Abstract
Inside-out signaling regulates the ligand-binding function of integrins through changes in receptor affinity and/or avidity. For example, IIbβ3 is in a low-affinity/avidity state in resting platelets, and activation of the receptor by platelet agonists enables fibrinogen to bind. In addition, certain mutations and truncations of the integrin cytoplasmic tails are associated with a high-affinity/avidity receptor. To further evaluate the structural basis of integrin activation, stable Chinese hamster ovary (CHO) cell transfectants were screened for high-affinity/avidity variants of IIbβ3. One clone (AM-1) expressed constitutively active IIbβ3, as evidenced by (1) binding of soluble fibrinogen and PAC1, a ligand-mimetic antiIIbβ3antibody; and (2) fibrinogen-dependent cell aggregation. Sequence analysis and mutant expression in 293 cells proved that a single amino acid substitution in the cysteine-rich, extracellular portion of β3(T562N) was responsible for receptor activation. In fact, T562N also activated Vβ3, leading to spontaneous binding of soluble fibrinogen to 293 cells. In contrast, neither T562A nor T562Q activated IIbβ3, suggesting that acquisition of asparagine at residue 562 was the relevant variable. T562N also led to aberrant glycosylation of β3, but this was not responsible for the receptor activation. The binding of soluble fibrinogen to IIbβ3(T562N) was not sufficient to trigger tyrosine phosphorylation of pp125FAK, indicating that additional post-ligand binding events are required to activate this protein tyrosine kinase during integrin signaling. These studies have uncovered a novel gain-of-function mutation in a region of β3 intermediate between the ligand-binding region and the cytoplasmic tail, and they suggest that this region is involved in integrin structural changes during inside-out signaling.
THE β3 SUBFAMILY OF heterodimeric integrin receptors includes αIIbβ3, which is specific for platelets and critical for adhesive events in hemostasis, and αVβ3, which is more widely distributed and involved in regulation of cell growth, migration, and programmed death.1-4 The ligand binding function of these integrins is tightly regulated by two processes often referred to collectively as inside-out signaling: (1) affinity modulation, which involves structural changes intrinsic to the heterodimer; and (2) avidity modulation due to lateral diffusion and clustering of heterodimers into oligomers.5,6 Ligand binding to and clustering of β3 integrins in turn generate outside-in signals, such as activation of the protein tyrosine kinases Src and pp125FAK, which act in concert with signals from growth factor receptors to regulate many anchorage-dependent cell functions.6-8
The β3 integrin subunit consists of 762 amino acids encompassing a large extracellular domain, a single membrane-spanning region, and a 47 amino acid cytoplasmic tail.9 The extracellular domain is notable for an I-domain like ligand-binding region (residues 110-294)10 and a cysteine-rich repeat region (residues 423-622) that contains 31 of the subunit’s 56 cysteine residues.9,11 Studies of individuals with variant Glanzmann thrombasthenia, who bleed due to functional defects of αIIbβ3, and of recombinant integrins expressed in mammalian cells have focused attention on the ligand-binding region and the cytoplasmic tail as particularly relevant to the process of inside-out signaling.12-19 In addition, antibodies known as ligand-induced binding site (LIBS), which bind better to β3 after fibrinogen binds, can in some cases increase receptor affinity without the need for signals from inside the cells.20,21 Some of these antibodies recognize epitopes within the cysteine-rich repeats of β3,21 22raising the possibility that this region may also be involved in propagating activating signals from inside the cell to the ligand binding region of the receptor. In this report, we provide the first direct evidence for this idea by characterizing a single amino acid substitution in the cysteine-rich repeat region of β3that results in constitutive activation of both αIIbβ3 and αVβ3.
MATERIALS AND METHODS
Antibodies and plasmids.
PAC1, a ligand-mimetic mouse monoclonal IgM antibody, is specific for the activated αIIbβ3 complex.23Two antibodies to LIBS within β3, LIBS1 and LIBS6,20,24 were obtained from Dr Mark H. Ginsberg (The Scripps Research Institute, La Jolla, CA). AP5 (anti-β3LIBS),22 AP3 (anti-β3; non-function blocking),25 and AP2 (anti-αIIbβ3; function blocking)26 were obtained from Dr Thomas J. Kunicki (The Scripps Research Institute). PT25-2, an αIIbβ3 activating antibody, and its Fab fraction have been characterized.27 LM142 (anti-αV; non-function blocking)28 were obtained from Dr David A. Cheresh (The Scripps Research Institute). αIIb cDNA and β3 cDNA inserted in pCDM8 (resulting in the expression plasmids, CD2b and CD3a)17were obtained from Dr M.H. Ginsberg. αIIb cDNA and β3 cDNA inserted in pcDNA3 (αIIb/pcDNA3, β3/pcDNA3) were obtained from Dr Peter J. Newman (The Blood Center of Southeastern Wisconsin, Milwaukee, WI). αV cDNA inserted in pcDNA1 was obtained from Dr D.A. Cheresh.
Cell lines.
Chinese hamster ovary (CHO) and 293 cells were maintained as described.29,30 CD2b and CD3a were cotransfected into a CHO-AA8 cell line (Clontech, Palo Alto, CA), and stable transfectants expressing αIIbβ3 were obtained by single cell sorting by a FACStar flow cytometry (Becton Dickinson, San Jose, CA). As shown previously, αIIbβ3transfectants obtained in this manner do not bind PAC1 or fibrinogen and are in a low-affinity/avidity state.17 30 Then, one of these clones (24/12) was stained with PAC1 and subjected to another round of single-cell sorting to select for rare variants that now bound PAC1. 192 cells were sorted and PAC1 binding of 48 clones was reassessed. Only one clone, AM-1, showed constitutive binding of PAC1, as described below.
Nucleotide sequence analysis of αIIb and β3.
Genomic DNA from 24/12 and AM-1 cells was isolated (Qiagen DNA extraction kit; Qiagen Inc, Chatsworth, CA), and the entire coding regions of αIIb and β3 were amplified using 4 and 3 paired sets of primers, respectively.31 32Amplified DNA fragments were purified and subjected to direct cycle sequence analysis using an Applied Biosystems Automated sequencer (Perkin Elemer-Japan, Chiba, Japan) according to the manufacturer’s directions.
Site-directed mutagenesis of β3 cDNA.
To introduce the T562N mutation in β3 cDNA, two-step ligation was performed. First, β3/pcDNA3 was cut withBamHI and Afl II, yielding three fragments, Frag1 (BamHI and Afl II ends: 6 kb), Frag2 (BamHI andBamHI ends: 1.5 kb), and Frag3 (BamHI and AflII ends: 0.5 kb). After agarose gel electrophoresis, Frag1 and Frag2 were cut out with Gel extraction kit (Qiagen). The β3cDNA fragment from AM-1 cells including the mutated site (T562N) was amplified using primers, IIIa3 and IIIa4-AflII, and digested withBamHI and Afl II. The 0.5-kb fragment [Frag3(T562N)] was cut out and ligated to Frag1 with a ligation kit (Boehringer Mannheim, Mannheim, Germany). After transformation to JM109 competent cells (Takara, Shiga, Japan), miniprep DNA was cut withBamHI and Frag2 was inserted. After transformation, a clone containing Frag2 in the correct orientation was selected by polymerase chain reaction (PCR), amplified, and purified with a cesium chloride gradient. The entire coding region of the plasmid was sequenced to confirm aquisition of T562N mutation and absence of other nucleotide change.
To introduce T562A, T562Q, and T564A into β3 cDNA, PCR-based mutagenesis was performed as described.33 In brief, first-round PCR was performed using β3/pcDNA3 as a template with antisense primer containing mutated nucleotide(s) or sense primer containing mutated nucleotide(s) and IIIa4-AflII. The sequence of primers used is shown in Table1. After purification of PCR fragments with a purification kit (Qiagen), second-round PCR was performed using mixture of first-round PCR products as template with primers, IIIa3 and IIIa4-AflII. PCR products were cut with BamHI and Afl II, and the appropriate fragments were purified and ligated to Frag1 and Frag2. To make the β3(T562N, T564A) double mutant, β3(T562N)/pcDNA3 was used as a template instead of β3/pcDNA3. All plasmids were sequenced to confirm authenticity.
Oligonucleotides Used in the Construction of the β3 Mutants
| IIIa3 | GAG CTG ATC CCA GGG ACC AC |
| IIIa4-AfIII | GCA TCC TTG CCA GTG TCC TTA AG |
| T562A-sense | CTG CAA CTG TAC CGC GCG TAC TG |
| T562A-antisense | CAG TAC GCG CGG TAC AGT TGC AG |
| T562Q-sense | CTG CAA CTG TAC CCA GCG TAC TG |
| T562Q-antisense | CAG TAC GCT GGG TAC AGT TGC AG |
| T564A-sense | CGT GCT GAC ACC TGC ATG TC |
| T564A-antisense | GAC ATG CAG GTG TCA GCA CG |
| IIIa3 | GAG CTG ATC CCA GGG ACC AC |
| IIIa4-AfIII | GCA TCC TTG CCA GTG TCC TTA AG |
| T562A-sense | CTG CAA CTG TAC CGC GCG TAC TG |
| T562A-antisense | CAG TAC GCG CGG TAC AGT TGC AG |
| T562Q-sense | CTG CAA CTG TAC CCA GCG TAC TG |
| T562Q-antisense | CAG TAC GCT GGG TAC AGT TGC AG |
| T564A-sense | CGT GCT GAC ACC TGC ATG TC |
| T564A-antisense | GAC ATG CAG GTG TCA GCA CG |
The bold letters indicate base changes from the wild-type sequence.
Transient transfection.
A human embryonic kidney cell line, 293 (obtained from American Type Cell Culture, Rockville, MD), was used for transient transfection assays. Transfections were performed using the calcium phosphate method as described.29 Functional assays were performed 48 hours after transfection.
PAC1 binding and fibrinogen binding assay.
PAC1 binding to cells was assessed as described30 with minor modification. Cells (5 × 105) were preincubated in 45-μL aliquots containing Tyrode’s buffer supplemented with 2 mmol/L CaCl2 and 2 mmol/L MgCl2 in the presence or absence of 10 μmol/L FK633 (a peptidomimetic αIIbβ3 antagonist from Dr Jiro Seki, Fujisawa Pharmaceutical Co, Osaka, Japan)34 or 10 μg/mL PT25-2, an αIIbβ3 activating antibody. Then, 5 μL of a 1:25 dilution of PAC1 ascites was added to each tube, and incubations were performed for another 30 minutes at room temperature. After washing, cells were resuspended in a 1:20 dilution of fluorescein isothiocyanate (FITC)-conjugated antimouse IgM (μ-chain specific; Caltag Lab, Burlingame, CA) for 25 minutes on ice. Then, 1 μL of 1 mg/mL of propidium iodide (PI; Sigma, St Louis, MO) was added, and 5 minutes later the cells were washed and resuspended in 500 μL of ice-cold Tyrode’s and analyzed on a FACScan flow cytometer (Becton Dickinson). PAC1 binding (FL1) was analyzed on the gated subset of single, live cells (PI-negative, FL3). To compare the PAC1 binding results from one experiment with those from another, PAC1 binding was expressed as an activation index, defined as (Fx − Fi)/(Fm − Fi), where Fx is the median fluorescence intensity of PAC1 binding in the absence of inhibitor, FK633; Fi is the median fluorescence intensity of PAC1 binding in the presence of inhibitor, FK633; and Fm is median fluorescence intensity of PAC1 binding in the presence of the activating antibody, PT25-2.30
Fibrinogen was labeled with FITC as described.35 In the case of αIIbβ3-stable cell lines, the binding of FITC-fibrinogen (150 μg/mL) was assessed in the same manner as described for PAC1. In the case of 293 cells transiently transfected with αVβ3, cells were suspended in calcium-free Tyrode’s buffer supplemented with 1 mmol/L MgCl2 and pretreated for 30 minutes on ice with or without 1 mmol/L RGDW, an inhibitor of fibrinogen binding to αVβ3, or 1 mmol/L MnCl2, which induces a high-affinity state of αVβ3.2 LM142 (10 μg/mL), a non-function blocking anti-αV antibody, was added simultaneously to the tubes to monitor expression of αVβ3. After washing, cells were incubated with 150 μg/mL of FITC-fibrinogen and phycoerythrin (PE)-conjugated antimouse IgG (Serotec, Oxford, UK) for 25 minutes at room temperature. Then, after 5 minutes of incubation with PI, cells were washed and three-color analysis was performed on FACScan. In this case, fibrinogen binding (FL1) was analyzed on the gated subset of single, live cells (PI-negative, FL3) that stained positively for αVβ3 (FL2).
Cell aggregation.
Fibrinogen-dependent cell aggregation was monitored as described.36 In brief, 5 × 106/mL cells were added to wells of a 24-well culture dish precoated with 1 mg/mL of bovine serum albumin (BSA). Fibrinogen (300 μg/mL) was added with or without 20 μmol/L FK633 or 20 μg/mL PT25-2 and the dish was rotated at 100 rpm for 20 minutes on a horizontal rotator (Multi-Mixer; Lab-Line Instruments Inc, Melrose Park, IL). Aggregate formation was stopped by adding an equal volume of 0.5% formaldehyde, and aggregates were visualized and photographed under a phase-contrast microscope (Olympus, Tokyo, Japan).
Cell surface labeling and immunoprecipitation.
Cells were labeled with sulfo-NHS-biotin (Pierce, Rockford, IL) and lysed in Triton X-100 buffer (1% Triton X-100, 25 mmol/L Tris-Cl, 100 mmol/L NaCl, pH 7.4, 0.1 mg/mL leupeptin, 4 μg/mL pepstatin A, 1 mmol/L phenylmethylsulfonyl fluoride, and 10 mmol/L benzamide). Two hundred micrograms of protein from each sample was immunoprecipitated with an αIIbβ3 complex-specific antibody, AP2, as described.32 Immunoprecipitates were resolved in 6% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride (PVDF) membrane (Immobilon; Millipore, Bedford, MA). After incubation with peroxidase-conjugated avidin (Vectastain ABC kit; Vector Lab, Burlingame, CA), immunoreactive bands were visualized by enhanced chemiluminescence (Renaissance; NEN Life Science, Boston, MA).
Tyrosine phosphorylation of pp125FAK.
Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 0.5% fetal bovine serum for 18 hours and then resuspended to 3 × 106 cells/mL in DMEM. To test the effects of soluble fibrinogen to αIIbβ3, suspended cells were incubated at 37°C for 30 minutes in the presence or absence of 10 μg/mL of Fab fragments of PT25-2, and then 250 μg/mL of fibrinogen was added to the cells. After 15 minutes of incubation at 37°C, the cells were washed with phosphate-buffered saline (PBS) and lysed with Triton lysis buffer supplemented with 1 mmol/L sodium vanadate. For studies of adherent cells, 1 mL of suspended cells were seeded onto plastic dishes that had been precoated with 100 μg/mL fibrinogen or poly-L lysine (Iwaki Glass, Tokyo, Japan). After 30 minutes at 37°C, plates were washed twice with ice-cold PBS. Then adherent cells were lysed on the plates with Triton lysis buffer containing sodium vanadate and scraped into microcentrifuge tubes. Lysates were incubated on ice for 30 minutes and clarified supernatants were processed for immunoprecipitation.
pp125FAK was immunoprecipitated with 1 μg of rabbit polyclonal antibody, FAK(C903) (Santa Cruz Biotech, Santa Cruz, CA), and protein-G sepharose (Pharmacia, Uppsala, Sweden). Precipitates were separated on 7.5% SDS-PAGE and transferred to a PVDF membrane. Phosphotyrosine was detected with monoclonal antibody, 4G10. To monitor loading of gel lanes, the blots were stripped (2% SDS, 62.5 mmol/L Tris, pH 6.7, 100 mmol/L 2-mercaptoethanol for 30 minutes at 70°C) and reprobed with FAK(C903).
RESULTS
Establishment of the AM-1 cell line expressing a constitutively active form of αIIbβ3.
αIIbβ3 expressed in resting platelets or in a CHO cell model system exists in a low-affinity/avidity state and does not bind soluble fibrinogen or the ligand-mimetic antibody, PAC1.6,17 To better understand the structural basis of αIIbβ3 activation, a stable CHO cell line that expresses αIIbβ3 (24/12) was stained with PAC1 and analyzed by flow cytometry to screen for rare variants that might bind PAC1 constitutively. In so doing, a stable cell line, AM-1 (Activated Mutant-1), that expressed this activated phenotype was established. Parental 24/12 cells did not bind to PAC1 spontaneously, but they did do so as expected in response to activating antibody, PT25-2 (activation index [AI], 0.04 ± 0.02 [mean ± SD]; n = 5). In contrast, AM-1 cells bound PAC1 even in the absence of PT25-2 (AI, 0.67 ± 0.09; n = 5) and the binding was completely inhibited by incubation with an αIIbβ3 specific antagonist, FK633 (Fig 1A). The same results were obtained when another activating antibody (LIBS6) was used to induce ligand binding21 or when another specific antagonist (Integrilin) was used to block ligand binding37 (data not shown). Preincubation of AM-1 cells with 0.2% sodium azide and 4 mg/mL 2-deoxy-d-glucose had no effect on the increase in PAC1 binding, indicating that the high-affinity/avidity state of the αIIbβ3 in AM-1 cells was independent of metabolic energy (data not shown).
Assessment of affinity state of IIbβ3 by PAC1 (A) and fibrinogen (B) binding. In (A), 24/12 cells (a through c) or AM-1 cells (d through f) were preincubated with 10 μmol/L FK633 (a and d), 10 μg/mL PT25-2 (c and f), or buffer (b and e) for 30 minutes on ice. Then, 250× diluted PAC1 ascites were added and incubated for another 30 minutes at room temperature. After washing, cells were incubated with FITC-conjugated antimouse IgM for 25 minutes on ice. To exclude dead cells, PI were added to the cells and incubated for 5 minutes. After washing, cells were resuspended in buffer and flow cytometric analysis was performed. In (B), cells were preincubated with 10 μmol/L FK633 (dotted lines), 10 μg/mL PT25-2 (solid lines), or buffer (bold lines) for 30 minutes on ice. Cells were then incubated with 150 μg/mL of FITC-labeled fibrinogen for 25 minutes at room temperature and then with PI for 5 minutes. After washing, flow cytometric analysis was performed.
Assessment of affinity state of IIbβ3 by PAC1 (A) and fibrinogen (B) binding. In (A), 24/12 cells (a through c) or AM-1 cells (d through f) were preincubated with 10 μmol/L FK633 (a and d), 10 μg/mL PT25-2 (c and f), or buffer (b and e) for 30 minutes on ice. Then, 250× diluted PAC1 ascites were added and incubated for another 30 minutes at room temperature. After washing, cells were incubated with FITC-conjugated antimouse IgM for 25 minutes on ice. To exclude dead cells, PI were added to the cells and incubated for 5 minutes. After washing, cells were resuspended in buffer and flow cytometric analysis was performed. In (B), cells were preincubated with 10 μmol/L FK633 (dotted lines), 10 μg/mL PT25-2 (solid lines), or buffer (bold lines) for 30 minutes on ice. Cells were then incubated with 150 μg/mL of FITC-labeled fibrinogen for 25 minutes at room temperature and then with PI for 5 minutes. After washing, flow cytometric analysis was performed.
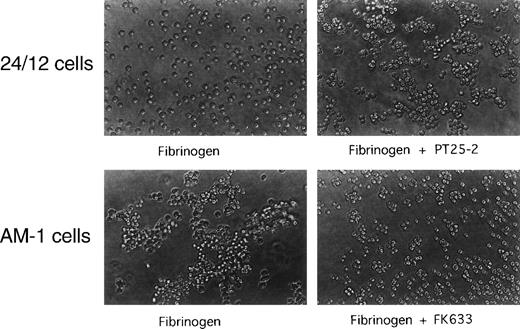
To establish whether AM-1 cells also bind a soluble physiological ligand in a constitutive fashion, binding of FITC-labeled fibrinogen was examined by flow cytometry. Consistent with the PAC1 results, fibrinogen bound to AM-1 in the absence of PT25-2 and the binding was completely blocked by FK633 (Fig 1B). To determine the functional relevance of this spontaneous fibrinogen binding, the ability of AM-1 cells to aggregate was assessed. The parental 24/12 cells exhibited fibrinogen-dependent aggregation only after addition of the activating antibody. In contrast, AM-1 cells exhibited fibrinogen-dependent and FK633-inhibitable aggregation even in the absence of the activating antibody (Fig 2). AP2, a function blocking anti-αIIbβ3 antibody,26 also inhibited the fibrinogen-dependent aggregation of AM-1 cells, and this aggregation was divalent cations dependent, because the aggregation was not observed when cells were resuspended in divalent cation free Tyrodes buffer with 2 mmol/L EDTA (data not shown). These results indicate that αIIbβ3 in AM-1 cells is in a constitutively activated state, fully capable of supporting spontaneous cell aggregation in the presence of fibrinogen.
Spontaneous, fibrinogen-dependent aggregation of AM-1 cells. Cells were resuspended in Tyrode’s buffer in the presence of 300 μg/mL of fibrinogen and with or without 20 μmol/L FK633 or 20 μg/mL PT25-2. Cells were then rotated for 20 minutes on a horizontal rotator. Aggregate formation was stopped by adding equal amount of 0.5% formaldehyde and incubating for 30 minutes. Shown are photographs representative of three independent experiments.
Spontaneous, fibrinogen-dependent aggregation of AM-1 cells. Cells were resuspended in Tyrode’s buffer in the presence of 300 μg/mL of fibrinogen and with or without 20 μmol/L FK633 or 20 μg/mL PT25-2. Cells were then rotated for 20 minutes on a horizontal rotator. Aggregate formation was stopped by adding equal amount of 0.5% formaldehyde and incubating for 30 minutes. Shown are photographs representative of three independent experiments.
αIIbβ3in AM-1 cells is in a high-affinity state.
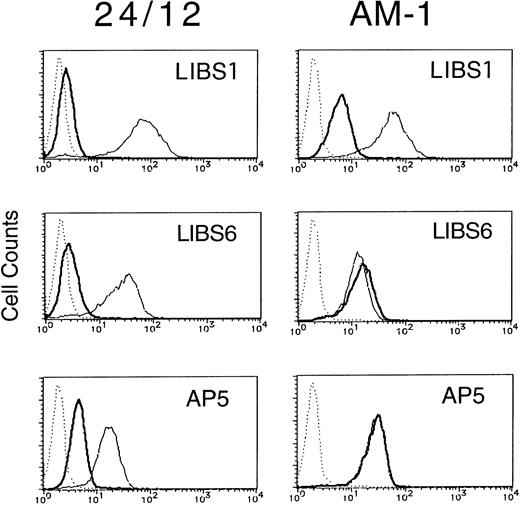
Integrins can become activated to bind ligands through conformational changes in the heterodimer (affinity modulation) and/or receptor clustering (avidity modulation).5-7 To determine whether the activation of αIIbβ3 in AM-1 cells was associated with conformational changes in the receptor, the binding of various anti-LIBS antibodies (LIBS1, LIBS6, and AP5) was examined. It has been shown that LIBS1, LIBS6, and AP5 preferentially recognize epitopes on β3 after ligand-binding, and the epitope of LIBS6 and AP5 has been defined within the cysteine-rich repeat region and the N-terminal region, respectively.20 22 FK633 (5 μmol/L) was used to induce LIBS expression, because preliminary studies showed that it could induce full expression of LIBS epitopes in platelets and CHO cell lines (Kashiwagi et al, unpublished observation). In parental 24/12 cells the LIBS antibodies bound to αIIbβ3 only after cell incubation with FK633. In contrast, LIBS6 and AP5 bound fully to AM-1 cells in the absence of FK633 and LIBS1 showed a slight increase in binding (Fig 3). Thus, some but not all LIBS epitopes are constitutively exposed on αIIbβ3 in AM-1 cells, even in the absence of added ligand.
LIBS expression of 24/12 (the left row) and AM-1 cells (the right row). Cells were preincubated with 5 μmol/L FK633 (solid lines) or buffer (bold lines) for 30 minutes on ice, and then 5 μg/mL of LIBS1 (upper row), LIBS6 (middle row), or AP5 (lower row) was added. After 30 minutes of incubation, cells were washed and then incubated with FITC-conjugated antimouse IgG for 30 minutes. After washing, flow cytometric analysis was performed. Dotted lines indicate MOPC21, a control mouse IgG antibody, binding.
LIBS expression of 24/12 (the left row) and AM-1 cells (the right row). Cells were preincubated with 5 μmol/L FK633 (solid lines) or buffer (bold lines) for 30 minutes on ice, and then 5 μg/mL of LIBS1 (upper row), LIBS6 (middle row), or AP5 (lower row) was added. After 30 minutes of incubation, cells were washed and then incubated with FITC-conjugated antimouse IgG for 30 minutes. After washing, flow cytometric analysis was performed. Dotted lines indicate MOPC21, a control mouse IgG antibody, binding.
Molecular analysis of αIIbβ3 in AM-1 cells.
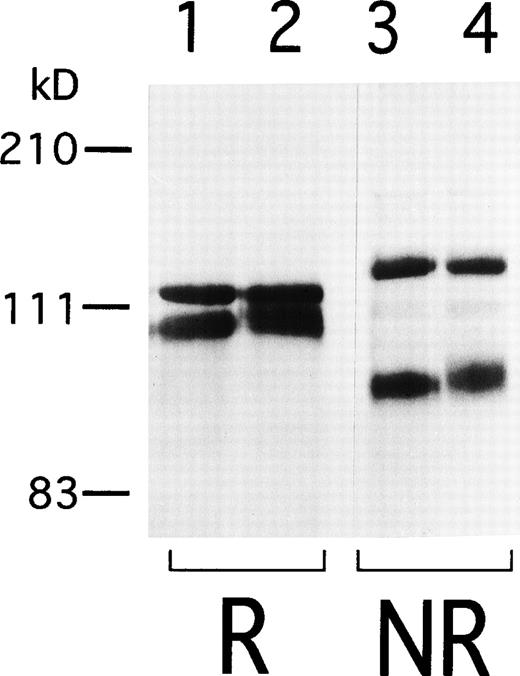
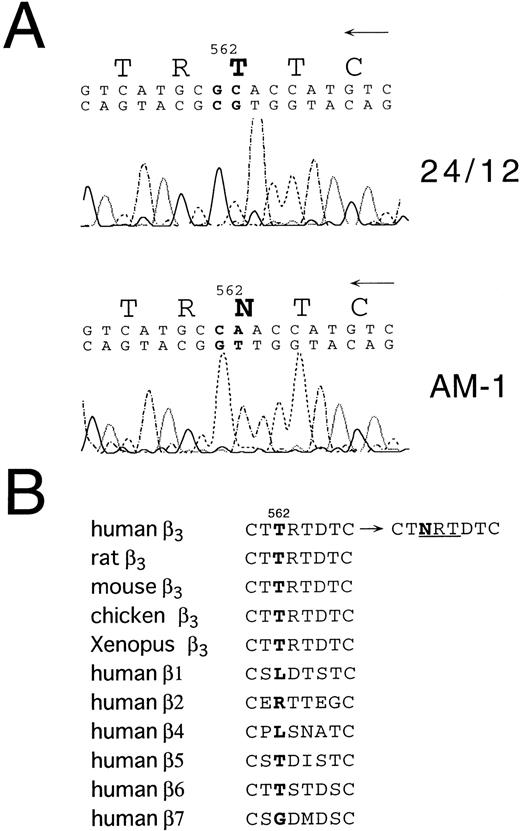
To begin to analyze the conformational change of αIIbβ3 in AM-1 cells, αIIbβ3 was immunoprecipitated with AP2. In contrast to β3 in the parental cells, β3 in AM-1 cells migrated more slowly and as a broad band under both reduced and nonreduced conditions, suggesting that mutation(s) and/or change of glycosylation state existed in β3 of AM-1 cells (Fig 4). Next, the sequence of the entire coding region of αIIb and β3 cDNA integrated in genomic DNA of AM-1 cells was determined. Amplification of genomic DNA using primer pairs that located in far separated exons excluded the possibility to amplify intrinsic hamster αIIb and β3 DNA. A consecutive dinucleotide change in β3 cDNA integrated in AM-1 cells (1732ACG→AAC) was discovered, leading to a single putative amino acid substitution, T562N (Fig 5A). No other sequence abnormalities were detected. This mutation is in the third of four cysteine-rich repeats in β3, and it would establish a new putative N-glycosylation site at the mutated residue (562TRT→NRT: NXT/S is a consensus sequence of N-glycosylation). Amino acid alignment around the mutated site shows that amino acid residues in C560-C567, that may make a small loop in the third repeat of the cysteine-rich repeat region,11 are completely identical in Xenopus, chicken, rodent, and human β3. By contrast, they are poorly conserved between human β3 and other human β integrins (Fig 5B).
Immunoprecipitation of IIbβ3from 24/12 and AM-1 cells. Cell surface was labeled with sulfo-NHS-LC-biotin and lysed with 1% Triton X-100 lysis buffer. Lysates from 24/12 cells (lanes 1 and 3) and AM-1 cells (lanes 2 and 4) were incubated with anti-IIbβ3 antibody, AP2, and immunoprecipitates were separated on 6% SDS-PAGE under reducing (lanes 1 and 2) or nonreducing conditions (lanes 3 and 4). After transfer, the membrane was incubated with peroxidase-conjugated avidin and developed with chemiluminescence.
Immunoprecipitation of IIbβ3from 24/12 and AM-1 cells. Cell surface was labeled with sulfo-NHS-LC-biotin and lysed with 1% Triton X-100 lysis buffer. Lysates from 24/12 cells (lanes 1 and 3) and AM-1 cells (lanes 2 and 4) were incubated with anti-IIbβ3 antibody, AP2, and immunoprecipitates were separated on 6% SDS-PAGE under reducing (lanes 1 and 2) or nonreducing conditions (lanes 3 and 4). After transfer, the membrane was incubated with peroxidase-conjugated avidin and developed with chemiluminescence.
Sequence analysis of β3 cDNA from 24/12 or AM-1 cells. (A) The results of sequencing using an antisense primer are shown. The same results were obtained using a sense primer for sequencing (data not shown). The mutated nucleotides and the changed amino acid are in bold. (B) Amino acid alignment around the mutated site is shown. T562 in β3 and corresponding amino acids in other β integrins are in bold. NRT (underlined) is a consensus sequence for N-glycosylation. Amino acid sequences were obtained from Wippler et al,44 Mimura et al,53and Ransom et al.54
Sequence analysis of β3 cDNA from 24/12 or AM-1 cells. (A) The results of sequencing using an antisense primer are shown. The same results were obtained using a sense primer for sequencing (data not shown). The mutated nucleotides and the changed amino acid are in bold. (B) Amino acid alignment around the mutated site is shown. T562 in β3 and corresponding amino acids in other β integrins are in bold. NRT (underlined) is a consensus sequence for N-glycosylation. Amino acid sequences were obtained from Wippler et al,44 Mimura et al,53and Ransom et al.54
Substitution of asparagine for threonine at position 562 of β3 causes a high-affinity state of αIIbβ3.
To prove that this single amino acid change was responsible for the constitutive activation of αIIbβ3, we introduced this mutation into wild-type β3 and transiently transfected it into 293 cells. Indeed, in the absence of activating antibody PT25-2, PAC1 bound spontaneously to cells transfected with αIIbβ3(T562N) (AI, 0.80 ± 0.10; n = 5), but not to cells transfected with wild-type αIIbβ3 (AI, 0.06 ± 0.02; n = 5), indicating that the T562N mutation is responsible for constitutive activation of the receptor (Fig 6A). When a D119Y mutation in β3 that abrogates fibrinogen binding to αIIbβ312 was introduced into β3(T562N), PAC1 failed to bind to αIIbβ3(D119Y, T562N) either in the absence or presence of PT25-2 (data not shown).
Assessment of the activation state of wild-type and mutant IIbβ3 in transiently transfected 293 cells. (A) Wild-type IIb cDNA was transfected with wild-type β3 cDNA (WT) (a through c) or mutant β3 cDNAs (T562N [d through f], T562A [g through i], T562Q [j through l]) to 293 cells, and PAC1 binding was determined. Plots in the upper row (a, d, g, and j) represent nonspecific PAC1 binding determined in the presence of FK633. Plots in the lower row (c, f, i, and l) represent maximal PAC1 binding in the presence of PT25-2. Plots in the middle row (b, e, h, and k) represent PAC1 binding in the absence of the antagonist and the activating antibody. (B) Wild-type IIbβ3, IIbβ3(T564A), or IIbβ3(T562N,T564A) transfected cells were incubated with PAC1 and biotinylated-AP3, a non-function blocking anti-β3 antibody, followed by incubation with FITC-conjugated antimouse IgM and PE-conjugated streptoavidin, and analyzed by flow cytometry. The overlay histogram represents PAC1 binding to cells expressing high levels of IIbβ3 determined by AP3 (denoted by the rectangle in the dot blots) [wild-type IIbβ3, dotted line; IIbβ3(T564A), solid line; IIbβ3(T562N,T564A), bold line]. (C) Wild-type and mutant IIbβ3 were surface-labeled with biotin, and immunoprecipitation was performed with AP2. Immnoprecipitates were electrophoresed on 6% polyacrylamide gel under reducing conditions. After transfer, membrane was incubated with peroxidase-conjugated avidin and developed with chemiluminescence.
Assessment of the activation state of wild-type and mutant IIbβ3 in transiently transfected 293 cells. (A) Wild-type IIb cDNA was transfected with wild-type β3 cDNA (WT) (a through c) or mutant β3 cDNAs (T562N [d through f], T562A [g through i], T562Q [j through l]) to 293 cells, and PAC1 binding was determined. Plots in the upper row (a, d, g, and j) represent nonspecific PAC1 binding determined in the presence of FK633. Plots in the lower row (c, f, i, and l) represent maximal PAC1 binding in the presence of PT25-2. Plots in the middle row (b, e, h, and k) represent PAC1 binding in the absence of the antagonist and the activating antibody. (B) Wild-type IIbβ3, IIbβ3(T564A), or IIbβ3(T562N,T564A) transfected cells were incubated with PAC1 and biotinylated-AP3, a non-function blocking anti-β3 antibody, followed by incubation with FITC-conjugated antimouse IgM and PE-conjugated streptoavidin, and analyzed by flow cytometry. The overlay histogram represents PAC1 binding to cells expressing high levels of IIbβ3 determined by AP3 (denoted by the rectangle in the dot blots) [wild-type IIbβ3, dotted line; IIbβ3(T564A), solid line; IIbβ3(T562N,T564A), bold line]. (C) Wild-type and mutant IIbβ3 were surface-labeled with biotin, and immunoprecipitation was performed with AP2. Immnoprecipitates were electrophoresed on 6% polyacrylamide gel under reducing conditions. After transfer, membrane was incubated with peroxidase-conjugated avidin and developed with chemiluminescence.
To determine the mechanism of activation by the T562N mutation, we introduced T562A and T562Q mutations into β3. The T562Q mutant was constructed because asparagine and glutamine have the same amide in their side chain. PAC1 did not bind to cells transfected with either αIIbβ3(T562A) or αIIbβ3(T562Q) in the absence of PT25-2 (AI, 0.08 ± 0.03 and 0.07 ± 0.06, respectively; n = 3; Fig 6A), indicating that it was the acquisition of asparagine at residue 562 that was important for spontaneous receptor activation. Next, we introduced the T564A mutation into β3 to disrupt the aberrant consensus sequence of N-glycosylation at residue 562. Immunoprecipitation of this form of β3 showed that β3(T564A) and β3(T562N,T564A) migrated on SDS gels similarly to the migration of β3(WT) (Fig 6C). Because the T564A mutation led to a mild reduction in the expression of αIIbβ3, we monitored αIIbβ3 expression by a non-function blocking anti-β3 antibody, AP3, and analyzed only cells expressing high levels of αIIbβ3 for PAC1 binding. Although αIIbβ3(T564A) showed a slight increase in the activation index (0.28 ± 0.03; n = 3), the activation of αIIbβ3(T562N, T564A) was even greater (AI, 0.46 ± 0.01; n = 3), and this difference was statistically significant (P < .001; Fig 6B). These results suggest that the glycosylation at N562 may not be essential for the constitutive activation of αIIbβ3 in AM-1 cells. A lack of relationship between the aberrant N-glycosylation of β3(T562N) and αIIbβ3activation was also suggested by the observation that 24 hours of incubation of the cells with 1 to 5 μg/mL of tunicamycin, a specific inhibitor of N-glycosylation,38 had no effect on the activation state of αIIbβ3 in AM-1 cells (AI, 0.67 ± 0.09, 0.67 ± 0.04, and 0.66 ± 0.02; 0, 1, and 5 μg/mL of tunicamycin, respectively; n = 3). On the other hand, the activation state of wild-type αIIbβ3 in 24/12 cells was slightly increased by tunicamycin treatment in a concentration-dependent manner (AI, 0.04 ± 0.02, 0.13 ± 0.04, and 0.18 ± 0.01; 0, 1, and 5 μg/mL of tunicamycin, respectively; n = 3).
The T562N mutation also leads to activation of αVβ3.
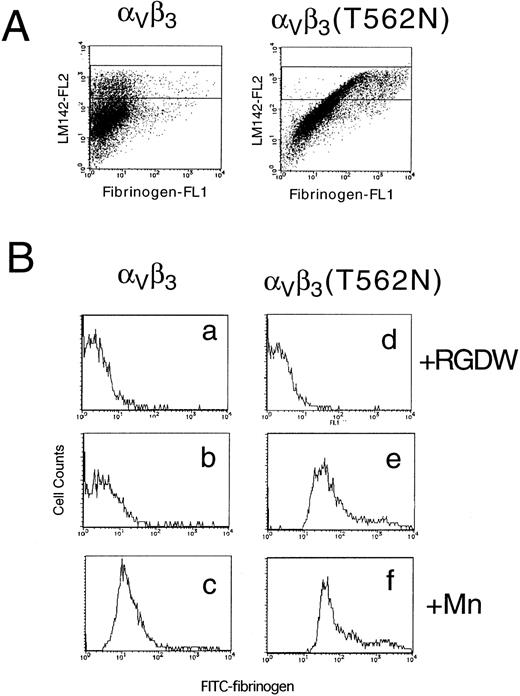
To determine whether the activating mutation in β3 would affect the affinity state of the related integrin αVβ3, this integrin was transiently expressed in 293 cells. In this case, FITC-labeled fibrinogen was used to determine the activation state of αVβ3and a non-function blocking anti-αV antibody, LM142, was used to monitor αVβ3 expression. When αV was transfected with wild-type β3, fibrinogen bound only if integrin affinity was upregulated by the addition of manganese. In contrast, fibrinogen binding to cells expressing αVβ3(T562N) could be detected even in the absence of manganese (Fig 7). These results indicate that the T562N mutation is capable of increasing the activation state of both αIIbβ3 and αVβ3.
Soluble fibrinogen binding to Vβ3 and Vβ3(T562N) transfected cells. Cells were preincubated with 1 mmol/L RGDW, 1 mmol/L manganese, or buffer with 10 μg/mL of anti-V antibody, LM142. After 30 minutes of incubation, cells were washed and then incubated with 150 μg/mL FITC-conjugated fibrinogen and PE-conjugated antimouse IgG for 30 minutes and analyzed by flow cytometry. (A) Dot blots represent FITC-fibrinogen (horizontal) and LM142 (vertical) binding in the absence of RGDW and manganese. (B) Fibrinogen binding to cells expressing high levels of Vβ3 (denoted by the rectangle in the dot blots) was analyzed on the histograms. (a through c) Wild-type Vβ3 transfected cells; (d through f) Vβ3(T562N) transfected cells. (a and d) With RGDW; (b and e) with buffer; (c and f) with manganese.
Soluble fibrinogen binding to Vβ3 and Vβ3(T562N) transfected cells. Cells were preincubated with 1 mmol/L RGDW, 1 mmol/L manganese, or buffer with 10 μg/mL of anti-V antibody, LM142. After 30 minutes of incubation, cells were washed and then incubated with 150 μg/mL FITC-conjugated fibrinogen and PE-conjugated antimouse IgG for 30 minutes and analyzed by flow cytometry. (A) Dot blots represent FITC-fibrinogen (horizontal) and LM142 (vertical) binding in the absence of RGDW and manganese. (B) Fibrinogen binding to cells expressing high levels of Vβ3 (denoted by the rectangle in the dot blots) was analyzed on the histograms. (a through c) Wild-type Vβ3 transfected cells; (d through f) Vβ3(T562N) transfected cells. (a and d) With RGDW; (b and e) with buffer; (c and f) with manganese.
Tyrosine phosphorylation of pp125FAK in AM-1 cells.
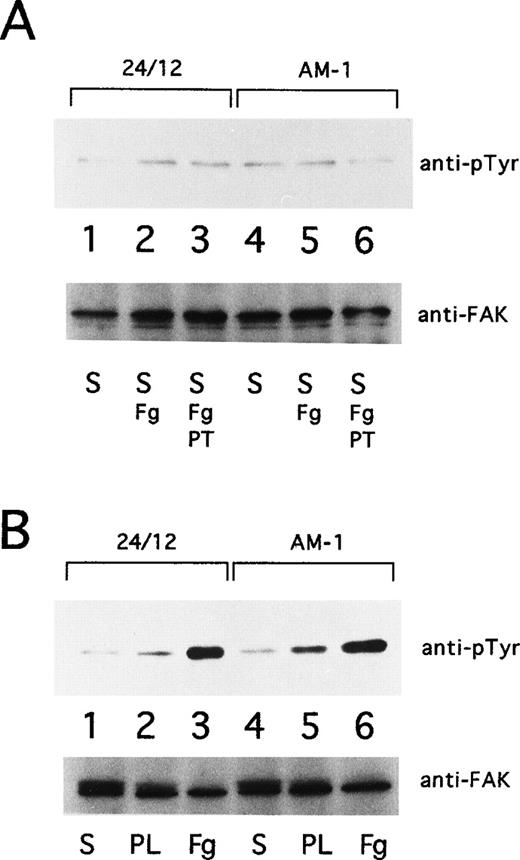
Ligand binding and clustering of integrins stimulate outside-in signaling, manifested by responses that include protein tyrosine phosphorylation and cytoskeletal reorganization.6-8 Focal adhesion kinase (FAK), a 125-kD cytoplasmic tyrosine kinase, is a component of focal adhesions and is a well-established component of integrin signaling pathways.39 40 Consequently, the tyrosine phosphorylation state of pp125FAK in AM-1 cells was studied. pp125FAK was not tyrosine-phosphorylated in AM-1 cells or in control 24/12 cells maintained in suspension for 15 minutes, either in the presence or absence of fibrinogen. Furthermore, neither cell type exhibited pp125FAK phosphorylation when adherent to plates coated with poly-L-lysine. On the other hand, both showed pp125FAK phosphorylation in response to cell adhesion to fibrinogen (Fig 8). These results indicate that receptor activation by the T562N mutation or the mere binding of soluble fibrinogen to the activated receptor is not sufficient to cause activation of pp125FAK; nonetheless, this mutant receptor is fully capable of mediating this outside-in signaling response upon cell adhesion.
pp125FAK phosphorylation. (A) 24/12 cells (lanes 1 through 3) or AM-1 cells (lanes 4 through 6) were maintained in suspension for 15 minutes with no addition (S; lanes 1 and 4), with the addition of 250 μg/mL of fibrinogen (S, Fg; lanes 2 and 5), and with addition of fibrinogen and 10 μg/mL of Fab fragments of PT25-2 (S, Fg+PT; lanes 3 and 6). (B) 24/12 cells (lanes 1 through 3) or AM-1 cells (lanes 4 through 6) were allowed to become adherent to immobilized fibrinogen (Fg; lanes 3 and 6) or poly-L lysine (PL; lanes 2 and 5) or were maintained in suspension (S; lanes 1 and 4) for 30 minutes. The cells were lysed and pp125FAK was immunoprecipitated with an anti-FAK polyclonal antibody. Phosphotyrosine was detected with 4G10 (upper panel), and blots were reprobed with anti-FAK to assess gel loading (lower panel).
pp125FAK phosphorylation. (A) 24/12 cells (lanes 1 through 3) or AM-1 cells (lanes 4 through 6) were maintained in suspension for 15 minutes with no addition (S; lanes 1 and 4), with the addition of 250 μg/mL of fibrinogen (S, Fg; lanes 2 and 5), and with addition of fibrinogen and 10 μg/mL of Fab fragments of PT25-2 (S, Fg+PT; lanes 3 and 6). (B) 24/12 cells (lanes 1 through 3) or AM-1 cells (lanes 4 through 6) were allowed to become adherent to immobilized fibrinogen (Fg; lanes 3 and 6) or poly-L lysine (PL; lanes 2 and 5) or were maintained in suspension (S; lanes 1 and 4) for 30 minutes. The cells were lysed and pp125FAK was immunoprecipitated with an anti-FAK polyclonal antibody. Phosphotyrosine was detected with 4G10 (upper panel), and blots were reprobed with anti-FAK to assess gel loading (lower panel).
DISCUSSION
In this report, we analyzed a cell line in which the ligand-binding function of integrin αIIbβ3 was constitutively activated, in contrast to the usual, default low-affinity/avidity state of this integrin in platelets and transfected tissue culture cells.6,17 We found the following: (1) A single amino acid change in the extracellular cysteine-rich repeat region of β3, T562N, is responsible for this constitutive activation. (2) The presence of the asparagine as opposed to the loss of the threonine appears responsible for this phenotype. (3) Although the T562N mutation leads to aberrant glycosylation, it is unlikely that this posttranslational modification actually causes the activated integrin phenotype. (4) The T562N mutation is capable of activating αVβ3 as well as αIIbβ3. (5) Activation of αIIbβ3 by T562N or binding of soluble fibrinogen to the mutant is not sufficient to trigger tyrosine phosphorylation of pp125FAK. Ligand binding as the result of the T562N mutation was similar to that induced through the more physiological process of inside-out signaling, because (A) it was completely blocked by synthetic αIIbβ3-specific antagonists, (B) it was abrogated by a mutation in β3(D119Y) associated with a variant form of thrombasthenia and clinical bleeding,12 and (C) it mediated fibrinogen-dependent cell aggregation.
Three types of activating mutations in αIIbβ3 have been described previously. (1) Bajt et al41 showed that swapping the ligand-binding site (residues 129-133) of β3 with the corresponding sequence in β1 led to a gain of function in αIIbβ3. (2) The proximal regions of cytoplasmic tails of αIIb and β3 are highly conserved, and deletions or amino acid changes that may break the interaction between them lead to activation of integrins.13,17,18 (3) Liu et al42 recently reported that disruption of the C5-C435 disulfide bond in β3 resulted in an increase in affinity of αIIbβ3. In this context, it has been shown that mild reducing agents, such as dithiothreitol, can increase the ligand-binding function of αIIbβ3.43 In addition, it has been shown that some LIBS antibodies that bind to epitopes within the cysteine-rich repeats of β3, such as LIBS2, LIBS3, and LIBS6, lead to activation of αIIbβ3 without ligand-binding.20-22 Wippler et al44 also demonstrated that recombinant αIIbβ3lacking the cysteine-rich repeats of β3 showed high-affinity binding to fibrinogen. Furthermore, sequence alignment of β3 integrins indicate that about 90% of noncysteine residues in the cysteine-rich repeats are conserved between rodents and human β3, whereas noncysteine residues in the region are poorly conserved among β1, β2, and β3 integrins (∼15%).45 These results suggest that the cysteine-rich repeats may have an important role for regulation of β3 integrin functions.
Our results provide the direct evidence that this region is involved in the activation of β3 integrins. Enhanced ligand binding to αIIbβ3(T562N) was observed both in CHO cells and 293 cells, suggesting that the functional effect by the mutation is not cell type-specific. Because neither the T562A nor the T562Q mutations caused activation of αIIbβ3, the side-chain of asparagine 562 is clearly an important variable contributing to induction of the activated receptor. The finding that the T562N mutation also led to activation of αVβ3 indicates that activation of αIIbβ3 by the mutation did not require a unique interaction of β3 with the αIIb subunit. Although T562N represents a new putative N-glycosylation site, it is unlikely that alternative glycosylation at this position is responsible for activation of αIIbβ3, because (1) αIIbβ3(T562N, T564A), which lacked the aberrant glycosylation observed with the single N562 mutation, showed an even more activated state than αIIbβ3(T564A); and (2) tunicamycin, an inhibitor of N-glycosylation,38 had no effect on the activated state of αIIbβ3(T562N) in AM-1 cells.
The binding of soluble ligands to integrins can be enhanced by two complementary mechanisms, conformational change within heterodimers (affinity modulation) and clustering of receptors into heteroligomers (avidity modulation).46,47 An understanding of the precise mechanism of integrin activation by the β3(T562N) mutation will require a level of understanding of integrin atomic structure that is not yet available. However, the predominant effect of the β3(T562N) mutation may be on β3integrin conformation. First, some LIBS epitopes are constitutively exposed on αIIbβ3 in AM-1 cells. Second, although affinity and avidity modulation both influence the functions of αIIbβ3, affinity modulation is the predominant regulator of ligand binding.47 Third, soluble fibrinogen binding to platelet or CHO cell αIIbβ3 activated through affinity modulation by means of an activating LIBS antibody is not sufficient to trigger tyrosine phosphorylation of FAK; rather, integrin clustering and other post-ligand binding events are also required.47-49 Similarly, fibrinogen binding induced by the β3(T562N) mutation was not sufficient to stimulate FAK phosphorylation, suggesting that this mutation was not primarily triggering receptor clustering.
It has been demonstrated that manganese,2β3-LIBS antibody,50 and purification of αVβ3 by affinity chromatography51 can induce a high-affinity state of αVβ3. However, the T562N mutation is the first one reported to induce spontaneous activation of αVβ3. A number of reports indicate that αVβ3 plays an important role in angiogenesis, tumor invasion, and bone absorption.2-4 One obvious question now being pursued is whether this activating mutation affects any of these functions of αVβ3. In any case, the current study serves to emphasize the possible involvement of residues in the cysteine-rich region of β3in affinity modulation of both αIIbβ3 and αVβ3. This will have to be taken into account in future refinements of models for integrin activation.52
ACKNOWLEDGMENT
The authors are grateful to Drs M.H. Ginsberg, T.J. Kunicki, J. Seki, D.A. Cheresh, and P.J. Newman for providing the materials.
Supported in part by grants from the Ministry of Education, Science and Culture of Japan; the Japan Society for the Promotion of Science; Research Foundation for Cancer and Cardiovascular Diseases, Osaka, Japan; and the National Institutes of Health (Bethesda, MD).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Hirokazu Kashiwagi, MD, The Second Department of Internal Medicine, Osaka University Medical School, B-5, 2-2 Yamada-oka, Suita, Osaka 565-0871, Japan; e-mail:kashi@hp-blood.med.osaka-u.ac.jp.


![Fig. 6. Assessment of the activation state of wild-type and mutant IIbβ3 in transiently transfected 293 cells. (A) Wild-type IIb cDNA was transfected with wild-type β3 cDNA (WT) (a through c) or mutant β3 cDNAs (T562N [d through f], T562A [g through i], T562Q [j through l]) to 293 cells, and PAC1 binding was determined. Plots in the upper row (a, d, g, and j) represent nonspecific PAC1 binding determined in the presence of FK633. Plots in the lower row (c, f, i, and l) represent maximal PAC1 binding in the presence of PT25-2. Plots in the middle row (b, e, h, and k) represent PAC1 binding in the absence of the antagonist and the activating antibody. (B) Wild-type IIbβ3, IIbβ3(T564A), or IIbβ3(T562N,T564A) transfected cells were incubated with PAC1 and biotinylated-AP3, a non-function blocking anti-β3 antibody, followed by incubation with FITC-conjugated antimouse IgM and PE-conjugated streptoavidin, and analyzed by flow cytometry. The overlay histogram represents PAC1 binding to cells expressing high levels of IIbβ3 determined by AP3 (denoted by the rectangle in the dot blots) [wild-type IIbβ3, dotted line; IIbβ3(T564A), solid line; IIbβ3(T562N,T564A), bold line]. (C) Wild-type and mutant IIbβ3 were surface-labeled with biotin, and immunoprecipitation was performed with AP2. Immnoprecipitates were electrophoresed on 6% polyacrylamide gel under reducing conditions. After transfer, membrane was incubated with peroxidase-conjugated avidin and developed with chemiluminescence.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/8/10.1182_blood.v93.8.2559/5/m_blod40812006ax.jpeg?Expires=1765927664&Signature=TjL-uc88XwIZvjEPo7ySxl2II0CUytYdqfYBrwSwjHvCvy2Ia2R5H-8QSpZKeWCLbwRLY7EGEBsvBtCHu5IMxlCOtEzrOyeBAgSvhTOkU0-6b2gXDkksLX9rgR0eC8u0r6ooSx3MbuxlcuKhZQfxWqvNIPev6xKYABW3Ovw2-WbxQchM2nqkrurOXNIiaTQwkOcWzJ57fsgKOPMG0CIMMbadXgo-SiijdYz1bIf-uSWv~UKHChFXN-OA4DcJivkp6lWRvmxc2w1X9py9-KNFOAQ8rxp59~JhmO7nvQ18q4gD3pBpOk~YH-WbJQJ9OY5hibK6wtnsdPdkmXITF8UIFw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. Assessment of the activation state of wild-type and mutant IIbβ3 in transiently transfected 293 cells. (A) Wild-type IIb cDNA was transfected with wild-type β3 cDNA (WT) (a through c) or mutant β3 cDNAs (T562N [d through f], T562A [g through i], T562Q [j through l]) to 293 cells, and PAC1 binding was determined. Plots in the upper row (a, d, g, and j) represent nonspecific PAC1 binding determined in the presence of FK633. Plots in the lower row (c, f, i, and l) represent maximal PAC1 binding in the presence of PT25-2. Plots in the middle row (b, e, h, and k) represent PAC1 binding in the absence of the antagonist and the activating antibody. (B) Wild-type IIbβ3, IIbβ3(T564A), or IIbβ3(T562N,T564A) transfected cells were incubated with PAC1 and biotinylated-AP3, a non-function blocking anti-β3 antibody, followed by incubation with FITC-conjugated antimouse IgM and PE-conjugated streptoavidin, and analyzed by flow cytometry. The overlay histogram represents PAC1 binding to cells expressing high levels of IIbβ3 determined by AP3 (denoted by the rectangle in the dot blots) [wild-type IIbβ3, dotted line; IIbβ3(T564A), solid line; IIbβ3(T562N,T564A), bold line]. (C) Wild-type and mutant IIbβ3 were surface-labeled with biotin, and immunoprecipitation was performed with AP2. Immnoprecipitates were electrophoresed on 6% polyacrylamide gel under reducing conditions. After transfer, membrane was incubated with peroxidase-conjugated avidin and developed with chemiluminescence.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/8/10.1182_blood.v93.8.2559/5/m_blod40812006cw.jpeg?Expires=1765927664&Signature=Hhh2p8f9qk6AxIbWMXp20bDYzONwrGm62863LNc-mWirGcgnjx-6zXVhAVEW6Jo~El~QKucNL-dytkwox7IyibDV11b5PiGTOlWSVZurIOGskh4jmckD9id51dZUstd7Nmgo5qzutR-oKuxrREHLOrQB3Hkr3BsvwDvYYmRscb9kcnyuDJkU3WgFnEiL2n208phOUpzCGADuejg5VFGW3Xw-i~bpE~qr65vMgE2WppCrkAkBu8tMxDmuTO8Lez-1YYqb1Xt2NLcRrDws~GzfNKb5NKopl3b-aife-k0d0KvhFa-K9wjnbILTzqmrztWFAMIW5aVJX85WZn~s22JORA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal