Abstract
The t(12;21) translocation resulting in TEL/AML1 gene fusion is present in approximately 25% of patients with precursor B-lineage pediatric acute lymphoblastic leukemia (ALL). Studies suggest an association with a good prognosis; however, relapse can occur. We studied the relation between t(12;21), determined by fluorescence in situ hybridization or polymerase chain reaction, and in vitro drug resistance, measured by the MTT assay, in childhood B-lineage ALL at diagnosis. A total of 180 ALL samples were tested, 51 (28%) of which were positive for t(12;21). The median LC50 values did not differ significantly between TEL/AML1-positive and -negative samples for prednisolone, dexamethasone, daunorubicin, thiopurines, epipodophyllotoxins, and 4-HOO-ifosfamide. However, the TEL/AML1-positive patients were relatively more sensitive tol-asparaginase (ASP; 5.9-fold; P = .029) and slightly but significantly more resistant to vincristine (1.5-fold;P = .011) and cytarabine (1.5-fold; P = .014). After matching for unevenly distributed patient characteristics—that is, excluding patients younger than 12 months, patients with CD10-negative immature B-lineage ALL, patients with Philadelphia chromosome, and patients who were hyperdiploid (more than 50 chromosomes) from the TEL/AML1 negative group—the only remaining difference was a relative sensitivity for ASP in the TEL/AML1-positive samples (10.8-fold; P = .012). In conclusion, the presence of TEL/AML1 gene fusion in childhood precursor B-lineage ALL does not seem to be associated with a high in vitro drug sensitivity, except for ASP, indicating that these patients could benefit from treatment schedules with significant use of this drug.
The prognosis of children with leukemia has improved dramatically in the past decades because of the development of effective combination chemotherapy. Nevertheless, approximately 25% of children with acute lymphoblastic leukemia (ALL) die of resistant or relapsed disease. Moreover, patients who may be cured with relatively mild chemotherapy might nowadays be overtreated and experience unnecessary side effects. It is, therefore, of great importance to identify children with highly sensitive leukemia who can be cured with minimal therapy and children who need specifically intensified therapy. Hence, clinical and biologic features of prognostic importance are sought to tailor therapy according to risk.1
Cytogenetic abnormalities of leukemic cells have proven to be valuable independent prognostic factors. For example, hyperdiploidy (more than 50 chromosomes) correlates with a favorable outcome, whereas translocations such as t(9;22) and t(4;11) are associated with a poor survival rate.1-6 It is thought that these aberrations reflect or cause differences in drug sensitivity. Since the improvement of in vitro drug sensitivity assays, the in vitro cellular drug resistance profile has itself also proven to be a strong independent prognostic factor.7-10
Recently, molecular techniques have identified the t(12;21)(p13;q22) translocation, occurring in approximately 25% of newly diagnosed precursor B-lineage childhood ALL,11-19 resulting in the fusion of TEL to the transactivation domain of AML1. The TEL gene is a member of the ETS-family of transcription factors.20,21AML1 knockout mice provide evidence that this gene is essential for hematopoiesis (for review, see Lococo et al22). The expression of the TEL/AML1 fusion gene appears to interfere with AML1-dependent gene regulation in a dominant-negative manner.23
In childhood ALL, patients with TEL/AML1 display a favorable phenotype, precursor B cell (CD10+, CD19+, HLA DR+), and a favorable age distribution, often between 2 and 10 years. Some studies have shown that patients with TEL/AML1 have excellent prognoses, ranging from 90% to 100% event-free survival rates.13,15,16,19,24,25However, Takahashi et al26 report that in the 8511/8610 Japanese TPOSG protocols, there is no significant difference in disease-free survival between TEL/AML1-positive and -negative B-precursor ALL. Furthermore, the frequency of TEL/AML1 gene fusion in relapsed Philadelphia (Ph1) chromosome-negative, precursor B-cell ALL has been reported to be 24% in patients treated with the Berlin–Frankfurt–Münster (BFM) group protocols.27,28 The latter study indicates that TEL/AML1-positive patients have relapses later and might only display better short-term outcomes. These results are in conflict with those of Loh et al,29 who report a very low incidence of TEL/AML1 gene fusion in relapsed patients treated with Dana–Farber Cancer Institute (DFCI) protocols, indicating that the prognostic impact of TEL/AML1 gene fusion might well be therapy dependent. Hence, the independent prognostic value of TEL/AML1 has to be further evaluated in large, prospective studies with sufficient lengths of follow-up. The question also arises whether this gene fusion leads to a specific in vitro drug resistance profile or phenotype and whether a more specific treatment can then be devised.
In this study using the MTT assay, we have investigated whether TEL/AML1 positive patients differ from other cases of B-lineage ALL in in vitro drug resistance profile, and compared to a matched precursor B-cell lineage TEL/AML1 negative group excluding Ph1 chromosome, infant and hyperdiploid (> 50) cases.
Materials and methods
Patients and leukemic cell samples
Freshly obtained cells from bone marrow or peripheral blood of 192 children with newly diagnosed, untreated ALL from the Dutch Childhood Leukemia Study Group, the German Co-operative ALL (COALL) study group, the Rigshospitalet in Copenhagen, Sophia Children's Hospital in Rotterdam, and the University Hospital Vrije Universiteit were used for this study.
Immunophenotyping and DNA index flow cytometry were performed at reference laboratories of the participating groups; patient characteristics (gender, age, white blood cell count at diagnosis) were collected by study centers. Karyotyping of the Dutch patients was performed by members of the Netherlands Working Party on Cancer Genetics and Cytogenetics at regional cytogenetics centers. Karyotyping of Danish patients was performed at the Rigshospitalet. B-lineage immunophenotype was defined as HLA-DR+/terminal deoxynucleotidyl transferase (TdT)+/CD19+ ALL and further differentiated as follows: proB-ALL (CD10−/cytoplasmic μ chain (cμ)−/surface immunoglobulin (sIg)−), common (c)-ALL (CD10+/cμ−/sIg−) and preB-ALL (CD10+ or CD10−/cμ+/sIg−). No B-ALL samples (CD10−/cμ−/sIg+) were present in this study.
Leukemic blast cells were isolated within 48 hours of sampling by density-gradient centrifugation (Lymphoprep, 1.077 g/mL; Nycomed Pharma, Oslo, Norway; at 480g for 15 minutes). After washing, the cells were resuspended in RPMI 1640 (Dutch modification; Gibco BRL, Breda, The Netherlands) containing 20% fetal calf serum (Gibco BRL) and other supplements.30 When necessary, contaminating normal cells were removed by monoclonal antibodies linked to magnetic beads, as described previously.31 All samples contained more than 80% leukemic cells, as determined by cytospin preparations stained with May–Grünwald–Giemsa (Merck, Darmstadt, Germany).
Reverse transcription–polymerase chain reaction assay
RNA isolation and reverse transcription (RT) were performed as described elsewhere.27 Amplification of the TEL/AML1 fusion gene was performed as published previously, using the following primers: TEL external sense, 5′-AGCCCCATCATGCACCCTCTGATCC-3′; TEL internal sense, 5′-GCAGAATTCCACTCCGTGGATTTCAAACAGTCC-3′; AML1 external antisense, 5′-GTGGTCGGCCAGCACCTCCACC-3′; AML1 internal antisense, 5′-AACGCCTCGCTCATCTTGCCTGGGCTC-3′.17,27 The ubiquitously expressed ABL gene was amplified in a separate polymerase chain reaction (PCR) as a control for the RNA isolation and subsequent cDNA synthesis.27
Fluorescence in situ hybridization
Dual-colored fluorescence in situ hybridization (FISH) experiments for the t(12;21) were performed with the cosmid probe 50F4 for the exon 2 of TEL32 and cosmid 664 containing the first 5 exons of the AML1 gene.33 Nick translation was used to label cosmid probe 664 with biotin-16-dUTP (Boehringer Mannheim, Amsterdam, The Netherlands) and cosmid probe 50F4 with digoxygenin-11-dUTP (Boehringer Mannheim).34
FISH analysis was performed on cytospins stored at −20°C as described previously.35 Briefly, slides were pretreated with RNase and pepsin solutions, followed by postfixation with acid-free formaldehyde and denaturation. Probes were denatured (4 minutes at 72°C in 70% formamide) in the presence of a 100-fold excess of Human-COT-1 DNA (Life Technologies, Breda, The Netherlands), and preannealed for 60 minutes at 37°C. Hybridization of the probes to the slides was allowed to proceed overnight at 37°C. The biotin hybridization signal (cosmid 664) was visualized using fluorescein avidin (Vector Laboratories, Burlingame, VT) and biotinylated anti-avidin D sandwich detection (affinity purified; Vector Laboratories). The digoxigenin hybridization signal (cosmid 50F4) was detected using anti-digoxigenin–rhodamine (Boehringer Mannheim) and donkey anti-sheep–Texas red (Jackson ImmunoResearch Laboratories, Westgrove, PA). Cells were counterstained with DAPI/Vectashield mounting medium (Vector Laboratories). Fluorescence signals were visualized on an Axioskope fluorescence microscope (Zeiss, Weesp, The Netherlands) with an Atto Arc 100-W lamp (Zeiss) equipped with double and triple bandpass filters for simultaneous visualization of rhodamine–TR/fluorescein isothiocyanate/DAPI. At least 200 nuclei were blindly scored by 2 independent observers. In t(12;21)-positive patients, the 2 probes coalesced to form a fusion spot (Slater et al, manuscript in preparation).
In vitro drug resistance assay
In vitro drug cytotoxicity was determined in the MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazoliumbromide) assay as described previously.30,36 Briefly, 100 μL cell suspension was cultured for 4 days in the absence (ie, control) or presence of 6 duplicate concentrations of each drug. During a final 6-hour incubation, the yellow MTT tetrazolium salt was reduced to formazan (purple-blue) crystals by the living cells only, resulting in an optical density linearly related to the number of viable cells.36,37 Drug sensitivity was assessed by the LC50, the drug concentration lethal to 50% of the cells. The assay was deemed evaluable when a minimum of 70% leukemic cells were present in the control wells after 4 days of incubation and when the control optical density was higher than 0.050.30 31
Drug concentration ranges were based on earlier studies and were aimed at obtaining LC50 values for highly sensitive and for resistant cases. The following drugs and concentration ranges were used: prednisolone disodium phosphate (PRED; 0.06-250 μg/mL; Bufa Pharmaceutical Products, Uitgeest, The Netherlands), dexamethasone sodium phosphate (DEX; 0.0002-6.0 μg/mL; Brocacef, Almere, The Netherlands), vincristine (VCR; 0.05-50 μg/mL; Oncovin; Eli Lilly, Amsterdam, The Netherlands), l-asparaginase (ASP; 0.003-10 IU/mL; Medac, Hamburg, Germany), daunorubicin hydrochloride (0.002-2.0 μg/mL; Cerubidine; Rhône–Poulenc Rorer, Amstelveen, The Netherlands), doxorubicin hydrochloride (0.008-8.0 μg/mL; Adriblastina; Pharmacia, Woerden, The Netherlands), mitoxantrone (0.001-1.0 μg/mL; Novantrone; Lederle, Etten–Leur, The Netherlands), 6-mercaptopurine (15.6-500 μg/mL; Sigma, St. Louis, MO), 6-thioguanine (1.6-50 μg/mL; Sigma), cytarabine (Ara-C; 0.002-2.5 μg/mL; Cytosar; Upjohn, Ede, The Netherlands), teniposide (0.003-8.0 μg/mL; Vumon; Bristol–Myers, Weesp, The Netherlands), etoposide (0.05-50 μg/mL; etoposide–TEVA; TEVA–Pharma, Mijdrecht, The Netherlands), and 4-hydroperoxy–ifosfamide (0.10-100 μg/mL; kindly provided by Asta Medica, Frankfurt am Main, Germany).
It has been shown that the source of the leukemic cells (bone marrow or peripheral blood) does not affect the drug resistance measured.36 Eleven of 180 samples were tested after cryopreservation (3 of 51 TEL/AML1-positive and 8 of 129 TEL/AML1-negative samples), which has been shown not to affect the drug resistance measured.37
Statistics
Distributions of clinical and biologic variables for patients with and without TEL/AML1 gene fusion were compared by the Mann–WhitneyU or χ2 test. LC50 values for the drugs tested were compared between TEL/AML1 gene fusion-positive and -negative samples by Mann–Whitney U test.
Results
For 192 patients in this study, the presence of TEL/AML1 gene fusion was successfully determined either by FISH (Dutch patients, n = 95) or by RT-PCR (Danish and German patients, n = 97). For the Dutch patients, whose diagnoses were made between November 1988 and June 1997, the FISH studies were carried out on cryopreserved cytospins. Some of these patients were selected because they had karyotypic abnormalities of chromosome 12p. Of the 95 patients with successful FISH results, 31 (32.6%) were positive for TEL/AML1. Using the TEL probe cos 50F4, 15 of 23 (65.2%) displayed loss of heterozygosity. The 97 German COALL study group and Danish patients with successful RT-PCR results received their diagnoses between April 1993 and April 1997 and were tested on a prospective basis. Twenty-one (21.6%) of these patients were positive for TEL/AML1 fusion (11 of 42, 26.2%, COALL samples; 10 of 55, 18.2%, Danish samples). In total, 52 TEL/AML1-positive patients and 140 TEL/AML1-negative patients were identified, yielding an overall prevalence of TEL/AML1 in the samples received for in vitro drug resistance testing of 27.1%.
All TEL/AML1-positive patients were older than 2 years of age and expressed the B-lineage common/precursor B (c/preB) immunophenotype. Patients whose DNA index or karyotype was available were not hyperdiploid (unknown, n = 12). Only 11.5% of the TEL/AML1-positive patients were older than 10 years of age, whereas 17.9% of the TEL/AML1-negative group were. In contrast, patients in the TEL/AML1-negative group had various immunophenotypes and displayed other high-risk parameters. For example, 6 patients were infants and 5 were positive for the Ph1 chromosome, as shown by cytogenetic analysis or PCR. The distribution of initial white blood cell count did not differ significantly between the 2 groups. Twenty-seven percent of the TEL/AML1-negative patients were hyperdiploid (more than 50 chromosomes).
Karyotypes were available for 29 (55.8%) TEL/AML1-positive patients and 89 (63.6%) TEL/AML1-negative patients; Table1 contains a summary for the positive patients. Seven of the TEL/AML1-positive samples had an apparently normal karyotype, no patient had hyperdiploidy, and no other specific translocations were identified in this group. Of the TEL/AML1-negative patients, 28 of 89 had a normal karyotype, and specific translocations included 2 t(9;22)(q34;q11), 2 involved 11q23 (t(6;11)(q27;q23) and t(1;11)(p32;q23), and 3 t(1;19)(q23;p13).
Available karyotypes of TEL/AML 1-positive patients described according to ISCN 1995
| Patient . | . |
|---|---|
| 1,2 | 46,XX |
| 3-7 | 46,XY |
| 8 | 45,X,−X,dup(1)(q21q32∼41),del(12)(p12p13)[13]/46,XX[20] |
| 9 | 45,XY,der(12)t(12;18)(p11∼12;q12),−18[22]/46,XY[3] |
| 10 | 95,XXYY,add(7)(q2?2)×2,del(9)(p21)×2,del(11)(q22)×2,add(12)(p12)×2, +mar[34]/46,XY[75] |
| 11 | 45,XY,t(1;6)(p21;q26∼27),add(2)(p?),−9,add(9)(p2?2),add(11)(q1?4), add(12)(p12)[37] |
| 12 | 45,X,−X,add(21)(p10)[12]/46,XX[1] |
| 13 | 45,X,−X,t(4;12)(q13;p11-p12),?der(21)[18]/46,XX[3] |
| 14 | 46,XX,del(6)(q14∼15q22∼23),del(14)(q21),−15,+?16[24]/46,XX[14] |
| 15 | 46,XX,?7,add(9)(q3?),del(12)(p11),?14,del(17)(q?),add(18)(q?)[6]/46, XX[4] |
| 16 | 46,XX,del(12)(p1?2)[12]/46,XX,t(3;19)(q1?3;p11)[3]/46,XX[11] |
| 17 | 46,XX,del(12)(p1?2)[18]/46,idem,del(6)(q15q22)[7]/47,idem,del(6)(q15q22), +21[3]/46,XX[7] |
| 18 | 46,XY,del(9)(p13)[12]/46,XY[15] |
| 19 | 46,XY,add(12)(p12)[5]/46,XY[45] |
| 20 | 46,XX,der(1)t(1;12)(q2?4;p13),der(12)t(1;12)(q2?4;p13)t(12;13)(q15∼21; q14∼21),der(13)t(12;13)(q15∼21;q14∼q21)[12]/46,XX[17] |
| 21 | 46,XX,del(12)(p1?),inc[11]/46,idem,dic(9;12)(p?;p?),inc[4]/46,idem, add(21)(q?),inc[3] |
| 22 | 46,XY,t(12;21)(p1?2;q2?2)[4]/47,XY,idem,+21[8]/46,XY[5] |
| 23 | 47,XY,+21[11]/47,idem,del(9)(p21p21)[6]/46,XY[26] |
| 24 | 47,XY,t(2;12)(p21;p13),+10der(21;22)(q10;q10),+der(21;22)(q10;q10)[48]/46, XY[6] |
| 25 | 47,XX,−10,−15,−17,+1-4mar[cp7]/46,XX[13] |
| 26 | 47,XY,del(9)(q21;q31),+21[8]/46,XY[34] |
| 27 | 48,XY,del(5)(q?),+10,−12,−15,+16,+mar1,+mar2[2]/48,idem,−8, +mar3[4]/46, XY[6] |
| 28 | 48,XY,t(12;15)(p10;q10),+21,+21[7]/46,XY[10] |
| 29 | 74-80,inc[10]/46,XX[5] |
| Patient . | . |
|---|---|
| 1,2 | 46,XX |
| 3-7 | 46,XY |
| 8 | 45,X,−X,dup(1)(q21q32∼41),del(12)(p12p13)[13]/46,XX[20] |
| 9 | 45,XY,der(12)t(12;18)(p11∼12;q12),−18[22]/46,XY[3] |
| 10 | 95,XXYY,add(7)(q2?2)×2,del(9)(p21)×2,del(11)(q22)×2,add(12)(p12)×2, +mar[34]/46,XY[75] |
| 11 | 45,XY,t(1;6)(p21;q26∼27),add(2)(p?),−9,add(9)(p2?2),add(11)(q1?4), add(12)(p12)[37] |
| 12 | 45,X,−X,add(21)(p10)[12]/46,XX[1] |
| 13 | 45,X,−X,t(4;12)(q13;p11-p12),?der(21)[18]/46,XX[3] |
| 14 | 46,XX,del(6)(q14∼15q22∼23),del(14)(q21),−15,+?16[24]/46,XX[14] |
| 15 | 46,XX,?7,add(9)(q3?),del(12)(p11),?14,del(17)(q?),add(18)(q?)[6]/46, XX[4] |
| 16 | 46,XX,del(12)(p1?2)[12]/46,XX,t(3;19)(q1?3;p11)[3]/46,XX[11] |
| 17 | 46,XX,del(12)(p1?2)[18]/46,idem,del(6)(q15q22)[7]/47,idem,del(6)(q15q22), +21[3]/46,XX[7] |
| 18 | 46,XY,del(9)(p13)[12]/46,XY[15] |
| 19 | 46,XY,add(12)(p12)[5]/46,XY[45] |
| 20 | 46,XX,der(1)t(1;12)(q2?4;p13),der(12)t(1;12)(q2?4;p13)t(12;13)(q15∼21; q14∼21),der(13)t(12;13)(q15∼21;q14∼q21)[12]/46,XX[17] |
| 21 | 46,XX,del(12)(p1?),inc[11]/46,idem,dic(9;12)(p?;p?),inc[4]/46,idem, add(21)(q?),inc[3] |
| 22 | 46,XY,t(12;21)(p1?2;q2?2)[4]/47,XY,idem,+21[8]/46,XY[5] |
| 23 | 47,XY,+21[11]/47,idem,del(9)(p21p21)[6]/46,XY[26] |
| 24 | 47,XY,t(2;12)(p21;p13),+10der(21;22)(q10;q10),+der(21;22)(q10;q10)[48]/46, XY[6] |
| 25 | 47,XX,−10,−15,−17,+1-4mar[cp7]/46,XX[13] |
| 26 | 47,XY,del(9)(q21;q31),+21[8]/46,XY[34] |
| 27 | 48,XY,del(5)(q?),+10,−12,−15,+16,+mar1,+mar2[2]/48,idem,−8, +mar3[4]/46, XY[6] |
| 28 | 48,XY,t(12;15)(p10;q10),+21,+21[7]/46,XY[10] |
| 29 | 74-80,inc[10]/46,XX[5] |
Of the 52 TEL/AML1-positive samples, 1 could not be used in the MTT assay because of a low percentage of blasts; of the remainder, 49 (96%) were successfully cultured in vitro. Of the 140 TEL/AML1-negative samples, 11 were not tested for in vitro drug sensitivity because of a low number of cells received or a low percentage of blasts in the sample. Of the remainder, only 70% (90 of 129) were successful. Most of the culture failures in the TEL/AML1-negative group resulted because of less than 70% leukemic blast cells in the control wells (n = 31); other causes of failure were insufficient cell survival (n = 4), contamination (n = 1), and technical reasons (n = 3). The failure rate was significantly higher in the TEL/AML1-negative group than in the positive group (Pχ2 < .001). Control cell survival, expressed as the average optical density of the control wells, was higher in the TEL/AML1-positive samples than in the TEL/AML1-negative samples (1.7-fold difference between averages;Pt test < .001). Because it has been described that hyperdiploid c/preB samples have a lower success rate in the MTT assay,38 only those known to be non-hyperdiploid were also compared. The success rate for the TEL/AML1-positive patients remained significantly higher (36 of 37, 97%) than the non-hyperdiploid TEL/AML1-negative patients (53 of 70, 76%;P = .005). The TEL/AML1-negative patients with successful MTT results did not differ significantly in presenting features from those with failed MTT assay results, except for age distribution (median age, 58 and 43 months, respectively; P = .048). The distribution of important clinical and biologic parameters within the TEL/AML1-positive and -negative groups with a successful MTT assay is summarized in Table 2. The event-free survival rate of the total group at the median follow-up time of 38 months was 81.9% ± 3.8%.
Clinical and biologic characteristics of 139 patients with TEL/AML1 gene fusion-negative and -positive B-lineage ALL with in vitro resistance data
| TEL/AML1 . | Negative n (%) . | Positive n (%) . | P . |
|---|---|---|---|
| Sex | |||
| Male | 59 (66) | 28 (57) | .327 |
| Female | 31 (34) | 21 (43) | |
| Median age (months, range) | 58 (3-191) | 57 (25-179) | .783 |
| WBC ×109/L | |||
| <50 | 76 (84) | 40 (82) | .670 |
| >50 | 14 (16) | 9 (18) | |
| Immunophenotype | |||
| c/preB | 85 (94) | 49 (100) | .161* |
| proB | 5 (6) | — | |
| DNA index | |||
| <1.16 | 50 (71) | 35 (97) | .001† |
| 1.16-1.35 | 18 (25) | — | |
| >1.35 | 3 (4) | 1 (3) | |
| Unknown | 19 | 13 |
| TEL/AML1 . | Negative n (%) . | Positive n (%) . | P . |
|---|---|---|---|
| Sex | |||
| Male | 59 (66) | 28 (57) | .327 |
| Female | 31 (34) | 21 (43) | |
| Median age (months, range) | 58 (3-191) | 57 (25-179) | .783 |
| WBC ×109/L | |||
| <50 | 76 (84) | 40 (82) | .670 |
| >50 | 14 (16) | 9 (18) | |
| Immunophenotype | |||
| c/preB | 85 (94) | 49 (100) | .161* |
| proB | 5 (6) | — | |
| DNA index | |||
| <1.16 | 50 (71) | 35 (97) | .001† |
| 1.16-1.35 | 18 (25) | — | |
| >1.35 | 3 (4) | 1 (3) | |
| Unknown | 19 | 13 |
Percentages expressed excluding the missing cases. Pvalues determined by χ2 test and Mann-Whitney U(age).
c/preB versus proB; Fisher exact test.
Non-hyperdiploid (DNA index <1.16 and >1.35) versus hyperdiploid.
For most of the 8 classes of drugs tested, there was a complete overlap of individual LC50 values between TEL/AML1-positive and -negative samples (Table 3). The only drug for which the TEL/AML1-positive group was significantly more sensitive was ASP (median, 5.9-fold; P = .029). The median LC50 values for the glucocorticoids PRED and DEX were, respectively, 4.5- and 1.3-fold lower in the TEL/AML1-positive group; however, this difference was not statistically significant (P = .297 and P= .324, respectively). The TEL/AML1-positive group was slightly but significantly more resistant to VCR (1.5-fold; P= .011) and Ara-C (1.5-fold; P = .014). Other analogues of anthracyclines, epipodophyllotoxins, and thiopurines showed similar results.
Relation between TEL/AML1 and in vitro drug resistance in 139 patients with B-lineage childhood ALL
| Drug . | TEL/AML1-negative median (25th-75th) . | TEL/AML1-positive median (25th-75th) . | P . |
|---|---|---|---|
| Prednisolone | 1.94 (0.30-24.7) | 0.43 (0.06-246) | .297 |
| Dexamethasone | 0.07 (0.03-4.11) | 0.06 (0.01-2.30) | .324 |
| Vincristine | 0.50 (0.16-1.52) | 0.72 (0.57-1.80) | .011 |
| L-Asparaginase | 0.31 (0.01-1.28) | 0.05 (0.01-0.38) | .029 |
| Daunorubicin | 0.09 (0.06-0.23) | 0.10 (0.07-0.20) | .461 |
| 6-Thioguanine | 6.03 (3.23-8.41) | 5.13 (3.45-7.99) | .314 |
| Cytarabine | 0.38 (0.15-1.14) | 0.59 (0.42-1.49) | .014 |
| Etoposide | 1.23 (0.54-2.50) | 1.53 (0.71-2.55) | .413 |
| Ifosfamide | 2.34 (0.83-5.16) | 3.29 (2.16-4.20) | .113 |
| Drug . | TEL/AML1-negative median (25th-75th) . | TEL/AML1-positive median (25th-75th) . | P . |
|---|---|---|---|
| Prednisolone | 1.94 (0.30-24.7) | 0.43 (0.06-246) | .297 |
| Dexamethasone | 0.07 (0.03-4.11) | 0.06 (0.01-2.30) | .324 |
| Vincristine | 0.50 (0.16-1.52) | 0.72 (0.57-1.80) | .011 |
| L-Asparaginase | 0.31 (0.01-1.28) | 0.05 (0.01-0.38) | .029 |
| Daunorubicin | 0.09 (0.06-0.23) | 0.10 (0.07-0.20) | .461 |
| 6-Thioguanine | 6.03 (3.23-8.41) | 5.13 (3.45-7.99) | .314 |
| Cytarabine | 0.38 (0.15-1.14) | 0.59 (0.42-1.49) | .014 |
| Etoposide | 1.23 (0.54-2.50) | 1.53 (0.71-2.55) | .413 |
| Ifosfamide | 2.34 (0.83-5.16) | 3.29 (2.16-4.20) | .113 |
Median LC50 values for TEL/AML1-negative (n = 90) versus TEL/AML1-positive patients (n = 49). All LC50values are expressed as μg/mL, except for L-asparaginase (IU/mL).P values were determined by the Mann-Whitney U test.
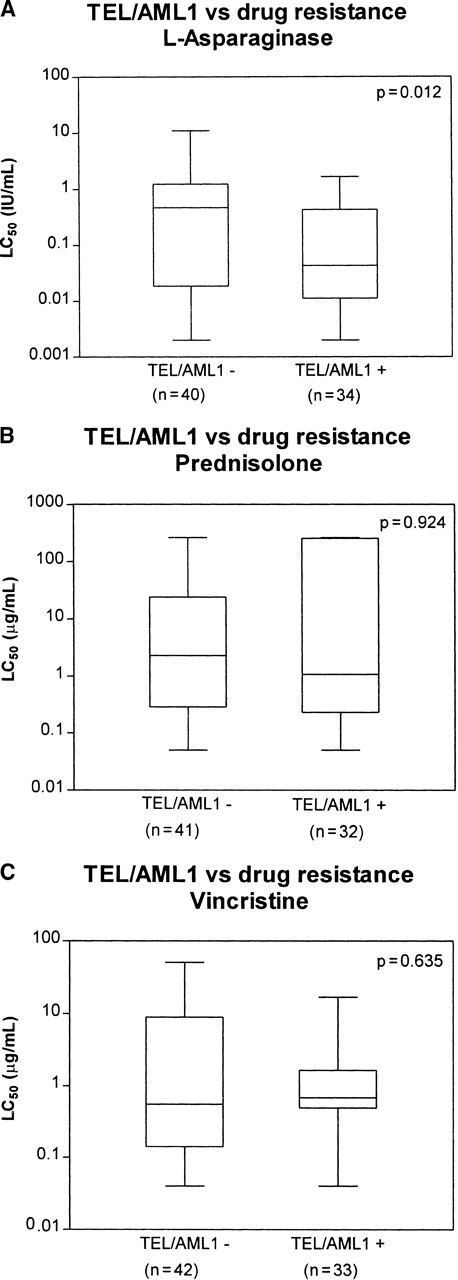
After comparing TEL/AML1-positive and -negative samples within the cohort of patients with CD10+ B-lineage phenotype, age older than 12 months, non-hyperdiploidy, and absence of the Ph1 translocation, the only remaining significant difference between the 2 groups was a relative sensitivity to ASP (10.8-fold; P = .012) in the TEL/AML1-positive patients (see Table 4for a summary of the drugs tested; Figure1). Sensitivity for ASP in the TEL/AML1-positive patients remained evident on removal of all patients older than 10 years from both groups (9.7-fold; P = .036).
Relation between TEL/AML1 and in vitro drug resistance in a cohort of 80 patients with childhood ALL with comparable risk factors
| Drug . | TEL/AML1-negative median (25th-75th) . | TEL/AML1-positive median (25th-75th) . | P . |
|---|---|---|---|
| Prednisolone | 2.30 (0.28-24.3) | 1.08 (0.22->250) | .924 |
| Dexamethasone | 0.08 (0.04->6.0) | 0.06 (0.02->6.0) | .455 |
| Vincristine | 0.55 (0.15-8.92) | 0.69 (0.49-1.65) | .635 |
| L-Asparaginase | 0.47 (0.02-1.25) | 0.04 (0.01-0.42) | .012 |
| Daunorubicin | 0.11 (0.07-0.28) | 0.10 (0.07-0.18) | .383 |
| 6-Thioguanine | 7.00 (5.06-10.05) | 5.80 (3.60-8.40) | .124 |
| Cytarabine | 0.40 (0.16-1.50) | 0.58 (0.34-1.44) | .181 |
| Etoposide | 1.37 (0.68-6.72) | 1.36 (0.68-2.26) | .825 |
| Ifosfamide | 3.89 (2.08-11.36) | 3.47 (1.86-4.39) | .666 |
| Drug . | TEL/AML1-negative median (25th-75th) . | TEL/AML1-positive median (25th-75th) . | P . |
|---|---|---|---|
| Prednisolone | 2.30 (0.28-24.3) | 1.08 (0.22->250) | .924 |
| Dexamethasone | 0.08 (0.04->6.0) | 0.06 (0.02->6.0) | .455 |
| Vincristine | 0.55 (0.15-8.92) | 0.69 (0.49-1.65) | .635 |
| L-Asparaginase | 0.47 (0.02-1.25) | 0.04 (0.01-0.42) | .012 |
| Daunorubicin | 0.11 (0.07-0.28) | 0.10 (0.07-0.18) | .383 |
| 6-Thioguanine | 7.00 (5.06-10.05) | 5.80 (3.60-8.40) | .124 |
| Cytarabine | 0.40 (0.16-1.50) | 0.58 (0.34-1.44) | .181 |
| Etoposide | 1.37 (0.68-6.72) | 1.36 (0.68-2.26) | .825 |
| Ifosfamide | 3.89 (2.08-11.36) | 3.47 (1.86-4.39) | .666 |
Median LC50 values for CD10+ B-lineage phenotype, older than 12 months, non-hyperdiploid (>50), non-Philadelphia translocation TEL/AML1-negative patients (n = 44) versus TEL/AML1-positive patients (n = 36). All LC50values are expressed as μg/mL, except for L-asparaginase (IU/mL).P values were determined by the Mann-Whitney U test.
TEL/AML1 and in vitro drug resistance for ASP, PRED, and VCR.
Relation between TEL/AML1 and in vitro drug resistance for ASP (A), PRED (B), and VCR (C) in childhood c/preB ALL. LC50 values for c/preB, older than 12 months, non-hyperdiploid (> 50 chromosomes), non-Philadelphia translocation TEL/AML1-negative versus TEL/AML1-positive patients. Boxes represent the 25th to the 75th percentiles with the median, shown as a horizontal bar. Whiskers give the minimum and maximum values. LC50 values are expressed as μg/mL; those for ASP are expressed as IU/mL. P values were determined by the Mann–Whitney U test.
TEL/AML1 and in vitro drug resistance for ASP, PRED, and VCR.
Relation between TEL/AML1 and in vitro drug resistance for ASP (A), PRED (B), and VCR (C) in childhood c/preB ALL. LC50 values for c/preB, older than 12 months, non-hyperdiploid (> 50 chromosomes), non-Philadelphia translocation TEL/AML1-negative versus TEL/AML1-positive patients. Boxes represent the 25th to the 75th percentiles with the median, shown as a horizontal bar. Whiskers give the minimum and maximum values. LC50 values are expressed as μg/mL; those for ASP are expressed as IU/mL. P values were determined by the Mann–Whitney U test.
Hyperdiploid patients with childhood ALL also have high in vitro sensitivity to l-asparaginase and other antimetabolites.38 Hence, the TEL/AML1-positive patients (with known DNA index; n = 36) were compared to the hyperdiploid TEL/AML1-negative patients (defined by DNA index 1.16-1.35; n = 18). Hyperdiploid patients were a median 3.5 times more sensitive to ASP (P = .057).
Within the TEL/AML1-positive group, the in vitro resistance profile was compared between the 7 patients with apparently normal karyotype versus the 22 patients with additional karyotypic abnormalities; there were no significant differences for any of the drugs tested.
Discussion
In the present study we have examined the relation between TEL/AML1 gene fusion and in vitro drug resistance. The prevalence of TEL/AML1 in this group of 192 children with B-lineage ALL was 27.1%. Clinical and biologic features of the TEL/AML1-positive patients agree with those of other reports in the literature in that they form a distinct group of exclusively c/preB phenotype, are mostly between 2 and 10 years of age, have non-hyperdiploid ALL, and are Ph1 chromosome negative.
We show that TEL/AML1 gene fusion in untreated childhood c/preB ALL is associated with relative sensitivity to ASP. For all other drugs tested, the TEL/AML1-positive patients did not differ from the TEL/AML1-negative patients except regarding VCR and Ara-C. When the overall group of TEL/AML1-negative patients was considered, including those with other immunophenotypes (proB), those with different ages (younger than 12 months; relatively more patients older than 10 years), and those with Ph1 positivity, the TEL/AML1-positive patients were unexpectedly resistant to VCR and Ara-C. However, the degree of relative resistance was small, with only a 1.5-fold difference in median LC50 for VCR and Ara-C.
In vitro drug resistance profiles have been shown to be related to immunophenotype, age, and hyperdiploidy in childhood ALL.38-41 Hence, to determine the effect of the t(12;21) on in vitro drug resistance, we compared the sensitivity profiles between TEL/AML1-positive and -negative patients in the cohort with CD10+ B-lineage phenotype, older than 12 months, no hyperdiploidy, and absence of high-risk t(9;22). Here again the TEL/AML1-positive patients were more sensitive to ASP, whereas the relative resistance to VCR and Ara-C disappeared. The degree of relative sensitivity to ASP is considerable; the TEL/AML1-positive patients were a median 10.8 times more sensitive. Because patients with hyperdiploid B-lineage childhood ALL have increased sensitivity to ASP,38 when this group was removed from the TEL/AML1-negative population the sensitivity of the TEL/AML1-positive patients to this drug became even more apparent. In this study, the hyperdiploid TEL/AML1-negative patients had even more in vitro sensitivity to ASP than the TEL/AML1-positive (non-hyperdiploid) patients did.
It is surprising that we find such a marked difference in culture success rates between the TEL/AML1-positive (96%) and TEL/AML1-negative (70%) patients. The average success rate of the MTT assay at our laboratory is approximately 76% to 80% for fresh ALL samples taken at diagnosis. Most of the failures in the present study resulted from low leukemic cell survival in the control wells, that is, non-drug–induced “spontaneous” cell death. We have no explanation for this higher proportion of autonomous cell survival in the TEL/AML1-positive patients.
After the first reports on the exceptionally good prognoses of TEL/AML1-positive patients,13,15,16,19,24,25 it has been questioned whether this could be partially dependent on the treatment protocol used. Lanza et al42 of the BFM-based Italian AIEOP study report that 2 of 11 TEL/AML1-positive ALL patients without any apparent poor-risk factors died after relapse (22 and 31 months after diagnosis). Takahashi et al26 report that 5 of 21 (24%) TEL/AML1-positive patients had relapses. Moreover, Harbott et al27 and Seeger et al28 of the BFM study group have found that the prevalence of TEL/AML1 gene fusion in c/preB non-Ph1+ ALL patients at relapse is, unexpectedly, equivalent to that at diagnosis (24%). In contrast to these reports, the incidence of TEL/AML1 in relapsed patients on DFCI protocols has been reported to be very low (1 of 32).29 Hence, the prognostic value of the TEL/AML1 gene fusion may not be as clear-cut as originally proposed.
In the present study, we show that the TEL/AML1-positive patients do not display an overall profile of relative in vitro drug sensitivity. It is interesting to note that the only drug for which TEL/AML1-positive patient samples are relatively sensitive is ASP. This may explain the remarkably good long-term prognosis of TEL/AML1-positive patients at St. Jude Children's Research Hospital and the DFCI,13,15,29 where the treatment protocols include a relatively intensive ASP-regimen, particularly when compared with the BFM-based protocols.43-46 As an example of the latter protocols, the DCSLG ALL-VII study prescribe 8 to 12× 10 000 IU/m2Erwinia ASP intravenously at 3- to 4-day intervals, depending on the risk group.47 In comparison, DFCI protocols 81-01 and 85-01 prescribe 20 to 28×Escherichia coli ASP 25 000 IU/m2 intramuscularly, depending on the risk group.43
It has been shown by 3 independent groups that in vitrol-ASP sensitivity is of prognostic value in patients with childhood ALL.7,9 48 It would be of great interest to study prospectively whether in vitro l-ASP sensitivity is predictive of outcome in TEL/AML1-positive patients in a group undergoing uniform treatment. The treatment received by the patients in the present study differed, and the follow-up was too short to address this or to analyze the event-free survival of the TEL/AML1-positive and the TEL-AML1-negative patients.
In conclusion, TEL/AML1-positive ALL patients are in vitro highly sensitive for ASP only. Our findings suggest that TEL/AML1-positive patients might benefit from treatment schedules with significant use ofl-ASP at doses that secure complete asparagine depletion.29
Acknowledgments
We thank the pediatric oncology centers participating in the DCLSG, the Netherlands, the German COALL study group, and the Rigshospitalet, Denmark. Board members of the DCLSG are H. van den Berg, M. V. A. Bruin, J. P. M. Bökkerink, P. J. van Dijken, K. Hählen, W. A. Kamps, F. A. E. Nabben, A. Postma, J. A. Rammeloo, I. M. Risseeuw-Appel, A. Y. N. Schouten-van Meeteren, G. A. M. de Vaan, E. T. van ‘t Veer-Korthof, A. J. P. Veerman, M. van Weel-Sipman, and R. S. Weening. Board members of the COALL–92/97 study group are U. Göbel, U. Graubner, R. J. Haas, G. E. Janka-Schaub, N. Jorch, H. Jürgens, H. J. Spaar, and K. Winkler. The cosmid probe 50F4 was developed by Dr M. Baens (Human Genome Laboratory, University of Leuven, Belgium), and the cosmid probe 664 was developed by Dr N. Sacchi (Medical School, University of Milan, Italy). Both probes were kindly provided by Prof A. Hagemeijer (European Concerted Action Coordinator, University of Leuven, Belgium). We also thank C. G. Beverstock, H. de France, A. Geurts van Kessel, A. Hagemeijer-Hausman, A. Hamers, B. de Jong, and R. M. Slater of the Netherlands Working Party on Cancer Genetics and Cytogenetics.
Supported by the Dutch Cancer Society grants IKA 89-06 and VU95-021; the Danish Cancer League and the Danish Cancer Society grant 96 144 10 9132.
Reprints:N. L. Ramakers-van Woerden, Department of Pediatric Hematology/Oncology, University Hospital Vrije Universiteit, PO Box 7057, 1007 MB Amsterdam, The Netherlands; e-mail:ramakersvanwoerden@azvu.nl.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal