Abstract
This study involved 12 patients with multiple myeloma (MM), in whom malignant plasma cells did not contain immunoglobulin heavy chain (IgH) protein chains. Southern blot analysis revealed monoallelic Jh gene rearrangements in 10 patients, biallelic rearrangement in 1 patient, and biallelic deletion of the Jh and Cμ regions in 1 patient. Heteroduplex polymerase chain reaction analysis enabled the identification and sequencing of 9 clonal Jhgene rearrangements. Only 4 of the joinings were complete Vh-(D)-Jhrearrangements, including 3 in-frame rearrangements with evidence of somatic hypermutation. Five rearrangements concerned incomplete Dh-Jh joinings, mainly associated with deletion of the other allele. Curiously, in at least 1 of these 5 cases the second allele seemed to be in germline configuration, whereas the in-frame Vκ-Jκgene rearrangements contained somatic mutations. The configuration of the IGH genes was further investigated by use of Ch probes. In 5 patients the rearrangements in the Jh and Ch regions were not concordant, probably caused by illegitimate IGH class switch recombination (chromosomal translocations to 14q32.3). These data indicate that in many IgH− MM patients illegitimateIGH class switch rearrangement or illegitimate deletion of the functional Vh-(Dh)-Jhallele are responsible for IgH negativity. For example, the exclusive presence ofDh-Jhrearrangements in combination with mutated IGK genes can only be explained in terms of normal B-cell development, if the second (functional) IGH allele is deleted, which was probably the case in most patients. Therefore, defects at the DNA level are responsible for the lack of IgH protein production in most IgH− MM patients.
Multiple myeloma (MM) is a clonal B-lineage malignancy affecting terminally differentiated bone marrow (BM) plasma cells bearing functional Vh-(Dh)-Jhgene rearrangements with somatic hypermutations.1,2 The lack of intraclonal diversity in the hypermutation pattern indicates that malignant transformation occurred after positive selection in germinal centers.1,2 It has been suggested that identical clonotypic cells but having the morphology and immunophenotype of mature B cells, can be detected in peripheral blood (PB) of patients with MM.3,4 Based on extensive studies, the pathogenesis of MM is believed to be a multistep transformation process (reviewed in Hallek et al5). One of the presumably earliest oncogenic events is a translocation involving the immunoglobulin heavy chain (IGH) gene locus (chromosome 14q32.3), which is the result of illegitimate IGH class switch processes.5,6Chromosome translocations involving IGH genes occur in most MM and several recurrent partner loci have been identified.6-15
Classical MM is characterized by the presence of osteolytic bone lesions and overproduction of structurally homogenous immunoglobulins (Ig), which can be detected as a monoclonal peak (M-protein) on serum or urine electrophoresis. Depending on the tumor mass in the BM, additional characteristic clinical features can be observed including anemia, hypercalcemia, and renal insufficiency.16 Besides the classical presentation of MM, several forms with atypical or absent M-protein are distinguishable. In up to 20% of MM cases only Ig light chain protein is detected in serum or urine without detectable Ig heavy chains (IgH): the so-called light chain (Bence-Jones) disease. In contrast, heavy chain disease is characterized by abnormally short monoclonal IgH proteins in serum without associated light chains. Nonsecretory MM without detectable M-protein in serum occurs in approximately 1% to 5% of MM patients.17-21 In about 85% of these cases intracellular Ig molecules are clearly detectable, suggesting an underlying defect in Ig excretion (nonexcretory MM), whereas the remaining 15% of cases have no detectable intracellular IgH and Ig light chains (true nonproducer MM), implying that the latter cases represent a rare subgroup (< 1% of all MM cases).18 21
The molecular background of the complete absence of IgH protein production in light chain disease and in true nonproducer MM (grouped together as IgH− MM) is not fully clarified. We aimed therefore at identifying the molecular features responsible for the absence of IgH protein synthesis in IgH− MM.
Patients, materials, and methods
Patients
Bone marrow (n = 9), PB (n = 1), or tissue biopsies (n = 2; 1 skin biopsy and 1 tonsil) were obtained from 12 MM patients, aged 35 to 78 years, without production of IgH protein chains at initial diagnosis or during the course of their disease. Morphologic examination of the cell samples revealed that the percentages of plasma cells ranged from 20% to 90%. In 9 patients the plasma cells produced Ig light chain proteins, whereas in the other 3 patients no evidence for Ig light chain production was found. The latter 3 patients (MM-10, MM-11, and MM-12) were diagnosed as true nonproducer MM. Immunoelectrophoresis demonstrated that in all 9 patients with monoclonal CyIgκ+ or CyIgλ+ plasma cells monoclonal Ig light chains were present in serum or urine or both at the time of investigation. Two patients (MM-4 and MM-8) showed high frequencies of plasma cells in their PB. In the other 10 patients no plasma cells were found in PB on cytomorphologic examination.
Immunophenotyping of mononuclear cells (MNC) and plasma cells in tissue biopsies
Mononuclear cells were isolated from BM or PB samples by Ficoll (density 1.077 g/mL; Pharmacia, Uppsala, Sweden) density centrifugation. In patients MM-10 and MM-11, snap frozen skin and tonsil biopsies, respectively, were used for immunohistology and molecular studies.
The MNC of the 10 BM or PB samples as well as the tissue biopsies from patients MM-10 and MM-11 were analyzed for surface expression of CD10 (VIL-A1), CD19 (Leu-12), CD38 (Leu-17) antigen, and HLA-DR (L243) as well as for cytoplasmic expression of IgM, IgD, IgG, IgA, IgE, Igκ, and Igλ. The Leu monoclonal antibodies and anti-HLA-DR antibody were obtained from Becton Dickinson (San Jose, CA) and VIL-A1 was a gift from Dr W. Knapp (Vienna, Austria). The antihuman Ig antibodies were polyspecific and obtained from Nordic Immunological Laboratories (Tilburg, The Netherlands). The monoclonal antibodies were used in indirect immunofluorescence assays with a fluorescein isothiocyanate (FITC)-conjugated goat antimouse Ig serum (Central Laboratory Blood Transfusion Service, Amsterdam, The Netherlands) as second-step reagent. The antihuman Ig antibodies were directly conjugated with either FITC or tetrahodamine isothiocyanate (TRITC) and used for IgH/Igκ or IgH/Igλ double stainings to confirm the absence of cytoplasmic IgM, IgD, IgG, IgA, and IgE protein chains in the Ig light chain-positive MM cells. Fluorescence stainings were evaluated using fluorescence microscopes (Zeiss, Oberkochen, Germany).22
Southern blot analysis
DNA was isolated from frozen MNC (n = 10) or from tissue samples (n = 2), digested, and blotted to nylon membranes as described previously.23
IGH gene rearrangements were studied by use of 32P labeled IGHJ6, Cμ, Cγ, Cα, and Cε probes.24-28 The IGHJ6 probe (DAKO Corporation, Carpinteria, CA) was used inBglII and BamHI/HindIII digests and inEcoRI, HindIII, or BamHI digests for confirmation. The Cμ probe was used in BamHI digests, whereas the other CH probes were used in EcoRI,HindIII, or BamHI digests.23
The IGK gene rearrangements were studied with the32P-labeled IGKJ5, IGKC, and IGKDE probes (DAKO).29 The IGKJ5 probe was used in EcoRI,HindIII, BglII, and BamHI digests, whereas the IGKC and IGKDE probes were used in BglII and BamHI digests.29
The IGL gene rearrangements were studied with the32P-labeled IGLC3 probe (DAKO), which detects 95% of all Jλ-Cλ gene rearrangements inEcoRI/HindIII digests.30
Polymerase chain reaction (PCR) amplification and heteroduplex analysis of PCR products
The PCR analysis was essentially performed as described previously.31,32 In each 50 μL PCR reaction 50 ng DNA sample, 6.3 pmol of the 5′ and 3′ oligonucleotide primers, and 0.5 U AmpliTaq Gold polymerase (PE Biosystems, Foster City, CA) were used. The sequences of the oligonucleotides used for amplification of complete Vh-Jh and incomplete Dh-Jh gene rearrangements as well as for Vκ-Jκrearrangements were published previously.33-36 PCR conditions were: preactivation of the enzyme for 10 minutes at 94°C, followed by 35 cycles of 45 seconds at 92°C, 90 seconds at 60°C, and 2 minutes at 72°C using a Perkin-Elmer 480 thermal cycler (PE Biosystems). After the last cycle an additional extension step of 10 minutes at 72°C was performed. Appropriate positive and negative controls were included in all experiments.32
To distinguish between polyclonal and monoclonal rearrangements we performed heteroduplex analysis of the obtained PCR products. In short, the PCR products were denatured at 94°C for 5 minutes to obtain single-stranded PCR products. Subsequently the single-stranded products were cooled to 4°C for 60 minutes to induce random renaturation (duplex formation).37 In case of monoclonal gene rearrangements homoduplexes are formed (identical junctional regions), whereas heteroduplexes are found in case of polyclonal gene rearrangements (heterogeneous junctional regions). The obtained duplexes were immediately loaded on 6% nondenaturing polyacrylamide gels in 0.5 × Tris-borate-EDTA (TBE) buffer, run at room temperature, and visualized by ethidium bromide staining to discriminate between the presence of rapidly migrating homoduplex bands or slowly migrating heteroduplexes smears.37 A 100-bp DNA ladder (Promega Corporation, Madison, WI) was used as size marker.
Sequence analysis of IGH and IGK gene rearrangements
Clonal PCR products as found by heteroduplex analysis were directly sequenced. Sequencing was performed using the dye-terminator cycle sequencing kit with AmpliTaq DNA polymerase FS on an ABI 377 sequencer (PE Biosystems) as described before.34Vh, Vκ, Dh, Jh and Jκ segments were identified using DNAPLOT software (W. Müller, H-H. Althaus, University of Cologne, Germany) by searching for homology with all known human germline Vh, Vκ, Dh,Jh, andJκsequences obtained from the VBASE directory of human Ig genes (http://www.mrc-cpe.cam.ac.uk/imt-doc/).38
Northern blot analysis
Total RNA was isolated with the LiCl/urea method39 from frozen MNC of BM samples from 6 patients with light chain MM of whom sufficient cells were available. Fifteen micrograms of total RNA was size-fractionated in 1.0% agarose gel containing formaldehyde and transferred to a Biodyne nylon membrane (Pall Ultrafine Filtration Corp, Glen Cove, NY). The above-mentioned Cκ, Cμ, Cγ, Cα, and Cε DNA probes were used for detection ofIGK and IGH transcripts. Total RNA from the human B-cell lines ROS-17, EB4B, ROS-15, and U266 were used as positive controls for the detection of IgM, IgG, IgA, and IgE transcripts, respectively.
Results
Immunophenotyping
Immunophenotyping of the MNC from the 9 light chain MM patients and the nonproducer patient MM-12 as well as immunohistology of the skin and tonsil biopsies from the other 2 nonproducer patients (MM-10 and MM-11) demonstrated that the malignant plasma cells in all 12 patients did not contain IgH chains, whereas Igκ chain and Igλ chains were found in the plasma cells from 8 patients (MM-1 to MM-8) and 1 patient (MM-9), respectively (Table 1). In 3 patients (MM-10, MM-11, and MM-12) neither Igκ nor Igλ chains were detected (Table 1). The plasma cells in all 12 patients were negative in CD10, CD19, and HLA-DR staining, but positive in CD38 staining.
IGH gene configuration in MM patients based on Southern blotting, heteroduplex PCR analysis, and sequencing
| Patients . | MM-1 . | MM-2 . | MM-3 . | MM-4 . | MM-5 . | MM-6 . | MM-7 . | MM-8 . | MM-9 . | MM-10 . | MM-11 . | MM-12 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MM phenotype | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igλ+ | True nonproducer | True nonproducer | True nonproducer |
| Tumor load (cell sample) | 35% (BM) | 25% (BM) | 20% (BM) | 90% (BM) | 80% (BM) | 80% (BM) | 30% (BM) | 80% (PB) | 60% (BM) | 25% (skin) | 70% (tonsil) | 35% (BM) |
| Southern blotting* | ||||||||||||
| Jh region | R/G | R/G | R/D | R/D | R/D | R/G | R/G or D | D/D | R/G | R/G or D | R/R | R/D |
| Cμ | RNC/G | D/G | RC/G | RC/D | D/G | RC/G | RC/G | D/G | RNC/G | RC/G | D/D | D/G |
| Cㆠ| G | D/G | G | G | D/G | G | G | G | G | G | G | G |
| Cα | G | RC/G | G | G | RNC/G | G | G | R/G | G | G | RNC/G | RC/G |
| Cε | G | G | RNC/G | G | D/G | G | G | G | G | G | D/G | G |
| Heteroduplex PCR and sequencing | ||||||||||||
| Gene segments | VH1-24/JH3 | negative | ND | DH3-16/JH4 | DH2-2/JH4 | DH2-2/JH5 | DH4-23/JH6 | NA | VH3-11/JH5 | DH1-26/JH5 | VH2-5/JH4 | VH1-18/JH4 |
| Reading frame | + | NA | ND | NA | NA | NA | NA | NA | + | NA | − | + |
| Somatic hypermutation | + | NA | ND | − | − | − | − | NA | + | − | − | + |
| Patients . | MM-1 . | MM-2 . | MM-3 . | MM-4 . | MM-5 . | MM-6 . | MM-7 . | MM-8 . | MM-9 . | MM-10 . | MM-11 . | MM-12 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MM phenotype | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igλ+ | True nonproducer | True nonproducer | True nonproducer |
| Tumor load (cell sample) | 35% (BM) | 25% (BM) | 20% (BM) | 90% (BM) | 80% (BM) | 80% (BM) | 30% (BM) | 80% (PB) | 60% (BM) | 25% (skin) | 70% (tonsil) | 35% (BM) |
| Southern blotting* | ||||||||||||
| Jh region | R/G | R/G | R/D | R/D | R/D | R/G | R/G or D | D/D | R/G | R/G or D | R/R | R/D |
| Cμ | RNC/G | D/G | RC/G | RC/D | D/G | RC/G | RC/G | D/G | RNC/G | RC/G | D/D | D/G |
| Cㆠ| G | D/G | G | G | D/G | G | G | G | G | G | G | G |
| Cα | G | RC/G | G | G | RNC/G | G | G | R/G | G | G | RNC/G | RC/G |
| Cε | G | G | RNC/G | G | D/G | G | G | G | G | G | D/G | G |
| Heteroduplex PCR and sequencing | ||||||||||||
| Gene segments | VH1-24/JH3 | negative | ND | DH3-16/JH4 | DH2-2/JH4 | DH2-2/JH5 | DH4-23/JH6 | NA | VH3-11/JH5 | DH1-26/JH5 | VH2-5/JH4 | VH1-18/JH4 |
| Reading frame | + | NA | ND | NA | NA | NA | NA | NA | + | NA | − | + |
| Somatic hypermutation | + | NA | ND | − | − | − | − | NA | + | − | − | + |
Gene configuration: G indicates germline allele or no rearrangements detectable; R, rearranged allele, RC, rearranged CH band, which comigrates with the rearranged JH band; RNC, rearranged CH band, which does not comigrate with the rearranged JH band; D, deleted allele.
It was not possible to determine the configuration of the Cγ gene regions, due to the complexity of the banding patterns (see text).
ND indicates not determined; NA, not applicable.
Configuration of IGH genes
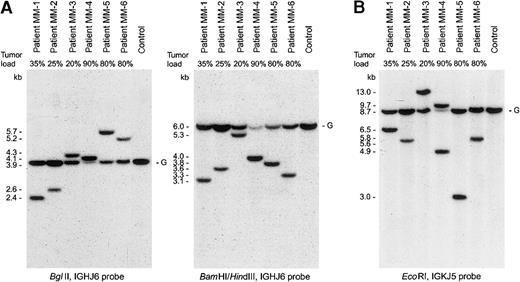
In 10 patients clonal rearrangements of JH gene segments were found on only 1 allele (MM-1 to MM-7, MM-9, MM-10, and MM-12), in 1 patient on both alleles (patient MM-11), and in 1 case (patient MM-8) the JH and Cμ regions were deleted on both alleles (Table 1, Figure 1). In some patients the presence of normal non-B cells with germline IGHgenes might hamper the detection of a monoallelic IGH gene deletion. However, based on the percentages of malignant plasma cells and the relative density of rearranged and germline bands, the absence or presence of a deleted second allele could be estimated in most cases. In fact, we concluded that in at least 4 of the 10 patients with monoallelic JH rearrangements the second allele was deleted, that in 2 additional patients the second allele might be deleted, whereas in the other 4 patients the second IGH allele seemed to be in germline configuration (Figure 1).
IGH and IGK gene configuration of 6 IgH−/Igκ+ MM patients.
(A) IGHJ6 probe hybridization to BglII digests orBamHI/HindIII digests. (B) IGKJ5 probe hybridization toEcoRI digests.
IGH and IGK gene configuration of 6 IgH−/Igκ+ MM patients.
(A) IGHJ6 probe hybridization to BglII digests orBamHI/HindIII digests. (B) IGKJ5 probe hybridization toEcoRI digests.
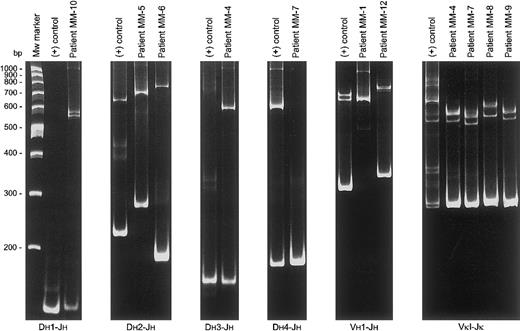
Detailed heteroduplex PCR analysis of the IGH locus was performed in 11 patients using 19 primer combinations (6 IGHframework-1 Vh-family specific primers, 6 Vh-leader primers, and 7 family-specific Dh primers in combination with 1 Jh consensus primer). In a total of 9 patients 9 monoclonal homoduplexes were found out of the total 12 JHgene rearrangements as identified by Southern blotting; in 1 patient (MM-3) insufficient DNA was available for detailed PCR studies, and PCR analyses in 2 other patients (MM-2 and MM-11) failed to detect a JH rearrangement (Table 1; Figure2). Only 4 of the detected JHrearrangements were complete Vh-(Dh)-Jhrearrangements and sequence analysis revealed 4 different functional Vh gene segments (Vh1-18, Vh1-24, Vh2-5, and Vh3-11). Three of these rearrangements were in a proper reading frame with evidence of somatic hypermutation (patients MM-1, MM-9, and MM-12), whereas the fourth rearrangement was out-of-frame and unmutated (patient MM-11). Five rearrangements concerned incomplete Dh-Jhjoinings, using 4 different Dhgene segments (Dh1-26, Dh2-2, Dh3-16, and Dh4-23). In 2 of these 5 cases the second IGH allele was deleted, in 2 other cases the second allele might be deleted, whereas in 1 case the second allele seemed to be germline. Based on the knowledge of the gene segments identified in the clonal PCR products together with the complete sequence of the human IGH locus,40 41 the theoretical sizes of restriction fragments containing the respective rearrangements were calculated and indeed found to be concordant with the sizes of the respective rearranged bands in Southern blot analysis.
Heteroduplex PCR analysis in several IgH−MM patients to distinguish between polyclonal and monoclonalIGH and IGK gene rearrangements.
Clonal homoduplexes found with particular primer combinations are illustrated as compared to positive controls. In patient MM-1, the size of the homoduplex found with the Vh1-Jh PCR was essentially larger than predicted. Sequence analysis showed a PCR product with a Vh1-24/Jh3 rearrangement extended to the Jh5 segment with extensive somatic hypermutation of the Jh3 gene (resulting in loss of the primer annealing site) and deletion of the Jh4 segment most probably owing to an abnormal somatic mutation process.48
Heteroduplex PCR analysis in several IgH−MM patients to distinguish between polyclonal and monoclonalIGH and IGK gene rearrangements.
Clonal homoduplexes found with particular primer combinations are illustrated as compared to positive controls. In patient MM-1, the size of the homoduplex found with the Vh1-Jh PCR was essentially larger than predicted. Sequence analysis showed a PCR product with a Vh1-24/Jh3 rearrangement extended to the Jh5 segment with extensive somatic hypermutation of the Jh3 gene (resulting in loss of the primer annealing site) and deletion of the Jh4 segment most probably owing to an abnormal somatic mutation process.48
The configuration of the IGH genes was further investigated by use of Cμ, Cγ, Cα, and Cε probes (Table 1). In 7 patients a rearranged Cμ gene band was found, which comigrated with the JH band in 5 of them (patients MM-3, MM-4, MM-6, MM-7, and MM-10), indicating that no IGH class switch had occurred; at least 4 of these 5 rearrangements concerned a Dh-Jhjoining (patient MM-3 was not studied by PCR), which explains the absence of IGH class switch. In the other 2 cases (patients MM-1 and MM-9) the rearranged JH and Cμregions were not present on the same restriction fragments, suggesting that illegitimate recombination with a breakpoint in the JH-Cμ area had occurred thereby making the in-frame Vh-Jh joining in these 2 patients nonfunctional. The Southern blot analyses with the Cγ probe did not provide reliable information concerning rearrangements and IGH class switch, due to the complex banding pattern, which is caused by the 5 Cγ gene segments and their genetic polymorphisms.23 Nevertheless, in patient MM-2 the Cγ probe allowed the detection of Cγ gene deletions, which could be explained by a presumably normal IGHclass switch to Cα, whereas in patient MM-5 extensive illegitimate Cγ gene deletions were found on 1 allele. Analyses with the Cα probe revealed a monoallelic rearrangement in 5 of the 12 patients. In patients MM-2 and MM-12, the rearranged bands comigrated with a rearranged JH gene band, suggesting that an IgA class switch had occurred. In the other 3 patients (patients MM-5, MM-8, and MM-11) no proof for close linkage between the JH region and the rearranged Cα region was found, which might be due to illegitimate IGH class switches. Finally, in 1 case (patient MM-3) a rearranged band was found with the Cε probe. This patient contained a JH-Cμrearrangement on 1 allele with deletion of the JH region on the other allele. The latter suggests that the detected Cε rearrangement might be caused by deletion of a large part of the IGH locus.
Configuration of IGK and IGL genes
In 6 of the 8 Igκ+ MM patients a monoallelic rearrangement in the Jκ-Cκ area was found (Figure 1), while the Jκ-Cκ region on the second allele was in germline configuration in 5 patients and deleted in the sixth patient (MM-2), owing to a Vκ to kappa deleting element (Kde) rearrangement (Table 2). In the remaining 2 Igκ+ patients (MM-3 and MM-4), Vκ-Jκ rearrangements were found on 1 allele; the other allele contained a Vκ-Jκrearrangement in combination with a Cκ deletion. BothIGL alleles were in germline configuration in all 8 Igκ+ patients. The patient with Igλ light chain MM had biallelic IGL gene rearrangements in combination with biallelic nonfunctional IGK gene rearrangements owing to intron RSS-Kde and Vκ-Kde recombinations.
Ig light chain gene configuration in MM patients based on Southern blotting, heteroduplex PCR analysis, and sequencing
| Patients . | MM-1 . | MM-2 . | MM-3 . | MM-4 . | MM-5 . | MM-6 . | MM-7 . | MM-8 . | MM-9 . | MM-10 . | MM-11 . | MM-12 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MM phenotype | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igλ+ | True nonproducer | True nonproducer | True nonproducer |
| Tumor load | 35% | 25% | 20% | 90% | 80% | 80% | 30% | 80% | 60% | 25% | 70% | 35% |
| IGK: Southern blot analysis* | ||||||||||||
| First allele | Vκ-Jκ | Vκ-Jκ | Vκ-Jκ | Vκ-Jκ | Vκ-Jκ | Vκ-Jκ | Vκ-Jκ | Vκ-Jκ | Vκ-Kde | Vκ-Jκ | Vκ-Jκ | Vκ-Jκ + intron-Kde |
| Second allele | G | Vκ-Kde | Vκ-Jκ + intron-Kde | Vκ-Jκ + intron-Kde | G | G | G | G | Vκ-Jκ + intron-Kde | G | Vκ-Kde | Vκ-Jκ + intron-Kde |
| IGK:PCR | ||||||||||||
| Gene segments | Vκ4-1/ Jκ2 | Vκ3-20/ Jκ4 | ND | Vκ1-12/Jκ4 Vκ2D-29/Jκ4 | Vκ1-5/ Jκ2 | Vκ3-20/ Jκ4 | Vκ1-33/ Jκ2 | Vκ1-5/ Jκ1 | Vκ1D-12/ Jκ4 | Negative | Vκ1-39/Jκ2 | Vκ1D-43/Jκ5 |
| Reading frame | + | + | ND | +/− | + | + | + | + | − | NA | + | − |
| Somatic hypermutation | + | + | ND | +/− | + | + | + | + | − | NA | + | − |
| IGL: Southern blot analysis | ||||||||||||
| Jλ-Cλ regions | G | G | G | G | G | G | G | G | Vλ-Jλ1/Vλ-Jλ2 | G | G | Vλ-Jλ1/Vλ-Jλ2 |
| Patients . | MM-1 . | MM-2 . | MM-3 . | MM-4 . | MM-5 . | MM-6 . | MM-7 . | MM-8 . | MM-9 . | MM-10 . | MM-11 . | MM-12 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MM phenotype | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igκ+ | Igλ+ | True nonproducer | True nonproducer | True nonproducer |
| Tumor load | 35% | 25% | 20% | 90% | 80% | 80% | 30% | 80% | 60% | 25% | 70% | 35% |
| IGK: Southern blot analysis* | ||||||||||||
| First allele | Vκ-Jκ | Vκ-Jκ | Vκ-Jκ | Vκ-Jκ | Vκ-Jκ | Vκ-Jκ | Vκ-Jκ | Vκ-Jκ | Vκ-Kde | Vκ-Jκ | Vκ-Jκ | Vκ-Jκ + intron-Kde |
| Second allele | G | Vκ-Kde | Vκ-Jκ + intron-Kde | Vκ-Jκ + intron-Kde | G | G | G | G | Vκ-Jκ + intron-Kde | G | Vκ-Kde | Vκ-Jκ + intron-Kde |
| IGK:PCR | ||||||||||||
| Gene segments | Vκ4-1/ Jκ2 | Vκ3-20/ Jκ4 | ND | Vκ1-12/Jκ4 Vκ2D-29/Jκ4 | Vκ1-5/ Jκ2 | Vκ3-20/ Jκ4 | Vκ1-33/ Jκ2 | Vκ1-5/ Jκ1 | Vκ1D-12/ Jκ4 | Negative | Vκ1-39/Jκ2 | Vκ1D-43/Jκ5 |
| Reading frame | + | + | ND | +/− | + | + | + | + | − | NA | + | − |
| Somatic hypermutation | + | + | ND | +/− | + | + | + | + | − | NA | + | − |
| IGL: Southern blot analysis | ||||||||||||
| Jλ-Cλ regions | G | G | G | G | G | G | G | G | Vλ-Jλ1/Vλ-Jλ2 | G | G | Vλ-Jλ1/Vλ-Jλ2 |
The presence of a Vκ-Jκ rearrangement was assumed from the finding of a rearranged band with the IGKJ5 probe. Rearranged bands found with the IGKDE probe were interpreted as Vκ-Kde joining, if accompanied by deletion of Jκ and Cκ segments, and as intron-Kde rearrangements, if they were found on the same restriction fragment as Vκ-Jκ rearrangements (IGKJ5 probe) together with a Cκ gene deletion (IGKC probe).
ND indicates not determined; NA, not applicable.
Two nonproducer MM patients (MM-10 and MM-11) had a monoallelic Vκ-Jκ gene rearrangement with theIGL genes in germline configuration. Interestingly, the third nonproducer MM patient (MM-12) had biallelic IGK rearrangements with deleted Cκ segments on both alleles (intron RSS-Kde recombinations) and biallelic IGL gene rearrangements.
Heteroduplex PCR analysis of Vκ-Jκ gene rearrangements was performed in 11 patients with 4 Vκ family-specific primers and 2 Jκreverse primers. Eleven monoclonal homoduplexes were found in 10 patients (Table 2 and Figure 2), including 8 in-frame rearrangements with moderate amounts of somatic mutations and 3 out-of-frame unmutated joinings. In the true nonproducer MM-10 no clonal Vκ-Jκ PCR products could be identified, whereas in nonproducer MM-11 the identified Vκ-Jκ gene rearrangement was in-frame.
Transcription of Ig genes
Six patients with Igκ+ light chain MM (patients MM-1, MM-2, MM-3, MM-4, MM-5, and MM-6) could be studied for the occurrence of Ig messenger RNA (mRNA) by use of Northern blot analysis. As expected, in all 6 patients high levels of IGK mRNA were present. In patient MM-1 a trace IGM mRNA, high levels ofIGG mRNA and low levels of IGA mRNA were found. In patient MM-2 no IGH transcripts were detected. In patient MM-3 we found trace levels of IGM and IGGtranscripts, which were essentially lower than the IGK mRNA transcription levels, thereby suggesting that these low IGHtranscript levels were probably derived from the background of normal B cells. In patients MM-4 and MM-6 low or moderate levels ofIGM transcripts were detected. Finally, in patient MM-5 a trace of IGG mRNA and moderate levels of truncatedIGA mRNA were found. IGE mRNA could not be detected in all 6 tested patients.
Discussion
Lack of IgH protein synthesis may be caused by abnormalities at several levels, for example, abnormalities at the DNA level (a defective gene with true nonsynthetic capability), aberrant transcription processes, aberrant translation processes, or rapid degradation of newly synthesized IgH protein.21 We mainly focused on detailed molecular analysis of the IGH genes. Although seemingly normal Jh gene rearrangements were found in all but 1 patient by Southern blotting, the further PCR-based identification of these rearrangements as well as Southern blot analysis of the downstream part of the IGH locus with probes for the various constant gene segments revealed distinct molecular abnormalities explaining the absence of IgH proteins (summarized in Table 3).
Molecular abnormalities responsible for IgH negativity in the 12 MM patients
| Abnormality . | MM-1 . | MM-2 . | MM-3 . | MM-4 . | MM-5 . | MM-6 . | MM-7 . | MM-8 . | MM-9 . | MM-10 . | MM-11 . | MM-12 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biallelic JH deletion | − | − | − | − | − | − | − | + | − | − | − | − |
| IllegitimateIGH class switch | + | − | (+) | − | + | − | − | − | + | − | + | − |
| No IGH mRNA | − | + | + | − | − | − | ND | ND | ND | ND | ND | ND |
| Missing (VH)-DH-JH PCR product3-150 | − | +3-150 | ND | − | − | − | − | − | − | − | +3-150 | − |
| Only DH-JH with second allele deleted (or germline?) | − | − | ND | + | + | + | + | − | − | + | − | − |
| Nonfunctional VH-DH-JH | − | − | ND | − | − | − | − | − | − | − | + | − |
| Unexplained | − | − | − | − | − | − | − | − | − | − | − | + |
| Abnormality . | MM-1 . | MM-2 . | MM-3 . | MM-4 . | MM-5 . | MM-6 . | MM-7 . | MM-8 . | MM-9 . | MM-10 . | MM-11 . | MM-12 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biallelic JH deletion | − | − | − | − | − | − | − | + | − | − | − | − |
| IllegitimateIGH class switch | + | − | (+) | − | + | − | − | − | + | − | + | − |
| No IGH mRNA | − | + | + | − | − | − | ND | ND | ND | ND | ND | ND |
| Missing (VH)-DH-JH PCR product3-150 | − | +3-150 | ND | − | − | − | − | − | − | − | +3-150 | − |
| Only DH-JH with second allele deleted (or germline?) | − | − | ND | + | + | + | + | − | − | + | − | − |
| Nonfunctional VH-DH-JH | − | − | ND | − | − | − | − | − | − | − | + | − |
| Unexplained | − | − | − | − | − | − | − | − | − | − | − | + |
Abbreviations: +, presence of abnormality; −, absence of abnormality; ND, not determined.
The single JH rearrangement in patient MM-2 and 1 of the 2 JH rearrangements in patient MM-11 could not be detected or identified by VH-JH or DH-JH PCR analysis, suggesting the presence of an aberrant JH rearrangement.
Southern blot analysis in 10 MM patients showed clonal JHrearrangements on only 1 allele. This is in striking contrast to normal B-cell development and most B-lineage malignancies, where IGHgene rearrangements are generally found on both alleles.42-44 Therefore, one would have expected biallelicIGH gene rearrangements in most MM cases. In patient MM-8 both Jhalleles were deleted, which is an obvious reason for IgH negativity in this case. In 5 patients (MM-1, MM-3, MM-5, MM-9, and MM-11) the rearranged Jhgene and Ch gene segments were not linked, suggesting that illegitimate class switch recombination had separated the rearranged (Vh)-Dh-Jhcomplex from a Ch gene segment. This phenomenon can explain the IgH negativity in patients MM-1 and MM-9 with functional (in-frame) somatically mutated Vh-(Dh)-Jhrearrangements. In patient MM-3 the illegitimate switch recombination resulted in the deletion of 1 Jhallele, whereas the second IGH allele, probably nonfunctional, did not undergo class switch. Unfortunately, we could not perform PCR/sequencing analysis due to insufficient DNA, but Northern blotting showed absence of the expected IGH mRNA levels in patient MM-3. In patients MM-5 and MM-11 additional reasons were found for IgH protein negativity, that is, an incomplete Dh-Jh rearrangement and an out-of-frame Vh-(Dh)-Jhrearrangement, respectively (Table 3). The causative mechanism underlying the aberrant Chrearrangements in the above 5 patients is most probably a translocation involving the IGH gene on chromosome 14q32.3, known to occur frequently in MM. Unfortunately, no cytogenetic data are available in our group of patients. Although chromosome aberrations with breakpoints in the Ch region of theIGH locus occur in the majority of MM patients, they seem to affect IgH protein production only in rare cases.6,8 In IgH+ MM cases such translocations presumably occur on the nonproductive allele or involve switch regions downstream of a functional Vh-(Dh)-Jh-Ccomplex.5 Even biallelic translocations involvingIGH genes may be accompanied by a normal IgH+phenotype.8 45 Nevertheless, our data clearly show that in at least 2 (but probably 3) of our patients an illegitimate class switch rearrangement seems to be the sole cause of the absence of IgH proteins, whereas in additional 2 cases, it appeared to be one of the causes for IgH negativity (Table 3).
In patient MM-2 extensive heteroduplex PCR analysis did not reveal any clonal IGH gene rearrangement and also Northern blotting showed no IGH gene transcription. This indicates that the Jh gene rearrangement found by Southern blotting in this patient might also reflect an illegitimate recombination. In fact, translocations to Jhgene segments (rather than to the switch regions) owing to abnormal V(D)J recombinase activity occur frequently in some subsets of non-Hodgkin lymphomas, but were previously also suggested to occur in MM and found in the MM-derived cell line FLAM-76.6,11 46
Remarkably, in 5 patients heteroduplex PCR analysis showed that the single IGH-Jh rearrangement detected by Southern blotting concerned an incomplete Dh-Jhjoining. In 2 of these patients (patients MM-4 and MM-5) the second allele (most probably having contained a functional Vh-(Dh)-Jhgene rearrangement) was deleted. However, in the remaining 3 patients Southern blot analysis suggested that the second allele might be in germline configuration, although we cannot exclude deletion of the second allele in 2 of the 3 cases because of the low tumor load (Table 1, Figure 1). Curiously, the 4 Igκ+ positive MM patients with incomplete Dh-Jhrearrangements had functional, somatically mutated IGKgene rearrangements. We cannot explain this finding in terms of normal B-cell maturation because exclusive Dh-Jhrearrangements, as found in our patients, are markers of the most immature B-cell precursors in BM and are in striking contrast with somatically mutated Vκ -Jκ gene rearrangements, which are typical for mature Ig+(post-)germinal B cells. In fact, from an immunobiologic point of view, the precursor of each plasma cell and each MM should originally have expressed a functional Ig molecule to reach its final stage of B-cell maturation. Therefore, the most likely explanation for the absence of a functional IGH gene rearrangement in the 5 patients with exclusive Dh-Jhrearrangements has to be that the second (functional) allele was deleted during or after the oncogenic process. This was indeed found in patients MM-4 and MM-5, might be true in patients MM-7 and MM-10, but seems not to be the case in patient MM-6 (Table 1). The high frequency of monoallelic IGH gene rearrangements (10 of 12 cases) already suggested that in IgH− MM deletion of 1 IGHallele is a frequent phenomenon.
Finally, we could not establish the reason for IgH negativity in 1 case (patient MM-12). Although on 1 allele Jh and Cμ gene segments were deleted, the second allele contained a functional, in-frame, somatically mutated Vh1-18/Jh4 rearrangement linked to a Cα gene. Unfortunately, Northern blot analysis could not be performed due to insufficient cell material.
One of the few studies addressing the potential mechanisms for inability of IgH protein production in Bence-Jones MM, demonstrated lack of IGH transcription in 6 IgH− MM cases and 3 IgH− MM cell lines.47 The authors suggested alterations of the transcriptional apparatus as a major cause of failure to produce IgH. However their analysis of the IGHgene configuration was too limited to exclude defects at the DNA level. Kuipers et al46 used several fluorescence in situ hybridization techniques for studying 19 MM cell lines, including 11 without IgH protein production. They concluded that the majority of IgH− cell lines contained abnormalIGH genes owing to illegitimate IGH class switch processes, presumably resulting from chromosome translocations involving 14q32.3. Curiously, 6 of 11 IgH− cell lines were derived from Ig-producing MM patients. Therefore, secondary genetic changes during culturing might have caused IgH negativity in these cell lines.
The combined Southern blot and PCR data presented here show that in the vast majority of IgH− MM patients defects at the DNA level are responsible for the lack of IgH protein production. We conclude that in at least 9 of the 12 patients (Table 3) these defects concern illegitimate IGH class switch rearrangements or illegitimate deletion of the functional Vh-(Dh)-Jhallele, which probably occurred during or after the malignant transformation process.
Acknowledgments
We are grateful to Prof dr R. Benner and Prof dr D. Sońta-Jakimczyk for their continuous support, Mr T. M. van Os for preparation of the figures and Mrs A. D. Korpershoek for her secretarial support.
Reprints:J. J. M. van Dongen, Department of Immunology, Erasmus University Rotterdam, PO Box 1738, 3000 DR Rotterdam, The Netherlands.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal