The chromatin reader protein SGF29 is a novel epigenetic vulnerability in AML.
SGF29 regulates AML oncogene transcription and KAT2A chromatin localization in AML.
Visual Abstract
Aberrant expression of stem cell–associated genes is a common feature in acute myeloid leukemia (AML) and is linked to leukemic self-renewal and therapy resistance. Using AF10-rearranged leukemia as a prototypical example of the recurrently activated “stemness” network in AML, we screened for chromatin regulators that sustain its expression. We deployed a CRISPR-Cas9 screen with a bespoke domain-focused library and identified several novel chromatin-modifying complexes as regulators of the TALE domain transcription factor MEIS1, a key leukemia stem cell (LSC)–associated gene. CRISPR droplet sequencing revealed that many of these MEIS1 regulators coordinately controlled the transcription of several AML oncogenes. In particular, we identified a novel role for the Tudor-domain–containing chromatin reader protein SGF29 in the transcription of AML oncogenes. Furthermore, SGF29 deletion impaired leukemogenesis in models representative of multiple AML subtypes in multiple AML subtype models. Our studies reveal a novel role for SGF29 as a nononcogenic dependency in AML and identify the SGF29 Tudor domain as an attractive target for drug discovery.
Introduction
Acute myeloid leukemia (AML) is a devastating blood cancer with a dismal survival rate.1 Current therapies often have strong toxicity and undesirable side effects, highlighting the need for safer, more effective alternatives. A major challenge in developing new drugs for AML is its molecular heterogeneity, comprising distinct mutational profiles and clinical characteristics.2 However, there are common molecular pathways dysregulated across different AML subtypes. Studies have shown that various upstream genetic alterations in AML, including DNMT3 or nucleophosmin 1 (NPM1) mutations and different chromosomal translocation fusion products, result in the activation of the clustered homeobox (HOX) genes and their cofactors, such as the 3 amino acid loop extension (TALE) homeobox gene MEIS1 (reviewed in3-11). Both loss-of-function and gain-of-function experiments have demonstrated the importance of HOX/MEIS expression in leukemia pathogenesis in diverse subsets of AML, including those bearing mixed lineage leukemia (MLL)-fusions,12,13 NUP98-fusions,14 and AF10-fusions.15,16 Importantly, ectopic HOXA9 overexpression in murine hematopoietic stem and progenitor cells (HSPCs) causes a myeloproliferative phenotype in mice that can progress to AML upon MEIS1 coexpression.17,18MEIS1 is a critical cofactor of leukemia-associated HOXA transcription factors and is required for their full leukemogenic capability,17,18 because it acts as a rate-limiting regulator of leukemia stem cells (LSCs).19 Taken together, there is compelling evidence implicating the HOX/MEIS oncoproteins as a critical node integrating a variety of functionally distinct oncogenic insults in AML.
Pharmacological targeting of the HOX/MEIS pathway may thus yield therapeutic benefits in multiple, genetically heterogeneous AML subtypes. Because the HOX/MEIS proteins are DNA-binding transcription factors, they have proven difficult to target directly using traditional drug-development methods. We reasoned that a detailed and systematic identification of epigenetic modulators critical for sustaining HOX/MEIS activation in AML would help identify nodes for targeted drug-discovery campaigns aimed at this clinically relevant pathway. To this end, we conducted pharmacological and CRISPR-based genetic screens using enhanced green fluorescence protein (eGFP) tagged MEIS1 AML cells.3 Our screens found several novel regulators of the expression of HOX/MEIS and other leukemia oncogenes in AML, including BMI1, SATB1, and MYC. Most notably, we identified a novel role for the Tudor domain–containing chromatin reader protein SGF29 in AML. We show that SGF29, through its Tudor domain, is a critical epigenetic regulator required for maintaining the transcription of key leukemia oncogenes and leukemogenesis of diverse AML subtypes.
Methods
Epigenetic CRISPR screen
The pooled epigenetics CRISPR library virus was produced using HEK293T cells transfected with polyethylimine, pMD2.G, and psPAX2 as previously described.6 Thirty million UB3 cells were transduced with the pooled library lentivirus in RPMI medium supplemented with 10% fetal bovine serum (FBS), antibiotics, and 0.8 μg/mL Polybrene. The medium was changed 24 hours after transduction to remove Polybrene, and cells were plated in fresh culture medium. Forty-eight hours after transduction, puromycin was added at a concentration of 1 μg/mL to select for cells expressing single-guide RNA (sgRNA) library and then removed after 72 hours. To ensure transduction of a single sgRNA per cell, the multiplicity of infection was set between ∼0.3 and 0.4. Adequate representation of sgRNAs during the screen was ensured by keeping >1000× cells in culture relative to the library size. Ten million cells per replicate were harvested 3 days after puromycin removal for an initial time point (T0). Cells were sorted using fluorescence-activated cell sorter 5 days later to collect the upper and lower quartiles on the basis of green fluorescent protein (GFP) expression using the FACSAria II Instrument (BD Biosciences) at the Sanford Burnham Prebys Flow Cytometry Core. Genomic DNA extraction from the T0 and GFP-low and -high cells was performed using the Zymo QuickDNA Midiprep Plus Kit (Zymo Research, catalog number D4068), according to the manufacturer’s instructions. A 2-step polymerase chain reaction (PCR)–amplification for sequencing library preparation was conducted with TaKaRa Ex Taq Polymerase (TaKara, catalog number RR001) and custom primers to achieve adequate sequencing coverage in 1 × 75 bp single-end reads, following published guidelines.5 Barcoded libraries were pooled and sequenced using an Illumina Hiseq 500.
CRISPR droplet sequencing (CROP-seq) experiment
Lentivirus was made from the CROP-seq pooled DNA by transfection of the CROP-seq pool together with polyethylimine, pMD2.G, and psPAX2 in HEK293T cells, as described previously.6 Viral supernatant was collected at 48 hours and filtered through a 0.45-μm filter. Viral supernatant was then concentrated by centrifugation (20 000 rpm, 2 hours, 4°C) and used for transduction of UB3 cells by incubation with 0.8 μg/μL of Polybrene overnight. Medium was changed after overnight incubation. At 48 hours after transduction, puromycin (1 μg/μL) was added to cell suspensions for 3 days of selection. Cells were pelleted for sequencing 3 days after removal of puromycin.
Cell culture
Human leukemia cell lines U937 (gift from Daniel Tenen) and MOLM13 (ACC-554; DSMZ) were cultured in RPMI-1640 medium with 2 mM L-glutamine and sodium pyruvate, 10% FBS and 50 U/mL penicillin/streptomycin (Thermo Fisher Scientific). Murine leukemia cells were cultured in Dulbecco modified Eagle medium with 2 mM L-glutamine, 15% FBS, and 50 U/mL penicillin/streptomycin, and 10 ng/mL murine interleukin-6, 6 ng/mL murine interleukin-3, and 20 ng/mL murine stem cell factor (Peprotech). HEK293T cells were cultured in Dulbecco modified Eagle medium with 2 mM L-glutamine and sodium pyruvate, 10% FBS and 50 U/mL penicillin/streptomycin. MLL-AF10 patient–derived xenograft (PDX) cells were cultured in Iscove modified Dulbecco medium supplemented with 20% BIT9500 (STEMCELL Technologies), human cytokines, and StemRegenin 1 44 (SR1), and UM171 as described earlier.9 All cells were incubated in 5% CO2 at 37°C.
Results
A high-density, domain–focused CRISPR screen identifies epigenetic regulators of MEIS1 expression
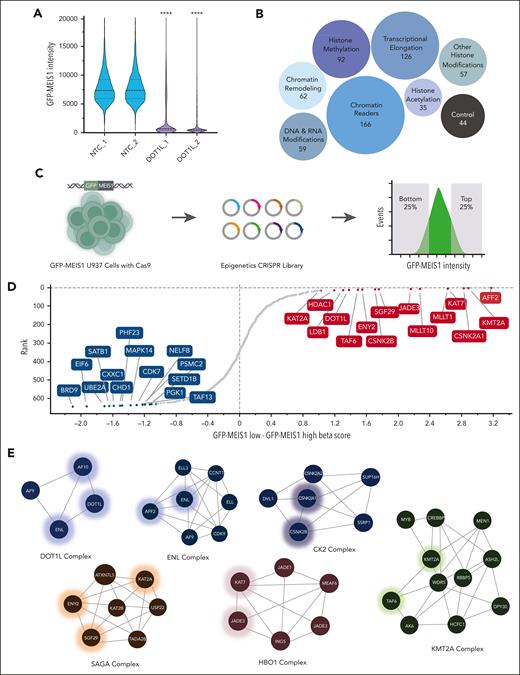
AML cases with the t(10;11)(p13;q14) translocation, which results in the CALM-AF10 fusion gene, display highly elevated expression of several stem cell–associated genes, including the HOXA cluster, MEIS1, and BMI1.20,21 Indeed, CALM-AF10–positive AML cell lines harbor some of the highest expression levels of these oncogenes (supplemental Figure 1A, available on the Blood website). We used a previously described3CALM-AF10–positive AML cell line, U937, with the eGFP gene knocked into the endogenous MEIS1 locus immediately upstream of the start codon. Leveraging this system, we used MEIS1 expression as a surrogate for identifying epigenetic regulators of the self-renewal–associated program in AML cells. First, we validated these U937-eGFP-MEIS1 cells (henceforth termed UB3) by perturbation with sgRNAs targeting DOT1L (Figure 1A). We performed chemical screens using a library of inhibitors of epigenetic pathways in the UB3 cells as well as in mouse GFP-MEIS1-tagged22 MLL-AF9 AML cells. Both screens only revealed the well-characterized MEIS1 regulators DOT1L and ENL/MLLT1 (supplemental Figure 1B-E; supplemental Tables 1 and 3). We reasoned that poorly studied epigenetic readers, writers, and eraser proteins are not adequately represented in currently available chemical epigenetic inhibitor libraries, precluding identification of their activities in phenotypic screens. Therefore, we sought to extend our investigation of MEIS1 epigenetic regulators using genetic screening. For this, we generated a CRISPR library targeting 645 genes, comprising all categories of epigenetic regulators (Figure 1B; supplemental Table 2). Given the demonstrated advantage of targeting functional domains,23 we designed 5 sgRNAs for every annotated domain, in addition to 5 sgRNAs for each early constitutive exon. We used this dense CRISPR library (total of 11 228 sgRNAs) to conduct an in vitro phenotypic enrichment screening in Cas9-expressing UB3 cells. We sorted the low- and high- eGFP fractions to infer positive or negative MEIS1 regulators, respectively (“Methods”; Figure 1C). Normalized read counts of nontargeting controls (NTCs) and polymerase-encoding genes were uniformly distributed in both eGFP fractions, indicating successful technical performance of controls (supplemental Figure 2A). MAGeCKFlute24,25 analysis of the screen revealed several novel hits, including AFF2, CSNK2A1, CSNK2B, SGF29/CCDC101, ENY2, TAF6, HDAC1, and LDB1, as well as regulators, such as DOT1L,4,26,27 MLLT1/ENL,12,13 MLLT10/AF10,28 KAT7/HBO1,29,30 JADE3,29 and KMT2A/MLL131,32 (Figure 1D). STRING analysis31 showed that multiple proteins from 6 chromatin complexes were among our top hits (Figure 1E). Thus, we identified distinct constituents of chromatin-modifying complexes with potential MEIS1 regulatory activity, offering independent, complementary nodes for therapeutic targeting.
Screening for epigenetic modulators identifies novel MEIS1 regulators in AML cells. (A) eGFP intensity values from monoclonal UB3 cells upon DOT1L CRISPR knockout are plotted on the y-axis (n ≥ 5000). Biological duplicates indicate independent sgRNAs. (B) Plot illustrating categories of chromatin modulators included in the CRISPR library design; the numbers indicate different genes under the category and the size of the bubble is proportional to the gene set size. Controls refer to pan-essential genes. (C) Strategy for phenotypic pooled CRISPR screening of epigenetic regulators for MEIS1 expression in the UB3 cell line. (D) Sorted gene hits based on differential beta scores for the eGFP-MEIS1 low minus eGFP-MEIS1 high fractions. Beta scores were calculated using MAGeCKFlute and plotted against rank of the genes on y-axis. Genes marked in red are potential MEIS1 activators while those in blue are candidate repressors. (E) Annotated protein complexes comprising top candidate hits identified in the epigenetic screen as identified using STRING database analysis. Hits from the epigenetics CRISPR library screen are marked with a glowing hue.
Screening for epigenetic modulators identifies novel MEIS1 regulators in AML cells. (A) eGFP intensity values from monoclonal UB3 cells upon DOT1L CRISPR knockout are plotted on the y-axis (n ≥ 5000). Biological duplicates indicate independent sgRNAs. (B) Plot illustrating categories of chromatin modulators included in the CRISPR library design; the numbers indicate different genes under the category and the size of the bubble is proportional to the gene set size. Controls refer to pan-essential genes. (C) Strategy for phenotypic pooled CRISPR screening of epigenetic regulators for MEIS1 expression in the UB3 cell line. (D) Sorted gene hits based on differential beta scores for the eGFP-MEIS1 low minus eGFP-MEIS1 high fractions. Beta scores were calculated using MAGeCKFlute and plotted against rank of the genes on y-axis. Genes marked in red are potential MEIS1 activators while those in blue are candidate repressors. (E) Annotated protein complexes comprising top candidate hits identified in the epigenetic screen as identified using STRING database analysis. Hits from the epigenetics CRISPR library screen are marked with a glowing hue.
Competition assays reveal the role of candidate hits in AML cell growth
Next, we tested the requirement of our top candidate hits for AML cell growth. We performed arrayed CRISPR competition assays in UB3 cells (Figure 2A for schematic). Specifically, we cloned 2 to 3 sgRNAs for each of the top candidate hits (SGF29, ENY2, AFF2, CSNK2A1, CSNK2B, MLLT1, KAT7, and JADE3), in addition to NTCs, KMT2A, and DOT1L as a positive control in a plasmid vector coexpressing a blue fluorescence protein (BFP). We transduced UB3 cells using lentivirus at an ∼50% rate and sampled the populations by flow cytometry for up to 14 days. We observed a significant (P < .05, n = 3) and progressive decline in the percentage of BFP-positive (BFP+) cells targeted by all the candidate hits compared with NTCs over time (Figure 2B). In addition, we observed a significant drop in eGFP fluorescence (P < .05, n = 3) for multiple sgRNAs targeting the top candidate hits, confirming a strong reduction in MEIS1 expression (supplemental Figure 2B). These results indicate that our phenotypic screening strategy uncovered high-confidence epigenetic regulators that affect the growth of U937 cells. To rule out that this antiproliferative effect was limited to the CALM-AF10–positive cell line U937, we tested our hits in a Cas9-expressing MLL-AF9 fusion-positive MOLM13 AML cell line. CRISPR deletion of these genes also led to a progressive decline in the percentage of BFP+ cells compared with NTC-transduced cells, indicating their importance for the proliferation of MOLM13 cells (supplemental Figure 2C, upper panel). Interestingly, deletion of the uncharacterized SAGA complex genes SGF29 and ENY2 had the most detrimental effects on the growth of both U937 as well as MOLM13 cells (Figure 2B; supplemental Figure 2C), and the strong antiproliferative effect was also observed in the MV-4-11 AML cell line (supplemental Figure 2C, lower panel).
Validation and characterization of candidate MEIS1 regulators. (A) Schematic of competition assay with cells transduced with sgRNAs expressed in a BFP-positive backbone. The proportion of BFP-positive, sgRNA-transduced cells (% BFP) is assessed over time using flow cytometry and compared to BFP negative, sgRNA nontransduced cells. (B) Bar graphs indicating flow cytometry measurements of the percentage of UB3 BFP-positive cells over time (y-axis), normalized to the baseline (time 0 or T “0”) measurement (n = 3); the x-axis indicates the sgRNA number for NTCs (gray bars) or each gene (n = 2-3 sgRNAs per gene). Each bar represents a time point measurement: baseline (T0), day 8, day 11, and day 14. (C) Uniform manifold approximation (UMAP) plots of synthetic gene expression profiles from CROP-seq perturbations in U937 GFP-MEIS1 cells. (D) Dot plot comparing the expression of self-renewal–associated and differentiation-associated genes in cells with each perturbation. Dot sizes scale with the percentage of cells per gene knockout. (E) A relative score of DepMap dependencies for leukemia (n = 41) compared with nonleukemia cell lines (labeled “other”; n = 979) is shown for candidate hits identified in our screen. The y-axis is the rank of AML selectivity (with a rank of 1 being the most selective), and the x-axis shows the negative log10 P values. The dotted line separates the significant (false discovery rate [FDR] P value <.05) from the nonsignificant (FDR P value >.05). The P value was calculated using the Wilcox test of Chronos scores in leukemia compared with other cell lines. (F) A sigmoid plot of DepMap data showing the dependency score (Chronos, x-axis), compared with the normalized dependency rank (y-axis) for SGF29 in 41 leukemia cell lines in blue compared to 979 nonleukemia cell lines labeled “other” (gray).
Validation and characterization of candidate MEIS1 regulators. (A) Schematic of competition assay with cells transduced with sgRNAs expressed in a BFP-positive backbone. The proportion of BFP-positive, sgRNA-transduced cells (% BFP) is assessed over time using flow cytometry and compared to BFP negative, sgRNA nontransduced cells. (B) Bar graphs indicating flow cytometry measurements of the percentage of UB3 BFP-positive cells over time (y-axis), normalized to the baseline (time 0 or T “0”) measurement (n = 3); the x-axis indicates the sgRNA number for NTCs (gray bars) or each gene (n = 2-3 sgRNAs per gene). Each bar represents a time point measurement: baseline (T0), day 8, day 11, and day 14. (C) Uniform manifold approximation (UMAP) plots of synthetic gene expression profiles from CROP-seq perturbations in U937 GFP-MEIS1 cells. (D) Dot plot comparing the expression of self-renewal–associated and differentiation-associated genes in cells with each perturbation. Dot sizes scale with the percentage of cells per gene knockout. (E) A relative score of DepMap dependencies for leukemia (n = 41) compared with nonleukemia cell lines (labeled “other”; n = 979) is shown for candidate hits identified in our screen. The y-axis is the rank of AML selectivity (with a rank of 1 being the most selective), and the x-axis shows the negative log10 P values. The dotted line separates the significant (false discovery rate [FDR] P value <.05) from the nonsignificant (FDR P value >.05). The P value was calculated using the Wilcox test of Chronos scores in leukemia compared with other cell lines. (F) A sigmoid plot of DepMap data showing the dependency score (Chronos, x-axis), compared with the normalized dependency rank (y-axis) for SGF29 in 41 leukemia cell lines in blue compared to 979 nonleukemia cell lines labeled “other” (gray).
CRISPR droplet sequencing reveals candidate hits regulating the leukemia oncotranscriptome
Although we used MEIS1 as a reporter, our overarching goal was to identify epigenetic regulators of genes associated with AML and LSC self-renewal. Thus, we tested the effect of deleting our top hits on global transcription using single-cell RNA sequencing (RNA-seq). To test several candidates in parallel, we generated a small pooled CRISPR library targeting our top hits with 2 to 3 sgRNAs per gene, along with NTCs (a total of 29 sgRNAs). We also included sgRNAs for control genes which are either chromatin readers without HOX/MEIS-specific gene regulatory activity in U937 cells (MLLT3) or have nonselective transcriptional effects (BRD4). We cloned this library for use in CROP-seq,32 which allows for matching the single-cell RNA-seq profile of each cell to the sgRNA expressed within it, and thus infer knockout signatures. After sgRNA assignment, unbiased clustering of whole-transcriptome single-cell RNA-seq data using the Ward.D2 minimum variance method (supplemental Methods) revealed the relatedness of each of the perturbations (Figure 2C; supplemental Figure 2D). Although the transcriptomes of CSNK2A1, KMT2A, and MLLT10 knockout cells clustered together, DOT1L, MLLT1, AFF2, and SGF29 deleted transcriptomes formed a distinct cluster, indicating transcriptional similarities.
Analysis of the CROP-seq data further showed that similar to other known targets, such as DOT1L and MLLT1, deletion of novel candidates SGF29 and AFF2 showed not only MEIS1 downregulation but also decreased the expression of other credentialed leukemia oncogenes, such as HOXA13, MYC, BMI1, and SATB1 (Figure 2D; supplemental Table 4). Moreover, deletion of the novel candidate hits SGF29 and AFF2 also concomitantly increased expression of myeloid differentiation-associated genes including lysozyme (LYZ), cathepsins CTSA and CTSD, and S100P (Figure 2D). The deletion of control genes included in our CROP-seq study, namely AFF4 and the chromatin readers BRD4 and MLLT3, did not have consistent effects on the expression of these AML oncogenes or on differentiation-associated signatures. Taken together, our studies identify novel epigenetic regulators important for sustaining key LSC signature genes and repressing the expression of differentiation-associated genes.
Analysis of whole-genome CRISPR screens in cancer cell lines earmarks SGF29 as a leukemia and AML-selective dependency
One of the challenges in developing epigenetic candidates as therapeutic targets is their potential for nonselective toxicity. One way of testing the selectivity of a candidate gene is to investigate the effects of its genetic deletion in a cancer subtype of interest compared with cancers of other lineages. For this, we used the dependency maps (DepMap) data set, containing genome-scale functional genetic screens across cancer cell lines.33 We assessed the lineage specificity of the candidate genes’ essentiality in leukemia by comparing the median CRISPR knockout fitness scores (Chronos) between leukemia (n = 41) and nonleukemia (n = 979) cell lines. In this analysis, depletion of SGF29, MLLT1/ENL, and DOT1L showed exceptionally high leukemia-selective fitness defects even more selective than KMT2A and MEN1 than nonleukemia cells (Wilcox test false discovery rate q-values of 3.75E−08, 1.40E−06, and 2.25E−07, respectively; Figure 2E-F; supplemental Figure 2E; supplemental Table 5). Because leukemia cell lines in DepMap include both AML and non-AML cell lines, we also tested the specificity of SGF29 deletion effects for AML in the DepMap data set. Again, SGF29, MLLT1/ENL, and DOT1L showed exceptional AML selectivity (supplemental Figure 2E-F; supplemental Table 6). We focused our attention on SGF29 because of its strong CRISPR deletion effects on U937, MOLM13, and MV-4-11 cell lines, high leukemia and AML selectivity in the DepMap data, strong effects on AML oncotranscriptome in CROP-seq studies, and most importantly—a yet uncharacterized role in AML. Interestingly, SGF29 transcripts were expressed at significantly higher levels in leukemia and AML cell lines than all other cancer cell lines in the DepMap data. This may help in explaining why SGF29 depletion has selective antiproliferative effects in these lineages (supplemental Figure 2G).
SGF29 deletion has marked antileukemia effects in human AML cell lines
Because our transcriptomic, arrayed CRISPR validation and computational studies showed SGF29 as one of the strongest dependencies from our hits across AML cell lines, we sought to directly assess its deletion on human AML cell growth and leukemogenesis. Consistent with our flow cytometry–based in vitro competition assays, we observed that SGF29 deletion using validated CRISPR sgRNAs (supplemental Figure 3A) significantly and progressively reduced the growth of U937 and MOLM13 AML cell lines (Figure 3A-B). SGF29 deletion led to increased retention of the CellTrace Far Red dye (“Methods”) in U937 cells, confirming diminished proliferation compared with that in cells treated with NTCs (Figure 3C). Furthermore, the antiproliferative effects of SGF29 deletion were accompanied by a significant increase in the proportion of cells in the G0/G1 fraction compared with that in NTC-sg1 cells (supplemental Figure 3B) as well as a substantial increase in the proportion of annexin V–positive apoptotic cells (supplemental Figure 3C). We then wanted to assess the effects of SGF29 deletion on AML cell differentiation. SGF29 deleted U937 cells showed a significantly increased uptake of fluorescence-labeled heat-inactivated Escherichia coli bioparticles compared to NTC-expressing cells, indicative of functional myeloid differentiation (Figure 3D-E; supplemental Figure 3D) as well as a significantly higher surface expression of CD11b, a marker associated with myeloid differentiation (Figure 3F; supplemental Figure 3E). Next, we performed bulk RNA-seq of MOLM13 and U937 cells with SGF29 CRISPR knockout compared with NTC cells. Our RNA-seq analysis showed that SGF29 deletion significantly decreased the expression of key AML oncogenes with established roles in leukemic self-renewal, including the HOXA cluster genes, MYC and BMI1 but not the SGF29 binding partner KAT2A in both U937 and MOLM13 cell lines (Figure 3G-H; supplemental Figure 3F-G). SGF29 deletion concomitantly increased the expression of differentiation-associated genes, such as LYZ (Figure 3G-H; supplemental Tables 7 and 8). We confirmed key genes by quantitative real-time PCR (supplemental Figure 3H). Interestingly, retroviral overexpression of HOXA9 and MEIS1 rescued the growth-inhibitory effects of SGF29 deletion (Figure 3I). Of note, in addition to HOXA and MEIS1 genes, the MYC gene consistently showed a highly significant reduction upon SGF29 knockout in both cell lines (supplemental Figure 3F-G) and several MYC signatures were among the top downregulated gene sets in both MOLM13 and U937 cell lines by gene set enrichment analysis (Figure 3J-K; supplemental Figure 3J). Importantly, gene signatures associated with LSCs were highly enriched in the NTC compared with the SGF29-deleted fraction (Figure 3K), and gene sets downregulated in hematopoietic stem cells (HSCs), associated with myeloid differentiation, or downregulated by HOX and MYC oncoproteins were significantly upregulated in SGF29 deleted cells in both MOLM13 and U937 cell lines (supplemental Figure 3I-J). We then assessed the in vivo impact of SGF29 ablation in cell line–derived xenograft models. SGF29-deficient U937 and MOLM13 cell lines showed a significantly increased disease latency (Figure 3L-M) compared with their wild-type counterparts. These results demonstrate that SGF29 regulates the expression of key AML oncogenes, including HOX/MEIS and especially MYC-target genes, as well as leukemogenesis of human AML cell lines.
Proliferative and transcriptional effects of SGF29 loss in AML. Proliferation assays in SGF29 wild-type or CRISPR knockout AML cell lines U937 (A) or MOLM13 (B) are shown over 10 days as indicated on the x-axis (n = 3). (C) Retention of the CellTrace Far Red dye in U937 cells transduced with an SGF29 sgRNA (bottom panel) compared with NTC (top panel), measured using flow cytometry over time (representative experiment from n = 3 is shown). (D) Schematic for differentiation assessment via phagocytosis of fluorescently labeled E coli bioparticles. (E) Histograms of U937 cells transduced with an NTC (gray) or an SGF29 sgRNA (blue) showing the fluorescence intensity of phagocytosed pHrodo Red E coli bioparticles (n = 3). (F) Histograms of U937 cells transduced with an NTC (gray) or an SGF29 sgRNA (blue) showing the normalized fluorescence intensity of CD11b on the x-axis (n = 3). (G-H) Volcano plot of genes differentially changed in SGF29 deleted, compared with NTC cells in U937 (G) or MOLM13 (H) is shown with key leukemia-associated and differentiation-associated genes marked. (I) Proliferation assays of SGF29 wild-type or deficient UB3 cells with or without retroviral overexpression of HOXA9-MEIS1 are shown over a period of 10 days (n = 3). (J) Select categories of gene sets showing significantly lower enrichment in SGF29 deleted MOLM13 cells compared with wild-type counterparts using gene set enrichment analysis (GSEA) are shown in the bubble plot. Normalized enrichment score (NES) is plotted on the x-axis, and bubbles are colored by FDR-q value and sized by gene set sizes. (K) Select GSEA plots with NES and FDR-q values are shown for gene sets associated with myeloid development, MYC-targets, and LSCs. (L-M) Kaplan-Meier survival curves for NTC vs SGF29 knockout in U937 (L) and MOLM13 (M) AML cell lines injected mice are shown. Five mice per group were injected with cells expressing NTC or SGF29 sgRNA.
Proliferative and transcriptional effects of SGF29 loss in AML. Proliferation assays in SGF29 wild-type or CRISPR knockout AML cell lines U937 (A) or MOLM13 (B) are shown over 10 days as indicated on the x-axis (n = 3). (C) Retention of the CellTrace Far Red dye in U937 cells transduced with an SGF29 sgRNA (bottom panel) compared with NTC (top panel), measured using flow cytometry over time (representative experiment from n = 3 is shown). (D) Schematic for differentiation assessment via phagocytosis of fluorescently labeled E coli bioparticles. (E) Histograms of U937 cells transduced with an NTC (gray) or an SGF29 sgRNA (blue) showing the fluorescence intensity of phagocytosed pHrodo Red E coli bioparticles (n = 3). (F) Histograms of U937 cells transduced with an NTC (gray) or an SGF29 sgRNA (blue) showing the normalized fluorescence intensity of CD11b on the x-axis (n = 3). (G-H) Volcano plot of genes differentially changed in SGF29 deleted, compared with NTC cells in U937 (G) or MOLM13 (H) is shown with key leukemia-associated and differentiation-associated genes marked. (I) Proliferation assays of SGF29 wild-type or deficient UB3 cells with or without retroviral overexpression of HOXA9-MEIS1 are shown over a period of 10 days (n = 3). (J) Select categories of gene sets showing significantly lower enrichment in SGF29 deleted MOLM13 cells compared with wild-type counterparts using gene set enrichment analysis (GSEA) are shown in the bubble plot. Normalized enrichment score (NES) is plotted on the x-axis, and bubbles are colored by FDR-q value and sized by gene set sizes. (K) Select GSEA plots with NES and FDR-q values are shown for gene sets associated with myeloid development, MYC-targets, and LSCs. (L-M) Kaplan-Meier survival curves for NTC vs SGF29 knockout in U937 (L) and MOLM13 (M) AML cell lines injected mice are shown. Five mice per group were injected with cells expressing NTC or SGF29 sgRNA.
Genetic Sgf29 inactivation impairs clonogenicity of transformed but not normal hematopoietic progenitors
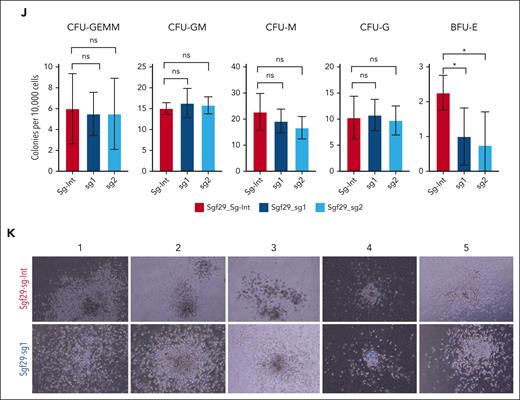
Given the role of SGF29 in the transcription of self-renewal–associated genes, we tested the effect of its inactivation on the clonogenic capability of myeloid bone marrow (BM) cells transformed by distinct AML driver oncoproteins. We performed colony-forming unit (CFU) assays on murine BM-derived HSPCs transformed with the CALM-AF10, MLL-AF10, or MLL-AF9 fusions. We observed a significant reduction in the number of colonies with a blast-like morphology in CALM-AF10 transformed cells upon Sgf29 deletion using exon-targeting sgRNAs, compared with Sgf29 intron-targeting controls (Figure 4A-C; supplemental Figure 4A-B). Interestingly, there was a dramatic decrease in the number of immature blast-like colonies but only modest effects on the differentiated colony numbers, an effect which persisted for at least 2 rounds of replating (supplemental Figure 4B). Wright-Giemsa–stained cytospins of Sgf29 knockout cells showed morphologically differentiating myeloid cells in contrast to the immature, blast-like morphology observed in Sgf29 wild-type counterparts (Figure 4C). Similar results were obtained from MLL-AF9 (Figure 4D-F; supplemental Figure 4C) and MLL-AF10 (Figure 4G-I; supplemental Figure 4D) transformed cells. Furthermore, Sgf29-deleted MLL-AF9 cells showed increase in Gr-1 as well as decrease in c-Kit expression (supplemental Figure 4E). Sgf29-deleted cells also showed increased apoptosis compared with wild-type cells, as measured using immunoblotting for cleaved poly (ADP ribose) polymerase (PARP) (supplemental Figure 4F). Next, we tested the effect of Sgf29 deletion on the colony-forming ability of normal HSPCs. In contrast to oncoprotein-transformed BM cells, CRISPR-mediated Sgf29 deletion in murine lineage-negative, Sca-1+, Kit+ (LSK) cells (supplemental Figure 8) did not significantly alter either the number or type of colonies observed in CFU assays, except for a small but significant decrease in burst-forming unit erythroid colonies (supplemental Figure 4G; Figure 4J-K). Lethally irradiated mice injected with these LSKs showed no statistically significant differences in 8-week-old BM BFP-positive cell engraftment when compared with those injected with control sgRNA-transduced LSK. Immunophenotypic analysis of the BM from the mice at 8 weeks after injection showed no differences in the contribution to the different lineages assessed using flow cytometry, except for a small but statistically significant increase in the percentage of CD11b-positive cells in the Sgf29-deleted fraction (supplemental Figure 4H).
Sgf29 deletion impairs the clonogenicity of transformed but not normal hematopoietic cells. (A) Number of CFU from CALM-AF10 transformed murine cells transduced with Sgf29 intron-targeting sgRNA (Sgf29-sg-Int) or 2 independent Sgf29 exon-targeting sgRNAs (Sgf29-sg1 & 2) are shown in the bar graphs (n = 2 replicates with 2 independent sgRNAs each). CFUs per 2000 plated cells at week 1 are plotted on the y-axis, and colonies are divided into those with a blast-like or differentiated colony morphology. (B) Pictures of representative colonies with each of the labeled sgRNA-transduced cells. Scale bar, 100 μm. (C) Wright-Giemsa–stained cytospins of representative cells from each of the CRISPR perturbations are shown. Scale bar, 10 μm. CFUs, colony morphology, and Wright-Giemsa stains are also shown for MLL-AF9 (D-F) and MLL-AF10 (G-I) transformed cells. (J) Number of CFUs per 10 000 LSK-sorted murine HSPCs expressing Cas9 and Sgf29-Intron (gray) or Sgf29-targeting (blue) sgRNAs, 10 days after seeding in methylcellulose media are shown. The y-axis shows the numbers of different types of colonies from cells expressing the indicated sgRNA and are categorized by their morphology as CFU-G (colony-forming unit-granulocyte), CFU-M (colony-forming unit-macrophage), CFU-GM (colony-forming unit-granulocyte monocyte) and CFU-Blast (blast-like colonies). (K) Representative images from each experimental condition (n = 4). Representative colonies are shown in bright field at 10× original magnification.
Sgf29 deletion impairs the clonogenicity of transformed but not normal hematopoietic cells. (A) Number of CFU from CALM-AF10 transformed murine cells transduced with Sgf29 intron-targeting sgRNA (Sgf29-sg-Int) or 2 independent Sgf29 exon-targeting sgRNAs (Sgf29-sg1 & 2) are shown in the bar graphs (n = 2 replicates with 2 independent sgRNAs each). CFUs per 2000 plated cells at week 1 are plotted on the y-axis, and colonies are divided into those with a blast-like or differentiated colony morphology. (B) Pictures of representative colonies with each of the labeled sgRNA-transduced cells. Scale bar, 100 μm. (C) Wright-Giemsa–stained cytospins of representative cells from each of the CRISPR perturbations are shown. Scale bar, 10 μm. CFUs, colony morphology, and Wright-Giemsa stains are also shown for MLL-AF9 (D-F) and MLL-AF10 (G-I) transformed cells. (J) Number of CFUs per 10 000 LSK-sorted murine HSPCs expressing Cas9 and Sgf29-Intron (gray) or Sgf29-targeting (blue) sgRNAs, 10 days after seeding in methylcellulose media are shown. The y-axis shows the numbers of different types of colonies from cells expressing the indicated sgRNA and are categorized by their morphology as CFU-G (colony-forming unit-granulocyte), CFU-M (colony-forming unit-macrophage), CFU-GM (colony-forming unit-granulocyte monocyte) and CFU-Blast (blast-like colonies). (K) Representative images from each experimental condition (n = 4). Representative colonies are shown in bright field at 10× original magnification.
The Tudor domain of SGF29 is essential for its role in myeloid transformation
The carboxy (C)-terminal tandem Tudor domain of SGF29 is important for recognizing H3K4 di/trimethylated chromatin and recruitment of KAT2A (GCN5)–containing chromatin-modifying complexes.34-36 We hypothesized that this domain may be required for the transcriptional activation of AML oncogenes. To assess the dependency on the Tudor domain for SGF29 localization, we cloned a Flag-tagged SGF29 gene and generated a stably transduced U937 cell line (supplemental Figure 5). We also generated cells expressing the SGF29D196R mutant, which disrupts H3K4me3 binding in vitro.34 Chromatin immunoprecipitation sequencing using a Flag antibody in the Flag-SGF29 U937 cells showed occupancy in regions enriched for H3K4me3-marked active promoters and H3K27-acetylated putative enhancers (Figure 5A-B; supplemental Table 9). SGF29 occupied the promoter and/or enhancer regions of HOX/MEIS and other AML oncogenes whose expression was dependent on SGF29, including BMI1 and MYC. The SGF29D196R mutant showed dramatically reduced binding to these loci (Figure 5C). Next, we tested the importance of the Tudor domain for clonogenicity of MLL-AF9 AML cells. We observed that the highly significant decrease in the immature blast-like colony numbers upon endogenous Sgf29 deletion could be rescued using retroviral overexpression of wild-type human SGF29 (impervious to the mouse Sgf29 sgRNAs). In contrast, the SGF29D196R mutant failed to rescue this defect in blast-like colony formation (Figure 5D-E). Notably, ectopic expression of the SGF29D196R mutant itself dramatically reduced the number of blast-like colonies even in MLL-AF9 cells with intact endogenous Sgf29 alleles, indicating that the SGF29D196R mutant has dominant negative activity (Figure 5D-E). These experiments highlight the importance of the Tudor domain in the leukemia-promoting activity of the SGF29 protein.
Evaluation of the activity of the SGF29 Tudor domain in AML cells. (A) Fraction of SGF29 chromatin immunoprecipitation (ChIP)-seq peaks in the promoter (black) and nonpromoter (gray) regions in U937 cells are shown in the donut plot. The number of peaks is indicated. (B) Association of SGF29 ChIP-seq peaks in promoter regions with H3K4me3 marked regions, and intergenic, intronic, and other peaks with regions marked by H3K27ac in U937 cells are shown in the bar graph as marked. The x-axis shows the number of peaks. (C) Genome tracks on the integrative genomics viewer (IGV) depicting normalized reads (y-axis) for wild-type (blue) or mutant SGF29D196R (orange) at the genomic loci of the HOXA/MEIS1, MYC, and BMI1 genes. (D) Number of CFU from MLL-AF9–transformed murine cells transduced with Sgf29 intron-targeting sgRNA (Sgf29-sg_Int) or Sgf29 exon-targeting sgRNA (Sgf29-sg1) (left panel) and the same group with wild-type SGF29 overexpression (middle panel) or SGF29 D196R mutant overexpression (right panel) are shown in the bar graphs. CFUs per 2000 plated cells at week 1 are plotted on the y-axis, and colonies are divided into those with a blast-like or differentiated colony morphology. (E) Representative images of CFUs in MLL-AF9 murine leukemias expressing: no construct (MLL-AF9, upper panel), SGF29 wild-type construct (middle panel), or SGF29D196R mutant (SGF29 mut, lower panel) and either of the indicated sgRNAs: none, Sgf29-sg_Int, or Sgf29-sg1. Representative colonies are shown in bright field at 10× original magnification.
Evaluation of the activity of the SGF29 Tudor domain in AML cells. (A) Fraction of SGF29 chromatin immunoprecipitation (ChIP)-seq peaks in the promoter (black) and nonpromoter (gray) regions in U937 cells are shown in the donut plot. The number of peaks is indicated. (B) Association of SGF29 ChIP-seq peaks in promoter regions with H3K4me3 marked regions, and intergenic, intronic, and other peaks with regions marked by H3K27ac in U937 cells are shown in the bar graph as marked. The x-axis shows the number of peaks. (C) Genome tracks on the integrative genomics viewer (IGV) depicting normalized reads (y-axis) for wild-type (blue) or mutant SGF29D196R (orange) at the genomic loci of the HOXA/MEIS1, MYC, and BMI1 genes. (D) Number of CFU from MLL-AF9–transformed murine cells transduced with Sgf29 intron-targeting sgRNA (Sgf29-sg_Int) or Sgf29 exon-targeting sgRNA (Sgf29-sg1) (left panel) and the same group with wild-type SGF29 overexpression (middle panel) or SGF29 D196R mutant overexpression (right panel) are shown in the bar graphs. CFUs per 2000 plated cells at week 1 are plotted on the y-axis, and colonies are divided into those with a blast-like or differentiated colony morphology. (E) Representative images of CFUs in MLL-AF9 murine leukemias expressing: no construct (MLL-AF9, upper panel), SGF29 wild-type construct (middle panel), or SGF29D196R mutant (SGF29 mut, lower panel) and either of the indicated sgRNAs: none, Sgf29-sg_Int, or Sgf29-sg1. Representative colonies are shown in bright field at 10× original magnification.
SGF29 regulates the chromatin localization of proteins with established roles in AML pathogenesis
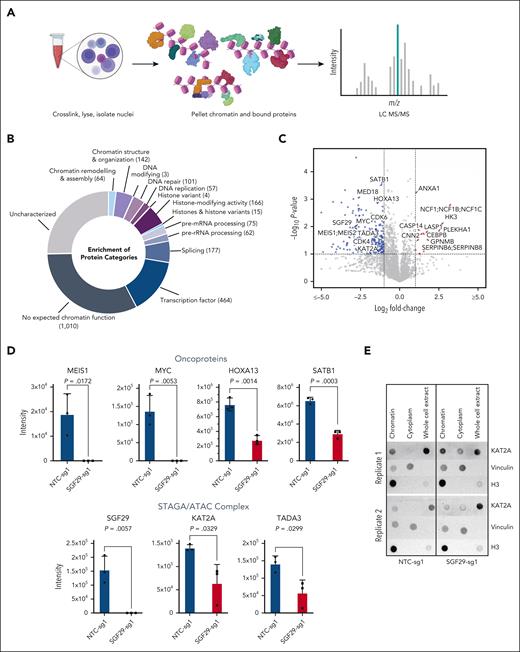
We sought to identify the molecular mechanism by which SGF29 affects the transcription of leukemia oncogenes. SGF29 participates in distinct chromatin regulatory complexes, including the SPT3-TAF9-GCN5-acetyltransferase complex (STAGA) complex, the mammalian homolog of the yeast SPT-ADA-GCN5-acetyltransferase (SAGA) complex and the ADA2A-containing (ATAC) complex. Both the SAGA and ATAC complexes harbor the KAT2A (GCN5) and KAT2B (PCAF) acetyltransferases with histone acetylating activity and have prominent roles in transcriptional activation.37 We hypothesized that SGF29 is important for tethering these epigenetic complexes to chromatin, explaining its role in the transcriptional activation of target genes. Thus, we asked which epigenetic regulators are evicted from chromatin upon SGF29 depletion in AML cells by performing chromatin enrichment proteomics (ChEP)33 (Figure 6A for schematic). This method allows an unbiased quantitative and qualitative assessment of the chromatin-associated proteome.33 Our ChEP purification yielded substantial enrichment of the chromatin fraction, as measured by the high proportional abundance of histones, histone modifying proteins, transcription factors, and other chromatin and nuclear-associated proteins in the enriched fractions (Figure 6B; supplemental Figure 6A). SGF29 deletion in U937 cells resulted in a significant decrease in the abundance of key AML oncoproteins, as measured through the intensity of chromatin-associated peptides using mass spectrometry (Figure 6C; supplemental Figure 6B). Specifically, the levels of HOXA13 and SATB1 were significantly reduced in the chromatin fraction of SGF29-deleted cells, compared with wild-type cells. We also observed a significant decrease in the abundance of CDK4 and CDK6 proteins, involved in cell-cycle regulation and known to play prominent roles in the proliferation of cancer cells, particularly in AML.38 Of note, the levels of MEIS1 and MYC were undetectable in the chromatin fraction of SGF29 deleted cells in contrast to their wild-type counterparts, in which these proteins were highly abundant (Figure 6D). Most importantly, our ChEP analysis demonstrated that SGF29 deletion significantly decreased the chromatin abundance of key STAGA complex components, including KAT2A and the transcriptional adapter protein TADA3 (Figure 6D; supplemental Figure 6B). Therefore, SGF29 deletion may lead to the eviction of the STAGA complex from the chromatin fraction. We confirmed our ChEP results using immunoblotting and observed that although KAT2A was mostly localized in the chromatin fraction of wild-type U937 cells, SGF29 deletion increased its abundance in the cytoplasmic fraction (Figure 6E; supplemental Figure 6C) without a concomitant decrease in total protein levels. Taken together, these results demonstrated that SGF29 is important for tethering key components of the STAGA complex on chromatin.
Changes in chromatin-enriched proteins upon SGF29 deletion. (A) Schematic for the ChEP sample preparation, as previously described.32 (B) A donut plot showing the functional protein categories identified in the chromatin-enriched proteome fractions. Proteins were annotated using the categories outlined in Datlinger et al.32 The number of proteins per category is shown in parentheses. (C) Volcano plot depicting −log(10) of the adjusted P value on the y-axis and log(2) of fold change (LogFC) of all proteins in the chromatin fractions for both SGF29 knockout vs NTC. Differentially abundant proteins are marked in blue for depletion and red for enrichment (absolute FC >2, adjusted P value < .05); n = 3 for every experimental condition. (D) Bar graphs depicting intensity values for ChEP-enriched key AML-associated oncoproteins (top panel) or STAGA complex members (bottom panel) in SGF29 knockout vs NTC cells (n = 3) are shown. (E) Dot blot representing 2 independent replicates for KAT2A expression in ChEP-enriched, cytoplasmic , and whole-cell fractions for NTC (left) and SGF29 knockout (right) U937 cells. Vinculin and histone H3 were included as cytoplasmic and chromatin protein controls, respectively.
Changes in chromatin-enriched proteins upon SGF29 deletion. (A) Schematic for the ChEP sample preparation, as previously described.32 (B) A donut plot showing the functional protein categories identified in the chromatin-enriched proteome fractions. Proteins were annotated using the categories outlined in Datlinger et al.32 The number of proteins per category is shown in parentheses. (C) Volcano plot depicting −log(10) of the adjusted P value on the y-axis and log(2) of fold change (LogFC) of all proteins in the chromatin fractions for both SGF29 knockout vs NTC. Differentially abundant proteins are marked in blue for depletion and red for enrichment (absolute FC >2, adjusted P value < .05); n = 3 for every experimental condition. (D) Bar graphs depicting intensity values for ChEP-enriched key AML-associated oncoproteins (top panel) or STAGA complex members (bottom panel) in SGF29 knockout vs NTC cells (n = 3) are shown. (E) Dot blot representing 2 independent replicates for KAT2A expression in ChEP-enriched, cytoplasmic , and whole-cell fractions for NTC (left) and SGF29 knockout (right) U937 cells. Vinculin and histone H3 were included as cytoplasmic and chromatin protein controls, respectively.
SGF29 deletion diminishes H3K9ac on the promoters of key leukemia oncogenes
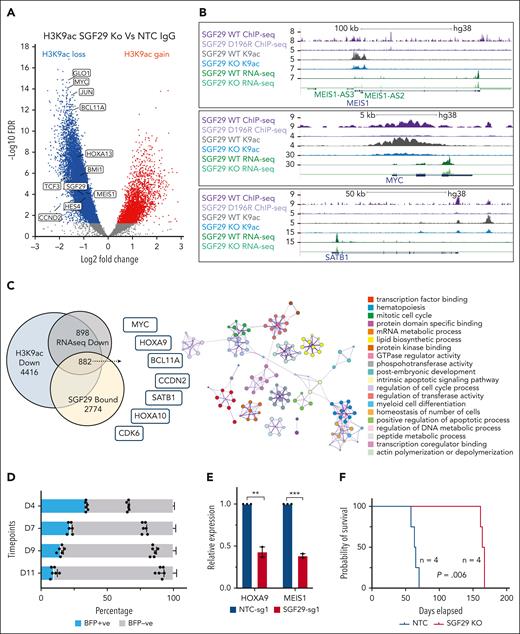
The STAGA complex member KAT2A is an acetyltransferase that plays important roles in leukemia and other cancers.37 Our results showing that SGF29 deletion led to decreased levels of KAT2A in the chromatin fraction led us to hypothesize that SGF29 recruits its activity on AML oncogene loci. We performed chromatin immunoprecipitation sequencing for H3K9ac, the major histone modification deposited by KAT2A and associated with transcriptional activation.39 SGF29 deletion led to significant changes in H3K9ac, with 10 834 peaks showing reduced acetylation and 3119 peaks showing increased acetylation in SGF29 knockout compared with wild-type U937 cells reflecting an overall decrease in global H3K9 acetylation (Figure 7A; supplemental Figures 6C and 7A; supplemental Table 10). Importantly, there was a pronounced decrease in acetylation levels at the promoters of several SGF29-bound AML oncogenes that were also transcriptionally downregulated in the SGF29 knockout RNA-seq data (Figure 7B-C). Consistent with the RNA-seq and ChEP data, the proto-oncogene MYC had one of the most profound losses of promoter-associated H3K9 acetylation (Figure 7A-C), indicating that MYC is one of the top targets of SGF29 in AML cells.
Effect of SGF29 deletion on H3K9 acetylation and antileukemia activity in human AML cells. (A) Volcano plot indicating changes in H3K9 acetylation peaks in U937 cells upon SGF29 deletion. Dots represent loci with significantly decreased (blue), increased (red), or unchanged (gray) acetylation peaks. Key SGF29-dependent AML oncogenes are labeled and (B) IGV tracks are shown for a subset of loci for SGF29 and SGF29 D196R mutant binding, H3K9ac and normalized RNA-seq transcript reads in the SGF29 wild-type and knockout (KO) conditions. (C) Venn diagram showing the overlap between SGF29 bound genes (yellow), genes downregulated upon SGF29 KO in RNA-seq (gray), and those that also lost promoter-associated H3K9ac (blue) are shown, and key AML oncogenes in the overlap are listed in the left panel. Right panel shows Metascape enrichment of gene ontology categories of the 882 genes common to all 3 sets. (D) Percentages of PDX AML-393 cells (x-axis) expressing an SGF29-targeting sgRNA (blue, BFP+) compared with untransduced cells (BFP−) over time in vitro. Progressive change in the percentage of BFP+ cells is shown at indicated time points on the y-axis. (E) Relative expression, as measured by quantitative real-time-PCR, of HOXA9 and MEIS1 in SGF29-deleted PDX AML-393 cells (y-axis) normalized to the NTC control is shown (n = 3). P values: ∗∗P < .05, ∗∗∗P < .01. (F) Kaplan-Meier survival curves for mice injected with NTC-sgRNA1 (black) or SGF29-sgRNA1 (blue) expressing PDX AML-393 (MLL-AF10 positive). Four mice were injected per group.
Effect of SGF29 deletion on H3K9 acetylation and antileukemia activity in human AML cells. (A) Volcano plot indicating changes in H3K9 acetylation peaks in U937 cells upon SGF29 deletion. Dots represent loci with significantly decreased (blue), increased (red), or unchanged (gray) acetylation peaks. Key SGF29-dependent AML oncogenes are labeled and (B) IGV tracks are shown for a subset of loci for SGF29 and SGF29 D196R mutant binding, H3K9ac and normalized RNA-seq transcript reads in the SGF29 wild-type and knockout (KO) conditions. (C) Venn diagram showing the overlap between SGF29 bound genes (yellow), genes downregulated upon SGF29 KO in RNA-seq (gray), and those that also lost promoter-associated H3K9ac (blue) are shown, and key AML oncogenes in the overlap are listed in the left panel. Right panel shows Metascape enrichment of gene ontology categories of the 882 genes common to all 3 sets. (D) Percentages of PDX AML-393 cells (x-axis) expressing an SGF29-targeting sgRNA (blue, BFP+) compared with untransduced cells (BFP−) over time in vitro. Progressive change in the percentage of BFP+ cells is shown at indicated time points on the y-axis. (E) Relative expression, as measured by quantitative real-time-PCR, of HOXA9 and MEIS1 in SGF29-deleted PDX AML-393 cells (y-axis) normalized to the NTC control is shown (n = 3). P values: ∗∗P < .05, ∗∗∗P < .01. (F) Kaplan-Meier survival curves for mice injected with NTC-sgRNA1 (black) or SGF29-sgRNA1 (blue) expressing PDX AML-393 (MLL-AF10 positive). Four mice were injected per group.
SGF29 deletion impairs in vitro and in vivo leukemogenesis in a PDX model of AML
We then assessed the effect of SGF29 CRISPR knockout in human AML patient cells. For this, we used PDX AML-393-Cas9, a MLL-AF10 fusion bearing PDX model expressing Cas9.40 These cells were generated by transduction of split Cas9 containing viral supernatant followed by sorting of Cas9 expressing GFP+ cells. In these cells, SGF29 knockout using a BFP-coexpressing sgRNA showed a strong reduction in proliferation in a competition assay compared with non-SGF29-sgRNA cells (Figure 7D). In addition, quantitative real-time-PCR of SGF29-knockout compared with NTC–transduced PDX AML-393-Cas9 cells showed a significant reduction of HOXA9 and MEIS1 transcripts (Figure 7E). We then injected PDX AML-393 cells expressing SGF29 or NTC sgRNAs into NSG mice and monitored engraftment of human Cas9-GFP+ cells in peripheral blood using flow cytometry (supplemental Figure 7B). Mice injected with NTC sgRNA–expressing cells succumbed to AML with a median latency of 64 days after injection, compared with a median of 165 days after injection with SGF29-sgRNA cells (Figure 7F). Notably, we used ribonucleoprotein-mediated delivery of these same sgRNAs into human cord blood–derived CD34-positive cells and plated the cells for CFU assays. Assessment of different individually picked colonies showed that several colonies of diverse subtypes (including CFU-granulocyte monocyte, CFU-GEMM, and burst-forming unit erythroid) showed indels in the SGF29 locus as assessed through tracking of indels by decomposition (TIDE) analysis indicating that SGF29-deleted, CD34-positive cells may still form diverse types of hematopoietic CFUs (supplemental Figure 7C).
Taken together, our results demonstrate that SGF29 is important for sustaining critical transcriptional networks in AML, for chromatin tethering of key AML-associated proteins and for leukemogenesis. Our studies nominate this chromatin reader protein and specifically the Tudor domain as an attractive therapeutic target in AML.
Discussion
In recent years, mounting evidence has shown that the expression of cancer-promoting oncogenes is sustained by specific chromatin modulators that are often essential and/or rate-limiting for oncogenesis. Thus, these epigenetic regulators may act as nononcogene dependencies presenting attractive new therapeutic targets in cancer. The chromatin reader protein SGF29 was discovered in yeast as a component of the SAGA (Spt–Ada–Gcn5 acetyltransferase) complex41 and is highly conserved across species (reviewed in42,43). SGF29 contains 2 tandem Tudor domains at the C-terminus, which specifically bind di and trimethylated H3K4 residues.34,35 In mammalian cells, SGF29 is a component of diverse chromatin-modifying complexes with overlapping subunits, of which the ATAC and the SAGA complexes are the most well-characterized and have distinct chromatin targets.34 As part of the enzymatic histone acetyltransferase (HAT) module in SAGA (along with Gcn5, Ada3, and Ada2), SGF29 allows the complex to dock to preexisting trimethyl marks at promoters and stimulates subsequent processive acetylation.44 The SAGA complex is responsible for H3K9 acetylation, a modification that fine-tunes, rather than initiates, locus-specific transcriptional activity.39 SAGA integrates multiple coactivator functions, with distinct genetic requirements for each module in gene regulation.45 Data from studies in murine MLL-AF9 leukemia suggest that ATAC is a generic requirement in cancer, whereas SAGA is selectively important in AML.38 In Saccharomyces cerevisiae, Sgf29 is required for SAGA promoter recruitment and H3K9 acetylation in vivo34 and our results show that this is also true in AML cells in which the localization of human KAT2A is controlled by SGF29. This indicates that SGF29 may be rate-limiting for the oncogenic activities of SAGA in AML46 and perhaps in other cancers in which KAT2A activity is oncogenic.47,48
Interestingly, our results also show that SGF29 may regulate transcription and chromatin abundance of several leukemia-associated transcription factors in AML cells and sustain leukemogenesis driven by diverse AML oncoproteins. We showed that SGF29 may regulate the differentiation block in AML, because its deletion promoted the expression of several differentiation-associated genes, enhanced phagocytic uptake, and led to a significant increase in differentiation in models of AML with distinct driver oncogenes. One of the strongest SGF29 targets in AML was MYC, in agreement with observations in human hepatocellular carcinoma.49 SGF29 has also recently been identified as a negative regulator of antitumor immunity in a mouse model of adenocarcinoma,50 indicating that it may have both cell-intrinsic and nonintrinsic tumor-promoting activities.
In this study, we demonstrate that a phenotypic pooled CRISPR screen based on the expression of MEIS1 revealed multiple constituents of 6 chromatin-modifying complexes as regulators of oncogenic AML transcription. Among our top hits, we encountered “writers” and “readers” known to function in concert. These results validate the robustness of the screen and importantly, point to separate nodes for therapeutic targeting within the same complex. Within chromatin-modifying complexes, targeting chromatin readers may arguably present a better therapeutic safety profile compared with chromatin-writer proteins, given that known chromatin writers, such as DOT1L, KAT2A, and KAT7 are localized to different oncogene loci by separate chromatin readers. The paradigm of inhibiting reader proteins began with bromodomain tool compounds,51 which spurred translation to clinical candidates. Other efforts for reader targeting include plant homeodomain (PHD) fingers, WD40 repeat domains, Royal family methyl-lysine readers, chromodomains, Tudor domains, PWWP domains, and the YEATS domain.52-57 Our work provides a rationale for inhibitors targeting the chromatin reading activity of the Tudor domain, which is required for the chromatin occupancy of SGF29-associated complexes, including SAGA, and transcriptional coactivation of oncogenic gene expression programs. We believe that small-molecule inhibitors of the SGF29 Tudor domain will have potent antileukemia effects and may also be effective in other cancers driven by activated expression of the HOX/MEIS or MYC oncogenes.
Acknowledgments
The authors thank Adriana Charbono and Buddy Charbono for their invaluable assistance with mouse studies and Chih-Cheng Yang and Chun-Teng Huang from the Sanford Burnham Prebys Medical Discovery Institute (SBP) functional genomics core, Yoav Altman from the SBP Flow Cytometry Core, and Kang Liu from the Genomics Core for their excellent support. The authors acknowledge the help of Philip Koeffler, Gery Sigal, Sarah Parker, and Aleks Scotland for their help with chromatin proteomics at Cedars Sinai. Some schematic figures were created using Biorender.com.
This work was supported by National Institutes of Health, National Cancer Institute grants CA262746 and P30 CA030199, the Rally Foundation for Childhood Cancer Research and the Luke Tatsu Johnson Foundation grant (award number 22IC33), an Emerging Scientist Award from the Children’s Cancer Research Fund (award number 20IC17), the V Foundation for Cancer Research under award number DVP2019-015, and the Department of Defense Horizon Award (number W81XWH-20-1-0703).
Authorship
Contribution: K.B. conceptualized the study, curated the data, performed formal analysis, was responsible for funding acquisition, conducted the investigation, developed the methodology, administered the project, created software, supervised the team, validated the research, visualized the results, and wrote the original draft; A.D. conceptualized the study, curated the data, performed formal analysis, conducted the investigation, developed the methodology, administered the project, created software, supervised the team, validated the research, visualized the results, and wrote the original draft; M.P. conducted the investigation, visualized the results, and validated the research; P.X., R.K.H., D.A., I.J., and P.M. provided resources; R.M. curated the data, performed formal analysis, developed the methodology, and created software; A.B.P. curated the data, performed formal analysis, developed the methodology, and created software; N.R., F.S., A.M., and X.L. curated the data, performed formal analysis, developed the methodology, and created software; Y.S. conducted the investigation; A.B. and J.G.D. provided resources and created software; E.R. conceptualized the study, provided resources, and created software; J.S. conceptualized the study and provided resources; P.D.A. conceptualized the study and provided resources; A.J.D. conceptualized the study, provided resources, was responsible for funding acquisition, conducted the investigation, developed the methodology, administered the project, supervised the team, visualized the results, and wrote the original draft.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Aniruddha J. Deshpande, Cancer Genome and Epigenetics Program, Sanford Burnham Prebys Medical Discovery Institute, 10901 N. Torrey Pines Rd, La Jolla, CA 92037; email: adeshpande@sbpdiscovery.org.
References
Author notes
K.B. and A.D. contributed equally to this work.
Sequencing data for RNA sequencing, ChIP sequencing, high-throughput CRISPR screens, and CROP sequencing are deposited in the NCBI Gene Expression Omnibus under accession number: GSE243935 and GSE217829. The single-guide RNA sequences used for the Human Epigenetics CRISPR Library are deposited in: https://github.com/kobarbosa/HOX-Manuscript. The scripts used in this manuscript are available at: https://github.com/kobarbosa/HOX-Manuscript.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


![Validation and characterization of candidate MEIS1 regulators. (A) Schematic of competition assay with cells transduced with sgRNAs expressed in a BFP-positive backbone. The proportion of BFP-positive, sgRNA-transduced cells (% BFP) is assessed over time using flow cytometry and compared to BFP negative, sgRNA nontransduced cells. (B) Bar graphs indicating flow cytometry measurements of the percentage of UB3 BFP-positive cells over time (y-axis), normalized to the baseline (time 0 or T “0”) measurement (n = 3); the x-axis indicates the sgRNA number for NTCs (gray bars) or each gene (n = 2-3 sgRNAs per gene). Each bar represents a time point measurement: baseline (T0), day 8, day 11, and day 14. (C) Uniform manifold approximation (UMAP) plots of synthetic gene expression profiles from CROP-seq perturbations in U937 GFP-MEIS1 cells. (D) Dot plot comparing the expression of self-renewal–associated and differentiation-associated genes in cells with each perturbation. Dot sizes scale with the percentage of cells per gene knockout. (E) A relative score of DepMap dependencies for leukemia (n = 41) compared with nonleukemia cell lines (labeled “other”; n = 979) is shown for candidate hits identified in our screen. The y-axis is the rank of AML selectivity (with a rank of 1 being the most selective), and the x-axis shows the negative log10 P values. The dotted line separates the significant (false discovery rate [FDR] P value <.05) from the nonsignificant (FDR P value >.05). The P value was calculated using the Wilcox test of Chronos scores in leukemia compared with other cell lines. (F) A sigmoid plot of DepMap data showing the dependency score (Chronos, x-axis), compared with the normalized dependency rank (y-axis) for SGF29 in 41 leukemia cell lines in blue compared to 979 nonleukemia cell lines labeled “other” (gray).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/143/8/10.1182_blood.2023021234/1/m_blood_bld-2023-021234-gr2.jpeg?Expires=1767710916&Signature=H976ecAkSeaCZbmVcTqaBq96amCATT1hiEyGDWHzYYnBGgv5BUnS4-Q6of51CXN2FJcrrvcx~lSXpYZQxu4QP2ogKPCAG9cE91YRtroDFa8zUKbzxwWgSkXOCL6YW8A4TQJG4HQLeSpNDX21x93YCmwHA8d8VLJcXgMRvjpH9T9RnxUJDTJqBQQjeSq2Qdy06UcD0qgFilqdf5A5qyJb0Gg55FoWhj2gaHKzHbXrad5sZUF2p-bKNZRZhf9VKW~Ku9kC-835ztlbw43yCvfTUc5bPVfzdET4-sykDWmge9bR9kOn~GHpue4GzQVQwiY3HmcO7yR5XtW~0ZvtZ9X2pw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal