Posttranslational modification of the histone proteins is a central component to timely activation and inhibition of genes important to cell growth and survival. Such modifications occur on the tail of histone proteins and include acetylation, phosphorylation, methylation, ubiquitination, and adenosine 5′-diphosphate (ADP) ribosylation. In this study, we have demonstrated that depsipeptide-induced inhibition of histone deacetylase and subsequent increased histone acetylation appear to be lysine specific, as illustrated by varied acetylation patterns of distinct lysine residues. The most notably acetylated lysine residues were those associated with chromatin formation and assembly (H4 K5 and K12, and H3 K9).30 Hyperacetylation of the K9 residue on H3 may prevent the silencing of genes by preventing methylation of H3 K9, which contributes to gene repression.31,32 In addition, changes in histone acetylation and other modifications can influence posttranslational modifications at other sites. To date, studies examining the direct consequences of lysine-specific modifications (acetylation, methylation, and phosphorylation) in neoplastic cells have been limited. The lysine residue pattern of histone acetylation induced by depsipeptide is similar to that resulting from mutation of the yeast histone deacetylase RPD3.33-35 RPD3 is the prototype of the human class I histone deacetylase class (HDAC 1-3), suggesting that depsipeptide may target these enzymes.36-39 Understanding the role of specific histone deacetylase enzymes on these lysine residues and which of these are affected by specific histone deacetylase inhibitors will be important for eventual clinical exploitation of these agents in CLL and other diseases.
The mechanism by which depsipeptide and other histone deacetylase inhibitors induce cytotoxicity in CLL cells and other hematologic malignancies is still uncertain but may involve down-regulation of cytokines necessary for survival40,41 or differentiation,42-44 or induction of genes that promote apoptosis. Indeed, much research on histone deacetylase inhibitors has focused on the ability of these agents to promote differentiation,42-44 presumably as a result of transcriptional activation of several genes. This increase in acetylation provides enhanced DNA access by transcription factors. Preliminary studies by our group have not demonstrated expression changes in B-cell differentiation markers such as CD22 and CD25 induction as observed by others with the differentiating agent bryostatin (data not shown). Others have demonstrated that histone deacetylase inhibitors such as trichostatin A and sodium butyrate cause diminished interleukin 2 (IL-2)–mediated gene expression prior to induction of apoptosis.41,42 Furthermore, in cytokine-dependent and independent hematopoietic cell lines, acetylation of histone proteins was always noted, while apoptosis was observed only in cell lines dependent upon IL-2 for growth.40,41 Such pathways are currently under investigation in our laboratory.
Other histone deacetylase inhibitors, including butyrate, suberoylanilide hydroxamic acid (SAHA), trichostatin, and apicidin, have been investigated in either myeloid or lymphoblastic cell lines,44-50 with varied results suggesting extrinsic, intrinsic, and caspase-independent apoptosis as the relevant death pathway. Only one of these studies, by Amin and colleagues,50 included primary tumor cells. This study investigated the affect of trichostatin, SAHA, and sodium butyrate on acute promyelocytic leukemia (APL) cells and in several APL cell lines. In these experiments, trichostatin, SAHA, and butyrate were demonstrated to induce caspase-dependent apoptosis, but results of investigation of the relevant upstream initiator caspase were not conclusive, leaving the importance of caspase 8 or 9 activation in this process unknown. The relevance of these previous studies to our own are limited, as the experiments reported herein were carried out with a histone deacetylase inhibitor in primary nonproliferating human tumor cells, as opposed to actively proliferating transformed cell lines.
In this study, we have demonstrated that depsipeptide induces caspase-dependent apoptosis in human CLL cells that involves relatively selective activation of the same apoptosis pathway used by the TNF-family receptors. Our studies have shown that activation of this process in CLL does not involve induction of Fas with subsequent activation of the Fas–Fas ligand signaling pathway, as observed in APL cell lines with apicidin.49 Alternative pathways of activating this pathway through death receptor 4 (DR4), DR5, and TNF receptors that could explain this were not examined, and these are currently under study in our laboratory. However, it is of interest that depsipeptide down-regulated c-FLIP concurrently with the processing of caspase 8. FLIP blocks death receptor–mediated signaling by preventing caspase 8 activation at the death-inducing signaling complex and/or release from the death-inducing signaling complex.51,52 The extrinsic pathway of apoptosis is generally not functional in patients with CLL, even when expression of CD95 occurs on the surface of CLL cells following treatment with bryostatin or CD40 ligand.12 Lack of apoptosis in this setting may occur as a consequence of CD40 ligand–induced up-regulation of c-FLIP or bryostatin-induced up-regulation of XIAP, which are effective inhibitors of caspase 8 and caspase 3, respectively.12 However, in this manuscript we have provided evidence that depsipeptide treatment results in a decrease in c-FLIP in the majority (7 of 9) patient samples assessed. Reasons for not observing a decrease in c-FLIP in these other 2 patients may reflect alternative regulatory mechanisms of c-FLIP in different genetic subtypes of CLL or possibly altered cellular uptake of depsipeptide due to varied presence of multidrug resistance (MDR) expression/efflux.53 Effective activation of this apoptosis pathway in CLL may explain previous work by our group that demonstrated no difference of in vitro CLL cell sensitivity to depsipeptide relative to previous treatment status.15 Exploitation of the ability of depsipeptide to activate the caspase 8 pathway in combination with other therapies such as bryostatin, CD40 ligand, IL-12, and CD154 adenovirus gene transfer, which increase CLL cell Fas expression, may offer a new treatment strategy for this incurable disease.
Virtually all agents used in the treatment of cancer are most effective when administered in combination with other treatments. Depsipeptide targets a specific class of enzymes whose activity can be followed to prevent unnecessary dose escalation that may enhance toxicity without additional therapeutic benefit. Here, we have provided in vitro validation of using the early pharmacodynamic end point of change in histone acetylation and inhibition of histone deacetylase, with later reductions in c-FLIP in human CLL cells. This provides a strategy to proceed with the clinical development of depsipeptide, targeting the minimally effective pharmacologic dose in vivo in patients that promotes acetylation of H3 and H4 histone proteins in primary CLL tumor cells. This will be most relevant to studies that seek to combine depsipeptide with therapies that have demonstrated cytotoxic and differentiation synergy in other diseases, but also produce both medullary and extramedullary toxicity. Using the biologic end points described in this report to target the minimally effective pharmacologic dose in initial clinical trials with depsipeptide will avoid exceeding the favorable therapeutic index of depsipeptide with normal immune effector cells and bone marrow progenitor cells, and will facilitate effective combinations with other therapies. On the basis of these data, such trials are currently underway in patients with CLL.54
Prepublished online as Blood First Edition Paper, March 20, 2003; DOI 10.1182/blood-2002-12-3794.
Supported in part by the CLL Research Consortium (P01 CA81534-02) (J.C.B., M.R.G., J.C.R., and S.K.); The Sidney Kimmel Cancer Research Foundation (J.C.B.); The Leukemia and Lymphoma Society of America (J.C.B.); The D. Warren Brown Foundation (J.C.B.); the National Cancer Institute CA96323-01 (J.C.B., M.R.P., G.M.); and P30 CA16058 and RPG-00-340-01-CSM from The American Cancer Society (M.R.P.). J.C.B. is a Clinical Scholar of the Leukemia and Lymphoma Society of America.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.