Key Points
Response associates with higher T cell to AML ratio, whereas resistance is marked by insufficient clearing of diseased progenitor cells.
Distinct T-cell phenotypes of bone marrow and extramedullary AML are linked to differential efficacy of ipilimumab treatment.
Abstract
The challenge of eradicating leukemia in patients with acute myelogenous leukemia (AML) after initial cytoreduction has motivated modern efforts to combine synergistic active modalities including immunotherapy. Recently, the ETCTN/CTEP 10026 study tested the combination of the DNA methyltransferase inhibitor decitabine together with the immune checkpoint inhibitor ipilimumab for AML/myelodysplastic syndrome (MDS) either after allogeneic hematopoietic stem cell transplantation (HSCT) or in the HSCT-naïve setting. Integrative transcriptome-based analysis of 304 961 individual marrow-infiltrating cells for 18 of 48 subjects treated on study revealed the strong association of response with a high baseline ratio of T to AML cells. Clinical responses were predominantly driven by decitabine-induced cytoreduction. Evidence of immune activation was only apparent after ipilimumab exposure, which altered CD4+ T-cell gene expression, in line with ongoing T-cell differentiation and increased frequency of marrow-infiltrating regulatory T cells. For post-HSCT samples, relapse could be attributed to insufficient clearing of malignant clones in progenitor cell populations. In contrast to AML/MDS bone marrow, the transcriptomes of leukemia cutis samples from patients with durable remission after ipilimumab monotherapy showed evidence of increased infiltration with antigen-experienced resident memory T cells and higher expression of CTLA-4 and FOXP3. Altogether, activity of combined decitabine and ipilimumab is impacted by cellular expression states within the microenvironmental niche of leukemic cells. The inadequate elimination of leukemic progenitors mandates urgent development of novel approaches for targeting these cell populations to generate long-lasting responses. This trial was registered at www.clinicaltrials.gov as #NCT02890329.
Introduction
Despite advances in the cancer therapeutic landscape, myeloid neoplasms, ranging from myelodysplastic syndrome (MDS) to acute myelogenous leukemia (AML), have remained difficult-to-treat.1 Long-standing treatment approaches including allogeneic hematopoietic stem cell transplantation (HSCT) can achieve remissions in most patients, but relapse still occurs frequently.2 Newer agents for AML/MDS include epigenetic modifiers, Bcl-2 inhibitors and immune-checkpoint–blocking antibodies, each showing promising disease activity,3-6 suggest combinatorial approaches incorporating different pharmacologic classes may provide therapeutic synergy.7 Indeed, preclinical studies have demonstrated that the DNA methyltransferase inhibitor decitabine can increase CTLA-4 expression on AML cells,8 thereby providing a rationale for strategic partnering with the CTLA-4 blocking antibody ipilimumab.
Based on this concept, we recently reported the clinical results of the phase I ETCTN/CTEP 10026 study, which tested combined decitabine with ipilimumab both in relapsed AML following HSCT (arm A) and in transplant-naïve AML/MDS (arm B).9,10 Arm A was motivated by clinical responses in extramedullary AML post-HSCT treated with ipilimumab monotherapy (ETCTN/CTEP 9204).5,11 The objective response rates of ETCTN/CTEP 10026 revealed 5 of 25 (arm A) and 12 of 23 (arm B) patients achieving CR/CRi. This was consistent with other recent studies that reported encouraging response rates with the combination of DNA methyltransferase inhibitors and PD-1/PD-L1 inhibition,6,12-15 reinforcing interest in this approach. Nevertheless, several questions remain including the reasons for higher response rates in transplant-naïve patients and the short duration of most clinical responses, as well as delineating the precise pharmacodynamics of decitabine or ipilimumab and their contribution to therapeutic activity.16,17 Based on our observations of CD8+ T-cell recruitment to extramedullary sites in posttransplant responders to CTLA-4 blockade,11 we hypothesized that bone marrow responses would also be driven by reinvigorated T-cell responses.
Given the availability of longitudinally collected paired blood and bone marrow samples from study subjects, we had an opportunity to perform in-depth analyses of response determinants and pharmacodynamics of combined decitabine and ipilimumab. Through analysis of 304 961 single-cell transcriptomes from 64 marrow samples and 18 patients before and during therapy, we report the distinct biological characteristics of responding patients (higher baseline T-to-myeloid cell ratios) and of resistant patients (long-term persistence of malignant progenitor populations despite treatment). Decitabine and ipilimumab exposure were associated with divergent effects on leukemic and immune cell populations, including modulation of cell metabolism and increased frequency of regulatory T cells (Tregs), respectively. Our data support the notion that decitabine and ipilimumab provide an active combination for the treatment of AML/MDS and a platform for further complementary antileukemic agents.
Methods
Bulk RNA sequencing
Sequencing was performed with a capture method utilizing DNA oligonucleotide probes to enrich mRNA transcript fragments as previously described.5 RNA was extracted from decalcified formalin-fixed paraffin-embedded (FFPE) bone marrow core biopsies using the High Pure FFPET RNA Isolation Kit (Roche; catalog number 06650775001). Paired-end sequencing reads were imported and processed with a standardized RNA-seq Immune Analysis pipeline (https://liulab-dfci.github.io/RIMA). Read pairs were aligned (hg38, NCI Genomic Data Commons) by STAR118 v2.6.1d. RSeQC19 was used for quality control of aligned BAM files. Transcripts per million were quantified by SALMON20 v0.13.1. Differential expression analysis was performed by DESeq221 v1.26.0 with FDR <0.05. Gene set–enrichment analysis was processed using ClusterProfiler22 v3.14.0 against Hallmark gene sets from Molecular Signature Database (MSigDB).23 Immune cell composition was estimated by Immunedeconv24 v2.0.2.
Whole exome sequencing
After verification of matched tumor and germline samples using 95 common single nucleotide polymorphisms (SNPs) with Fluidigm genotyping, sequencing was performed at the Broad Institute as previously described.25 A matched normal for post-HSCT samples was created by merging reads from patient and donor germline samples. Reads were aligned to the GDC reference (GRCh38.d1.vd1, https://gdc.cancer.gov/about-data/gdc-data-processing/gdc-reference-files) using bwa-mem.26 Aligned reads were deduplicated and underwent base quality score recalibration. Sequenza27 was used to call CNV segments on recalibrated bam files with a window size of 50bp. Resulting CNV segments were filtered for regions >5MB and plotted using a bin size of 5MB.
Single cell RNA/T-cell receptor (TCR) sequencing
After resuspension at 1000 cells/μl, 17 000 cells per sample were loaded onto a Chromium Chip K (10× Genomics, catalog number 1000286). Single-cell gene expression was obtained using the Chromium Next GEM Single Cell 5' Kit v2 (catalog number 1000263). For single cell–T-cell receptor sequencing the V(D)J Chromium Single Cell Human TCR Amplification Kit (catalog number 1000252) was used. Library preparations were performed according to manufacturer’s instructions. After quality control with a Bioanalyzer High Sensitivity DNA Kit (Agilent), pooled libraries were sequenced on an Illumina NovaSeq 6000 with 26/28 bp read1, 90 bp read2, 10 bp for index 1, and 10 bp for index 2.28 Demultiplexing of raw sequencing reads was followed by alignment using CellRanger v6.2.0 against GRCh38-2020-A. Low quality cells were excluded from downstream analyses (RStudio with Seurat package v4.1.0.) based on percentage of mitochondrial reads (<20), features per cell (>200 and <4000), and number of reads per cell (<20 000).
Results
Baseline features predictive of response to decitabine/ipilimumab
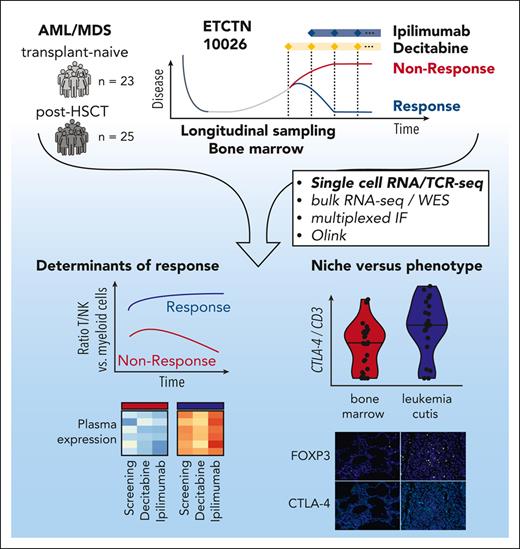
Treatment on ETCTN 10026 consisted of 1 year of planned therapy including 1 priming cycle of decitabine monotherapy (20 mg/m2 on days 1-5) followed by treatment using a combination of decitabine and ipilimumab (3-10 mg/m2 on day 1) (Figure 1A). Patients were stratified in 2 cohorts based on transplant status: arm A with morphologic AML/MDS relapse post-HSCT and arm B with transplant-naïve relapsed/refractory AML/MDS. Overall, response rates (defined as CR/CRi at any timepoint during the study) were 20% (5/25, arm A) vs 52% (12/23, arm B).10 Longitudinal sampling of peripheral blood and bone marrow was obtained at study entry (‘Screening’), following decitabine priming (end of lead-in, ∼day 30) after introduction of ipilimumab (Cycle 1, ∼day 60) and at predefined later timepoints including at the end of treatment (days 48-531).
Study overview and predictive markers of response to decitabine and ipilimumab treatment. (A) The ETCTN/CTEP 10026 study consisted of a cohort of relapsed AML after HSCT (arm A) and transplant-naïve AML/MDS (arm B) with morphologic disease. Following baseline assessment at study entry (Screening), a priming cycle of decitabine monotherapy (Lead-in) was administered. Subsequent cycles of therapy were given as a combination of decitabine and ipilimumab (C1, C2, …). Biopsies were also acquired at end of treatment. The swimmer plots indicate clinical course and duration of treatment for responders (blue) and nonresponders (red). Performed assays are indicated. (B) Median variant allele frequency of recurrent somatic mutations in bone marrow aspirates at screening grouped by response status across both arms (response [n = 16] and non-response [n = 25] according to study protocol) and study arm. Statistical testing using 2-sided t-test. (C) Changes of HSC score (van Galen et al31) throughout the study. Blue indicates responders; red indicates nonresponders. Statistical testing using Wilcoxon rank-sum test. (D,E) Differential gene-expression analysis of bone marrow core biopsies between responders (n = 6) and NRs (n = 18) across both arms according to study protocol (D) and gene set enrichment analysis (E) FDR, false discovery rate.
Study overview and predictive markers of response to decitabine and ipilimumab treatment. (A) The ETCTN/CTEP 10026 study consisted of a cohort of relapsed AML after HSCT (arm A) and transplant-naïve AML/MDS (arm B) with morphologic disease. Following baseline assessment at study entry (Screening), a priming cycle of decitabine monotherapy (Lead-in) was administered. Subsequent cycles of therapy were given as a combination of decitabine and ipilimumab (C1, C2, …). Biopsies were also acquired at end of treatment. The swimmer plots indicate clinical course and duration of treatment for responders (blue) and nonresponders (red). Performed assays are indicated. (B) Median variant allele frequency of recurrent somatic mutations in bone marrow aspirates at screening grouped by response status across both arms (response [n = 16] and non-response [n = 25] according to study protocol) and study arm. Statistical testing using 2-sided t-test. (C) Changes of HSC score (van Galen et al31) throughout the study. Blue indicates responders; red indicates nonresponders. Statistical testing using Wilcoxon rank-sum test. (D,E) Differential gene-expression analysis of bone marrow core biopsies between responders (n = 6) and NRs (n = 18) across both arms according to study protocol (D) and gene set enrichment analysis (E) FDR, false discovery rate.
To identify predictive baseline characteristics of leukemic cells, we evaluated the distribution of somatic mutations with amplicon sequencing (supplemental Figure 1A-B; available on Blood website). Responders had lower variant allele frequencies of recurrent AML/MDS-associated somatic mutations (P < .05) (Figure 1B), consistent with lower disease burden at study entry. Recent studies have linked more primitive stem cell signatures to poorer clinical outcomes.29,30 We thus calculated a hematopoietic stem cell (HSC) gene score from bulk RNA sequencing (RNA-seq) based on published data of leukemia-associated genes,31 which was highly concordant with blast counts obtained from clinical routine diagnostics (r = 0.53) (supplemental Figure 1C). Although responders did not have a lower HSC score at study entry (P = .31), they saw greater reduction after combined treatment with decitabine and ipilimumab (P = .019; Figure 1C). Differential gene expression analysis of screening samples showed upregulation of pathways associated with cell proliferation in non-responders (NRs), consistent with higher disease activity (Figure 1D-E; supplemental Table 1).
Single cell map of longitudinally collected bone marrow-derived cell populations
To define differences in cellular composition of marrow-infiltrating cells of responders and nonresponders, we performed single-cell RNA sequencing (scRNA-seq) on 64 samples from 18 patients with AML/MDS, yielding a total of 304 961 cells (supplemental Table 2). To ensure consistent annotation of immune cell types, we mapped cells to a healthy human bone marrow reference (Methods, Figure 2A, top).32 Compared with 8 healthy bone marrow samples, screening samples demonstrated a left-shifted myeloid compartment consistent with AML (Figure 2B-C). The cell annotation did not allow immediate distinction of malignant from non-malignant myeloid cells, nor did subclustering of myeloid cells. Nonetheless, subclustered myeloid cell types segregated into the annotated myeloid cell types of the reference map (Figure 2A, inset top), which was highly concordant with previously reported AML subtypes (supplemental Figure 1D).31 Further differences in clustering were driven by interindividual heterogeneity (Figure 2A, inset bottom).
Single cell bone marrow map of AML. (A) Uniform manifold approximation and projection (UMAP) embedding of single cell transcriptomic profiles of 64 AML bone marrow samples of 18 AML patients (2-5 samples per patient) (refer to supplemental Figure 2-3 for details). The annotated cell types are indicated by the color code. The inset UMAP embeddings show the subclustered myeloid cell compartment at screening colored by annotated cell types (top) and individual patients (bottom). (B-C) Distribution of cell types in screening samples of AML and healthy bone marrow. Differential abundance of myeloid cell types between AML and healthy donors (HD) (C). Statistical testing using Wilcoxon rank-sum test. (D) Genetic deconvolution of 145,595 donor (29% total) and recipient-derived (71%) single cells determines donor chimerism of individual cell types across baseline samples posttransplant (arm A). The bars above the heatmap indicate the total number of donor-derived cells across all myeloid subsets per study subject. ∗Sample at Lead-in. (E) Copy number changes across myeloid cell types identified through inferCNV in AML1012 and AML1016. (F) Comparison of distribution of T/NK cell subsets in AML (n = 18) vs healthy donor (n = 8) bone marrow. Statistical testing using Wilcoxon rank-sum test. (G) Comparison of distribution of T/NK cell subsets in AML posttransplant vs AML/MDS transplant-naïve bone marrow. (H) Clonal T-cell expansion (fraction of T cells sharing the same TCR) across CD4+ and CD8+ T cell subsets with most clonal expansion in effector and memory CD8+ T cells.
Single cell bone marrow map of AML. (A) Uniform manifold approximation and projection (UMAP) embedding of single cell transcriptomic profiles of 64 AML bone marrow samples of 18 AML patients (2-5 samples per patient) (refer to supplemental Figure 2-3 for details). The annotated cell types are indicated by the color code. The inset UMAP embeddings show the subclustered myeloid cell compartment at screening colored by annotated cell types (top) and individual patients (bottom). (B-C) Distribution of cell types in screening samples of AML and healthy bone marrow. Differential abundance of myeloid cell types between AML and healthy donors (HD) (C). Statistical testing using Wilcoxon rank-sum test. (D) Genetic deconvolution of 145,595 donor (29% total) and recipient-derived (71%) single cells determines donor chimerism of individual cell types across baseline samples posttransplant (arm A). The bars above the heatmap indicate the total number of donor-derived cells across all myeloid subsets per study subject. ∗Sample at Lead-in. (E) Copy number changes across myeloid cell types identified through inferCNV in AML1012 and AML1016. (F) Comparison of distribution of T/NK cell subsets in AML (n = 18) vs healthy donor (n = 8) bone marrow. Statistical testing using Wilcoxon rank-sum test. (G) Comparison of distribution of T/NK cell subsets in AML posttransplant vs AML/MDS transplant-naïve bone marrow. (H) Clonal T-cell expansion (fraction of T cells sharing the same TCR) across CD4+ and CD8+ T cell subsets with most clonal expansion in effector and memory CD8+ T cells.
Donor and recipient SNPs provided us with an opportunity to definitively distinguish malignant (recipient) from normal (donor) myeloid cells in cohort A (total 145 595 cells).33 By applying souporcell34 (Figure 2D, left) on 62 673 myeloid cells from the post-HSCT baseline time point from 8 patients (median 296 days post-HSCT, range 151-1290), we identified 1422 cells (2.3%) as donor-derived. Donor-derived myeloid cells were present at higher percentage in responders (7% to 73%) and consisted predominantly of differentiated monocytes and dendritic cells, thereby linking higher myeloid engraftment with improved response (Figure 2D, right). NRs had <7% detectable donor myeloid cells, representing minimal normal myelopoiesis, which was supported by an analysis of copy number aberrations (inferCNV)35, demonstrating that most progenitor myeloid cells harbored malignancy-associated genetic markers. For example, in AML1012, del(5) was detectable across all myeloid subsets and in AML1016 del(5q) was detectable in HSC and LMPP cells but not in exclusively donor-derived CD16+ monocytes (Figure 2E). Likewise, the transplant-naïve responders AML1002 and AML8007 had detectable del(7q) or del(3p) across almost all progenitor cells at the time of screening (supplemental Figure 2A-E).
The distribution of T-cell populations among patients was markedly skewed: healthy donor bone marrow had more naïve CD4+ and CD8+ T cells whereas memory CD4+ T cells, natural killer (NK) cells, and Tregs were more abundant in AML/MDS (Figure 2F). Although the distribution of T/NK cell types at screening was not predictive of response (supplemental Figure 2F), post-HSCT and transplant-naïve cases showed subtle differences, including lower numbers of naïve and memory CD4+ and effector CD8 T cells posttransplant (P < .05; Figure 2G). Analyses of clonal T-cell expansion (fraction of T cells sharing the same TCR) for post-HSCT and transplant-naïve patients revealed effector and memory CD8+ T cells to have the highest levels of TCR skewing, whereas CD4+ and naïve T cells had lower levels, consistent with previous reports36-40 (Figure 2H). Altogether, both the myeloid and the immune cell compartment in bone marrow of participants from the ETCTN 10026 study was highly altered compared with that of healthy donors.
Stably higher T cell-to-myeloid ratio in responders
To address whether CTLA-4 blockade reshaped T-cell clonality, we investigated dynamics of T-cell chimerism and TCR repertoire. Single-cell donor chimerism for T/NK subsets was lowest in CD4+ and naïve CD8+ T cells and highest in NK cells, reflecting known kinetics of T-cell reconstitution posttransplant.41 The lower engraftment of CD4+ and CD8+ T cells remained unaffected by ipilimumab treatment (Figure 3A). Comparison of clonal T-cell expansion in both posttransplant and transplant-naïve patients at screening and after decitabine priming or ipilimumab revealed only minimal changes (167/29 268 [0.6%] and 250/38 678 clones [0.7%]), which were equivalently distributed among responders and NRs (Figure 3B), indicating overall stability of the TCR repertoire throughout treatment. These results, within the confines of the shallow TCR coverage provided by scTCR-seq, imply that most detected BM-associated T-cell clones did not mediate clinical activity of ipilimumab.
Stability of T cell compartment during combined decitabine and ipilimumab treatment. (A) Donor chimerism across T/NK cell subsets (left) and longitudinal chimerism of CD4+, CD8+ and NK cells throughout treatment (right) for 8 posttransplant patients (arm A). Statistical testing using Wilcoxon rank-sum test. Shaded areas indicate 95% confidence interval. (B) Changes in TCR repertoire in transplant-naïve and posttransplant samples after decitabine (left) and ipilimumab treatment (right). Break-down by responders (Rs) and NRs shown in inset. Statistical testing using Fisher exact test and FDR correction for individual T-cell clones and Wilcoxon rank-sum test between responders and NRs (FDR < 0.05). (C) Ratio of CD8+ T cell to myeloid cell infiltration in bone marrow core biopsies estimated using quanTIseq from available bulk RNA-seq data of patients across both study arms. (D) Percentage of myeloid cells in screening bone marrow samples of NRs (n = 10) and responders (n = 8) using scRNA-seq data (left). T/NK cell to myeloid ratio for the same patients detailed throughout treatment (right). (E) Differential expression analysis of screening bone marrow plasma profiles between responders (n = 13) and NRs (n = 25) across both study arms. Statistical testing using Wilcoxon rank-sum test and FDR correction for multiple hypothesis testing (FDR < 0.05). (F) Principal component analysis of bone marrow plasma profiles from all available samples (n = 185) colored by clinical response (top) and study arm (bottom). (G) Expression of proteins associated with clinical response throughout treatment in responders and NRs. (H) Decrease in the number of cell-cell interactions detected using CellPhoneDB from scRNA-seq data across different cell subsets following decitabine and ipilimumab in responders (n = 8) compared with NRs (n = 10) and healthy donors (n = 8).
Stability of T cell compartment during combined decitabine and ipilimumab treatment. (A) Donor chimerism across T/NK cell subsets (left) and longitudinal chimerism of CD4+, CD8+ and NK cells throughout treatment (right) for 8 posttransplant patients (arm A). Statistical testing using Wilcoxon rank-sum test. Shaded areas indicate 95% confidence interval. (B) Changes in TCR repertoire in transplant-naïve and posttransplant samples after decitabine (left) and ipilimumab treatment (right). Break-down by responders (Rs) and NRs shown in inset. Statistical testing using Fisher exact test and FDR correction for individual T-cell clones and Wilcoxon rank-sum test between responders and NRs (FDR < 0.05). (C) Ratio of CD8+ T cell to myeloid cell infiltration in bone marrow core biopsies estimated using quanTIseq from available bulk RNA-seq data of patients across both study arms. (D) Percentage of myeloid cells in screening bone marrow samples of NRs (n = 10) and responders (n = 8) using scRNA-seq data (left). T/NK cell to myeloid ratio for the same patients detailed throughout treatment (right). (E) Differential expression analysis of screening bone marrow plasma profiles between responders (n = 13) and NRs (n = 25) across both study arms. Statistical testing using Wilcoxon rank-sum test and FDR correction for multiple hypothesis testing (FDR < 0.05). (F) Principal component analysis of bone marrow plasma profiles from all available samples (n = 185) colored by clinical response (top) and study arm (bottom). (G) Expression of proteins associated with clinical response throughout treatment in responders and NRs. (H) Decrease in the number of cell-cell interactions detected using CellPhoneDB from scRNA-seq data across different cell subsets following decitabine and ipilimumab in responders (n = 8) compared with NRs (n = 10) and healthy donors (n = 8).
To define dynamics of immune cell subsets after therapy, we deconvoluted the bulk RNA-seq data with quanTIseq24 which estimates T-cell and myeloid fractions with reasonable correlation to scRNA-seq data from the same biopsies (supplemental Figure 3A-B). This revealed an increased ratio of T cells relative to myeloid cells in responders (Figure 3C). We confirmed these results with scRNA-seq data, in which we observed increased T cell-to-myeloid ratios in responders that persisted throughout the treatment (P = .027) (Figure 3D).
Given potential crosstalk between leukemic and immune cells, we evaluated soluble factors associated with response to therapy. Plasma abundance from 13 responders and 25 NRs revealed strikingly similar expression levels of most analytes between peripheral blood and bone marrow (supplemental Figure 3C). Patients who underwent a transplant had higher expression of CXCL9, CXCL10, and PD-1 that have been associated with chronic graft-versus-host disease42-44 (supplemental Figure 3D-E). Responders of both study arms had higher circulating expression of CCL17, CXCL1, CXCL5, EGF, LAMP3, and PDGF subunit B at screening and after initiation of treatment (Figure 3E-G), in agreement with findings from our previous study of ipilimumab monotherapy (ETCTN/CTEP 9204).5,11 Expression of 4 of these cytokines (CCL17, CXCL5, EGF, and PDGF subunit B) negatively correlated with blast counts, suggesting reduced production or depletion by malignant cell populations (supplemental Figure 4A-B). Responding study subjects thus entered the study with lower disease burden than NR and displayed distinct cytokine expression profiles.
To explore how combined decitabine and ipilimumab treatment could affect interactions within the bone marrow microenvironment, we calculated predicted interactions for each cell type (CellphoneDB).45 Although no differences in the number of interactions were detected between responders and NRs at screening, this number remained stable in NRs but decreased in responders during treatment to a level similar to that in bone marrow of healthy donors, suggesting that effective therapy led to a normalization of interactions between AML and the immune cell microenvironment (Figure 3H).
Decitabine and ipilimumab preferentially act on myeloid and CD4+ T cells, respectively
The pharmacodynamics of decitabine has been intensely studied, including its rate-limiting enzyme deoxycytidine kinase (DCK), the inhibition of DNMT1 and subsequent DNA hypomethylation.16,46 The pyrimidine metabolism enzyme cytidine deaminase (CDA) inhibits decitabine activity through degradation of intermediate metabolites and has been implicated in resistance to decitabine.47 Our single cell transcriptomic profiles revealed DCK as expressed by HSC, LMPP, and dendritic cells, whereas DNMT1 expression was minimal in HSC but high in LMPP, GMP, and progenitor dendritic cells. Expression of CDA was undetectable in most cell types; differences between responders and NRs were not detectable for all 3 genes (supplemental Figure 5A-C). The high expression of DNMT1 in GMP and LMPP was consistent with their high proliferative activity and was preserved throughout treatment (supplemental Figure 5D-E). Although hypomethylating agents have been implicated in upregulation of major histocompatibility complex class I and II expression,48,49 such changes at transcript level across myeloid subsets after decitabine treatment were not detected in our dataset (supplemental Figure 5F-G).
From unbiased analyses of the bulk RNA-seq data, we detected global transcriptional changes after a single cycle of decitabine (supplemental Figure 6A). By scRNA-seq, the most differentially expressed genes were detected in myeloid populations and were implicated in basic cellular mechanisms such as protein translation, metabolism, and apoptosis (Figure 4A-B). Consistent with gene expression changes in myeloid cells, soluble IL-8 (encoded by CXCL8) was upregulated, and IL-12 was downregulated in blood and bone marrow plasma (Figure 4C; supplemental Figure 6B-D). CXCL8 was mainly expressed by CD14+ monocytes and in myeloid progenitor cell types (Figure 4D). IL-12 is known to be secreted by myeloid cell populations.50
Pharmacodynamics of decitabine and ipilimumab. (A) Number of differentially expressed genes across cell types between before decitabine treatment (Screening) and after one cycle of decitabine (Decitabine, Lead-in) from scRNA-seq dataset shows predominantly myeloid-specific effect of decitabine (pink) (Log2FC > 0.25, −log10FDR > 10). (B) Gene set enrichment analysis of differentially expressed genes in myeloid cells. (C) Soluble IL8 in peripheral blood plasma quantified using Olink assay throughout treatment. Statistical testing using Wilcoxon rank-sum test. NPX – Normalized protein expression. (D) Mean gene expression of CXCL8 (encoding IL8) across cell subsets shows preferential expression in CD14+ monocytes and other myeloid cells, while expression is absent in T and NK cells. (E) Number of differentially expressed genes across cell types after 1 cycle of decitabine (Decitabine) and after combined treatment of decitabine and ipilimumab (Ipilimumab) shows ipilimumab-specific effect on CD4+ T cells (blue box) (log2FC > 0.25, −log10FDR > 10). (F) Gene set enrichment analysis of differentially expressed genes in CD4+ T cells. (G) Soluble CXCL13 in peripheral blood plasma throughout treatment quantified using Olink assay. Statistical testing using Wilcoxon rank-sum test. (H-I) Percentage of Tregs in bone marrow aspirates measured using single cell sequencing (H) and multiplexed immunofluorescence (I) shows ipilimumab-induced increase of Tregs. Data from ETCTN 9204 were obtained before (Pre-Ipi) and after (Post-Ipi) ipilimumab monotherapy. Tissue biopsies from ETCTN 9204 were obtained from bone marrow (n = 17) and extramedullary AML sites (skin n = 3; breast n = 2; soft tissue n = 1), while tissue biopsies from ETCTN 10026 were exclusively bone marrow (n = 36). Statistical testing using Wilcoxon rank-sum test. (J) Validation of ipilimumab-induced increase in Tregs (CD3+ FOXP3+) in tissue biopsies from ETCTN/CTEP 10026 study using multiplexed immunofluorescence staining at screening, after 1 cycle of decitabine monotherapy (Decitabine, Lead-in) and after combination treatment (Ipilimumab). NPX, normalized protein expression.
Pharmacodynamics of decitabine and ipilimumab. (A) Number of differentially expressed genes across cell types between before decitabine treatment (Screening) and after one cycle of decitabine (Decitabine, Lead-in) from scRNA-seq dataset shows predominantly myeloid-specific effect of decitabine (pink) (Log2FC > 0.25, −log10FDR > 10). (B) Gene set enrichment analysis of differentially expressed genes in myeloid cells. (C) Soluble IL8 in peripheral blood plasma quantified using Olink assay throughout treatment. Statistical testing using Wilcoxon rank-sum test. NPX – Normalized protein expression. (D) Mean gene expression of CXCL8 (encoding IL8) across cell subsets shows preferential expression in CD14+ monocytes and other myeloid cells, while expression is absent in T and NK cells. (E) Number of differentially expressed genes across cell types after 1 cycle of decitabine (Decitabine) and after combined treatment of decitabine and ipilimumab (Ipilimumab) shows ipilimumab-specific effect on CD4+ T cells (blue box) (log2FC > 0.25, −log10FDR > 10). (F) Gene set enrichment analysis of differentially expressed genes in CD4+ T cells. (G) Soluble CXCL13 in peripheral blood plasma throughout treatment quantified using Olink assay. Statistical testing using Wilcoxon rank-sum test. (H-I) Percentage of Tregs in bone marrow aspirates measured using single cell sequencing (H) and multiplexed immunofluorescence (I) shows ipilimumab-induced increase of Tregs. Data from ETCTN 9204 were obtained before (Pre-Ipi) and after (Post-Ipi) ipilimumab monotherapy. Tissue biopsies from ETCTN 9204 were obtained from bone marrow (n = 17) and extramedullary AML sites (skin n = 3; breast n = 2; soft tissue n = 1), while tissue biopsies from ETCTN 10026 were exclusively bone marrow (n = 36). Statistical testing using Wilcoxon rank-sum test. (J) Validation of ipilimumab-induced increase in Tregs (CD3+ FOXP3+) in tissue biopsies from ETCTN/CTEP 10026 study using multiplexed immunofluorescence staining at screening, after 1 cycle of decitabine monotherapy (Decitabine, Lead-in) and after combination treatment (Ipilimumab). NPX, normalized protein expression.
To evaluate the pharmacodynamics of ipilimumab, we compared gene expression changes after 1 cycle of decitabine with the next available sample following combined decitabine and ipilimumab. We detected differentially expressed genes in CD4+ T cells enriched for pathways involved in T-cell differentiation, activation, and adhesion (Figure 4E-F), consistent with known effects of ipilimumab.51 Addition of ipilimumab led to increased circulating CD27 and CXCL13 (Figure 4G; supplemental Figure 6E), which is produced by T follicular helper cells and has been linked to immunosuppressive effects of Tregs.52,53 Indeed, scRNA-seq revealed an increase in marrow-infiltrating Tregs after infusion of ipilimumab (Treg-specific markers shown in supplemental Figure 7). We used multiplexed immunofluorescence (mIF) to validate this finding on tissue biopsies collected before and after ipilimumab, including from the ETCTN/CTEP 9204 study.11 We indeed detected increased density of CD3+ FOXP3+ cells, a phenotype associated with increased T-cell activation and infiltration by Tregs (Figure 4H-J; supplemental Figure 6F). Altogether, the effects of decitabine and ipilimumab were cell type–specific, with decitabine preferentially acting on myeloid cells and ipilimumab acting on CD4+ T cells.
Longitudinal persistence of malignant cell clones
Despite encouraging response rates of decitabine and ipilimumab in the ETCTN/CTEP 10026 study, most remissions only lasted from a few weeks to a few months.9 To determine whether treatment would lead to emergence of resistant AML subclones, we evaluated the genetic profiles of leukemic cells during therapy. Most somatic mutations remained detectable at the time of response (Figure 5A; supplemental Figure 8A-B). Similarly, cancer-cell fractions and copy number changes calculated from whole-exome sequencing data showed overall stability irrespective of response status (Figure 5B; supplemental Figure 9A-C). In 2 cases (AML1016 [nonresponder] and AML1019 [responder]), we tracked AML subclones at single-cell resolution based on distinct copy-number changes and gene-expression differences. In agreement with the genetic analyses, both AMLs displayed subclonal stability (Figure 5C; supplemental Figure 9D-E).
Longitudinal tracking of malignant cell clones reveals insufficient clearing from progenitor cell populations. (A) Median variant allele frequency of recurrent somatic mutations in bone marrow aspirates of responders (blue) and NR (red) at screening, after 4 cycles of decitabine and ipilimumab (C4) and at end of treatment. Statistical testing using Wilcoxon rank-sum test. (B) Cancer cell fractions calculated from whole exome sequencing of bone marrow aspirates at screening and C4 in AML1007, AML1008 and AML1019 (all responders). (C) Identification of 2 distinct AML subclones defined by amp(8) in pink and amp(21) in cyan using inferCNV. Longitudinal tracking of both clones within the HSC-like compartment (right). (D) Longitudinal tracking of donor chimerism across myeloid cell subsets of responders (blue) and NRs (red). Disease burden (% blasts) obtained from routine clinical diagnostics with 5% mark indicated by horizontal bar. Grey indicates no data available. (E) Longitudinal detection of copy number changes across myeloid subsets. Grey indicates no data available or too few cells to perform analysis of copy number changes.
Longitudinal tracking of malignant cell clones reveals insufficient clearing from progenitor cell populations. (A) Median variant allele frequency of recurrent somatic mutations in bone marrow aspirates of responders (blue) and NR (red) at screening, after 4 cycles of decitabine and ipilimumab (C4) and at end of treatment. Statistical testing using Wilcoxon rank-sum test. (B) Cancer cell fractions calculated from whole exome sequencing of bone marrow aspirates at screening and C4 in AML1007, AML1008 and AML1019 (all responders). (C) Identification of 2 distinct AML subclones defined by amp(8) in pink and amp(21) in cyan using inferCNV. Longitudinal tracking of both clones within the HSC-like compartment (right). (D) Longitudinal tracking of donor chimerism across myeloid cell subsets of responders (blue) and NRs (red). Disease burden (% blasts) obtained from routine clinical diagnostics with 5% mark indicated by horizontal bar. Grey indicates no data available. (E) Longitudinal detection of copy number changes across myeloid subsets. Grey indicates no data available or too few cells to perform analysis of copy number changes.
For post-HSCT samples, we used chimerism measurements to track response dynamics. We did not detect conversion to donor chimerism in HSC or LMPP cells in any of the patients, reflecting the observations of disease stability from targeted and whole-exome sequencing. Some differentiated myeloid cells (GMP and CD14+ monocytes) showed transient increase of donor chimerism (AML1019 and AML1026), however all NRs had near-complete absent donor myelopoiesis (Figure 5D). Similarly, copy number changes continued to be detectable in progenitor cell populations, including transplant-naïve responders AML1002 and AML8007 (Figure 5E). These results demonstrate that combined treatment of decitabine and ipilimumab was cytoreductive but unable to restore physiologic myelopoiesis.
Low frequency of exhausted T cells in AML bone marrow suggests low immune pressure
Given the overall stable T-cell compartment after CTLA-4 blockade, we wondered whether phenotypes of CD8+ T cells in the bone marrow of AML differed from tumor-infiltrating T cells (TILs) in solid tumor malignancies, in which ipilimumab monotherapy has displayed higher clinical activity. We calculated a memory and an exhaustion gene score associated with tumor-specific reactivity for CD8+ T cells from our data set. Compared with TILs from basal cell carcinoma, bladder cancer, and metastatic melanoma, recently characterized by single-cell transcriptome sequencing,36,54-56 the single cell profiles of T cells from healthy and leukemic human bone marrow57,58 demonstrated dramatic differences. Only few (0.9%) CD8+ T cells from healthy bone marrow had a high exhaustion score, and this was likewise low in people with AML (2%) and chronic myeloid leukemia (2.8%) (Figure 6A). In contrast, 5.8% to 24% CD8+ tumor-infiltrating T cells had an exhausted phenotype. Further, metastatic melanoma cases had high expression of CTLA-4, PDCD1 (encoding PD-1), and ENTPD1 (CD39) on exhausted T cells, whereas in AML bone marrow CTLA-4 and ENTPD1 were mainly expressed by Tregs, and PDCD1 by a few effector memory CD8+ T cells (Figure 6B). Nevertheless, CD8+ T cells in AML demonstrated TIGIT and LAG3 expression (supplemental Figure 10A-B). CD4+ T cells also had higher exhaustion gene scores in solid tumors than bone marrow (supplemental Figure 11A-B). In contrast to a chronic myeloid leukemia dataset where shifts in T-cell exhaustion after weekly-dosed donor-lymphocyte infusion (DLI) were detectable,58 demonstrating that such dynamics are in principle detectable, no changes were found in marrow-infiltrating T cells from AML subjects after decitabine and ipilimumab treatment (supplemental Figure 12A-B). These analyses show that AML bone marrow is characterized by comparatively low levels of T-cell exhaustion, which were likely insufficient for any measurable ipilimumab-induced modulation.
Comparative analysis of T-cell exhaustion in hematologic and solid malignancies. (A) T-cell exhaustion and memory scores calculated for CD8+ T cells from healthy bone marrow, AML and chronic myeloid leukemia bone marrow and different solid tumors including basal cell carcinoma, bladder cancer and metastatic melanoma. (B) Expression of CTLA-4, PDCD1 (encoding PD-1) and ENTPD1 (encoding CD39) across T-cell subsets in AML bone marrow (top, n = 18 patients) and in metastatic melanoma (bottom, n = 4 patients; Oliveira et al36). (C) Bulk RNA sequencing (RNA-seq) expression of ZNF683 and KLRB1 across 29 AML/MDS bone marrow biopsies with disease involvement and 21 extramedullary AML (eAML) biopsies from patients with sole extramedullary relapse. (D) Ratio of CTLA-4 and KLRB1 to CD3 expression obtained from bulk RNA-seq data across AML/MDS and eAML. Statistical testing using 2-sided t-test. (E) Percentage of CD3+ T-cell infiltrate (left), CD103+ T cells (middle) and FOXP3+ CTLA-4+ T cells (right) compared between bone marrow (BM; n = 10) and leukemia cutis (eAML; n = 10) before treatment. The exceptional responder with ongoing complete remission >3 years after treatment with decitabine and ipilimumab is indicated by the blue arrow. Statistical testing with 2-sided t-test. Medians are indicated for each group by the horizontal bar. (F) Representative single stains of CD3 (white), CD103 (red), CTLA-4 (cyan) and FOXP3 (yellow) for bone marrow (BM) and leukemia cutis (eAML) (top). Integrated staining of CD3, FOXP3 and CTLA-4 is shown for BM and eAML (bottom). The images were captured with 20× optical magnification and 250% zoom. Yellow bars indicate a distance of 50 μm.
Comparative analysis of T-cell exhaustion in hematologic and solid malignancies. (A) T-cell exhaustion and memory scores calculated for CD8+ T cells from healthy bone marrow, AML and chronic myeloid leukemia bone marrow and different solid tumors including basal cell carcinoma, bladder cancer and metastatic melanoma. (B) Expression of CTLA-4, PDCD1 (encoding PD-1) and ENTPD1 (encoding CD39) across T-cell subsets in AML bone marrow (top, n = 18 patients) and in metastatic melanoma (bottom, n = 4 patients; Oliveira et al36). (C) Bulk RNA sequencing (RNA-seq) expression of ZNF683 and KLRB1 across 29 AML/MDS bone marrow biopsies with disease involvement and 21 extramedullary AML (eAML) biopsies from patients with sole extramedullary relapse. (D) Ratio of CTLA-4 and KLRB1 to CD3 expression obtained from bulk RNA-seq data across AML/MDS and eAML. Statistical testing using 2-sided t-test. (E) Percentage of CD3+ T-cell infiltrate (left), CD103+ T cells (middle) and FOXP3+ CTLA-4+ T cells (right) compared between bone marrow (BM; n = 10) and leukemia cutis (eAML; n = 10) before treatment. The exceptional responder with ongoing complete remission >3 years after treatment with decitabine and ipilimumab is indicated by the blue arrow. Statistical testing with 2-sided t-test. Medians are indicated for each group by the horizontal bar. (F) Representative single stains of CD3 (white), CD103 (red), CTLA-4 (cyan) and FOXP3 (yellow) for bone marrow (BM) and leukemia cutis (eAML) (top). Integrated staining of CD3, FOXP3 and CTLA-4 is shown for BM and eAML (bottom). The images were captured with 20× optical magnification and 250% zoom. Yellow bars indicate a distance of 50 μm.
We previously reported that ipilimumab monotherapy (ETCTN/CTEP 92045) led to durable remissions in cases of sole leukemia cutis. We compared previously acquired bulk RNA-seq data from 21 FFPE biopsies of extramedullary sites with expression profiles from 29 AML/MDS bone marrow samples (all ETCTN 9204).11 Bone marrow samples segregated from eAML based on expression of KLRB1, mainly found in MAIT, NK, and γδ T cells, and ZNF683, a marker of tissue-resident T cells (Figure 6C; supplemental Figure 12C-E). Thus, while extramedullary sites harbored more antigen-experienced tissue-resident memory T cells, bone marrow was infiltrated by innate T/NK cells. Further, ratios of FOXP3, CTLA-4, HAVCR2 (encoding Tim-3), and PDCD1 to CD3 expression in eAML were higher than in bone marrow, suggesting a higher infiltration with Tregs and potential susceptibility to ICB (Figure 6D; supplemental Figure 12F).
We validated these findings using mIF staining on a series of bone marrow and leukemia cutis cases (both n = 10), which revealed that although eAML had similar amount of overall T-cell infiltration, the fraction of CD103+ (tissue-residency), FOXP3+, and CTLA-4+ T cells was markedly higher (P < .05; Figure 6E,F). In contrast, PD-1 expression was not consistently higher in eAML (supplemental Figure 13A-B). These data support CTLA-4 expression as predictive of response to ipilimumab-based therapy. Consistent with this finding, we observed that 1 exceptional responder with bone marrow disease and remission >3 years following decitabine and ipilimumab harbored a baseline T-cell infiltrate with high expression of FOXP3, CTLA-4, and PD-1 (supplemental Figure 13C).
Discussion
The clinical testing of combined decitabine and ipilimumab has been a logical next step for the treatment of AML given that prior in vitro and ex-vivo studies suggested immunomodulatory effects of hypomethylating agents59 and durable remissions after ipilimumab monotherapy in the setting of relapsed leukemia cutis after stem cell transplantation.60 Indeed, our current single-cell sequencing analysis of serially collected marrows from AML participants enrolled on the ETCTN/CTEP 10026 study showed evidence of decitabine-mediated myeloid cytoreduction.61 We further detected evidence of the pharmacodynamic impact of ipilimumab such as increase in marrow-infiltrating Tregs, in line with previous studies.11,51 Our analyses support the notion that decitabine and ipilimumab act on distinct cell types and modulate leukemic cell-immune cell interactions in responding patients.
Our in-depth transcriptomic characterization enabled us to address several outstanding questions regarding the treatment of AML. First, we wondered if determinants of response to combined decitabine and ipilimumab could be identified. Although we did not discover predictive T-cell subsets and did not detect CD8+ T-cell recruitment into bone marrow of responders, an increased T cells-to-AML ratio was associated with a more favorable response. Among posttransplant relapse cases, responding patients demonstrated greater donor-derived myelopoiesis before therapy, in line with observations that conditions of low disease burden provide the highest chance of long-term disease control through immunotherapeutic approaches and indicate that the local quantity of T cells relative to AML is a major determinant for response to ipilimumab-based therapy.62
Second, we explored the impact of alloreactivity given the dramatic responses previously reported for ipilimumab monotherapy post-HSCT.5,11 In the current study, we found a greater proportion of complete responses in transplant-naïve disease.9 Among posttransplant samples, we leveraged the repertoire of expressed germline SNPs to track donor- and recipient-derived single cells, which enabled us to dissect cellular composition and engraftment dynamics. Among T/NK cells, CD4+ T cells had the lowest engraftment consistent with flow cytometry-based studies of posttransplant immune reconstitution,41 followed by CD8+ and NK cells. These differences persisted throughout ipilimumab treatment, indicating that although CTLA-4 blockade drives T-cell differentiation,11 it was unable to convert T-cell subsets to full-donor chimerism. Our differential gene expression analysis of eAML vs bone marrow biopsies intriguingly indicated the higher infiltration of eAML with tissue-resident memory and CTLA-4+ T cells, raising a possible explanation for previously reported responses to ipilimumab monotherapy. Together with the higher response rate among transplant-naïve patients, our results suggest that combined decitabine and ipilimumab is unlikely to require an alloreactive environment for clinical activity. Rather, local microenvironmental niches, with their differences in expression states, seem to contribute to responses. Of note, host tissue–resident memory T cells survive to a substantial degree after allogeneic stem cell transplantation,63,64 but whether they were involved in responses of post-HSCT relapsed leukemia cutis cases to ipilimumab is unresolved.
Third, we evaluated potential mechanisms of escape from long-term responses to combined decitabine-ipilimumab. Notably, treatment failed to eradicate AML as our detailed tracking of donor- vs recipient-derived stem and progenitor-like cells revealed persistence of host cells and retained copy number changes, consistent with a lack of conversion to physiologic myelopoiesis. We also observed a reduced percentage of naïve CD4+ and CD8+ T cells in bone marrow of AML/MDS compared with healthy donors, which could impede ipilimumab-induced T-cell differentiation and proliferation. Counterintuitively, ipilimumab was associated with increases in Tregs and soluble CXCL13, intriguingly suggesting that CTLA-4 blockade can lead to longer term enhanced immunosuppression. These findings support its combination with Treg-depleting strategies, currently under investigation (NCT03912064).
Finally, the percentage of exhausted T cells in bone marrow of AML/MDS, a phenotype associated with tumor-reactive T cells in solid tumor malignancies, was dramatically lower than in tumor-infiltrating lymphocytes from external datasets. Although far more remains to be elucidated, we speculate that this relates in part to lower mutational burden in blood malignancies, and hence fewer opportunities for interactions with neoantigens that might elicit leukemia-reactive T cells.65 It may also relate to hemodilution and relatively high percentages of circulating T cells within bone marrow as opposed to more stationary tissue-resident memory T cells that likely have greater opportunities for sustained exposure to tumor antigens.66
Altogether, our analysis reveals that the activity of combined decitabine and ipilimumab in marrow-involved MDS/AML is determined by the quality and quantity of bone marrow–infiltrating T cells, suggesting that ipilimumab-based therapy may be most active in cases of low disease burden or in the setting of extramedullary niches (leukemia cutis). Our study opens numerous questions regarding immune phenotypes that characterize the marrow vs the extramedullary AML niches and the target antigens of ipilimumab-induced AML T-cell responses.67 Our findings further motivate continued therapeutic innovation to eradicate leukemic progenitor cells more durably. For example, as myeloid donor engraftment at relapse appears to impact the efficacy of immunotherapeutic salvage therapies, our findings support considerations for earlier post-HSCT treatment strategies before morphologic relapse.68,69
Acknowledgments
The authors thank the Cancer Moonshot CIMAC-CIDC Network for their tremendous support of the work and particularly appreciate the support and feedback from Holden Maecker, Ignacio Wistuba, Ethan Cerami, James Lindsay, Radim Moravec, and many others. The authors are grateful for support from Carol Reynolds and the members of the DFCI Flow Cytometry Core, DFCI Center for Immuno-Oncology, Doreen Hersey and the members of the Ted and Eileen Pasquarello Tissue Bank in Hematologic Malignancies for provision of samples; the patients who generously consented for the research use of these samples; the research coordinators, research nurses, advanced practice providers, and site staff for their support of the trial; Alicia Sabbio from the Translational Biomarker Core (CAMD TBC) at the Brigham and Women’s Hospital for isolation of RNA from FFPE samples; Fanny Dao, Sam Pollock, and Candace Patterson for excellent project management support; X. Shirley Liu for computational resources for analyses of bulk transcriptomic and whole exome sequencing data; and members of the Wu laboratory for their valuable feedback.
This work was financially supported by National Institutes of Health, National Cancer Institute grants P01CA229092 (C.J.W. and R.J.S.) and UM1CA186709 (principal investigator: Geoffrey Shapiro), National Cancer Institute Cancer Therapy Evaluation Program, Bristol-Myers Squibb, and LLS Therapy Accelerator Program. L.P. is supported by a research fellowship from the German Research Foundation (DFG, PE 3127/1-1) and is a scholar of the American Society of Hematology. M.S.D. is supported by the National Institutes of Health, National Cancer Institute grant R01CA266298-01A1, and is a clinical scholar of the Leukemia and Lymphoma Society. J.S.G. is supported by the Conquer Cancer Foundation Career Development Award, Leukemia and Lymphoma Society Translational Research Program Award, and NIH K08CA245209. NCI CTEP provided study drug (ipilimumab) support. S.L. is supported by the National Institutes of Health and National Cancer Institute Research Specialist Award (R50CA251956). S.Gnjatic is supported by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grant U01DK124165, and National Cancer Institute grant P30CA196521. N.Cieri is supported by an AACR-Incyte Immuno-oncology Research Fellowship (20-40-46-CEIR). The Human Immune Monitoring Center at ISMMS received support from the Cancer Center Support Grant CA196521. Scientific and financial support for the CIMAC-CIDC Network is provided through National Institutes of Health, National Cancer Institute Cooperative Agreements U24CA224319 (to the Icahn School of Medicine at Mount Sinai CIMAC), U24CA224331 (to the Dana-Farber Cancer Institute CIMAC), U24CA224285 (to the MD Anderson Cancer Center CIMAC), U24CA224309 (to the Stanford University CIMAC), and U24CA224316 (to the CIDC at Dana-Farber Cancer Institute). Additional support is made possible through the NCI CTIMS Contract HHSN261201600002C (to the Emmes Company, LLC). MDACC received support from NIH Cancer Center Support Grant P30CA016672 and the University of Texas SPORE NCI P50CA70907. Scientific and financial supports for the PACT project are made possible through funding support provided to the FNIH by AbbVie Inc, Amgen Inc, Boehringer-Ingelheim Pharma GmbH & Co KG, Bristol-Myers Squibb, Celgene Corporation, Genentech Inc, Gilead, GlaxoSmithKline plc, Janssen Pharmaceutical Companies of Johnson & Johnson, Novartis Institutes for Biomedical Research, Pfizer Inc, and Sanofi. The CIMAC-CIDC website is found at https://cimac-network.org/.
Authorship
Contribution: L.P. performed experiments, analyzed data, and designed figures; Y.L., L.Y., L.T., A.J., and J.A. analyzed WES and bulk RNA-seq data; J.S., S.L., and K.J.L. generated single-cell sequencing data; J.O.W., N.M.C., K.L.P., and S.J.R. provided multiplex immunofluorescence staining; N.C., G.O., D.S.N., F.S.H., P.B., and F.M. interpreted data; M.P., S.K.-S., and S. Gnjatic provided proximity extension assay data; S.R., H.X.C., M.T., H.S., R.L., M.H.S. and T.R. provided organizational oversight to the study; C.C. and S. Gabriel provided support with whole exome and bulk transcriptome sequencing; E.A.M., J.R., M.S.D., and R.J.S. provided clinical data and samples; J.S.G. is the principal investigator of the ETCTN/CTEP 10026 study; C.J.W. supervised the study; and L.P. and C.J.W. wrote the manuscript.
Conflict-of-interest disclosure: S.R. receives research support from Bristol-Myers-Squibb and KITE/Gilead, and is a member of the SAB of Immunitas Therapeutics. P.B. reports equity in Agenus, Amgen, Breakbio Corp, Johnson & Johnson, Exelixis, and BioNTech. and receives research support from Allogene Therapeutics. S.Gnjatic reports consultancy and/or advisory roles for Merck, Neon Therapeutics and OncoMed and research funding from Bristol-Myers Squibb, Genentech, Boehringer-Ingelheim, Takeda, and Regeneron. J.R. receives research funding from Kite/Gilead, Oncternal, and Novartis, serves on a Data Safety Monitoring Committee for AvroBio and on the Scientific Advisory Boards for Akron Biotech, Clade Therapeutics, Garuda Therapeutics, LifeVault Bio, Novartis, Smart Immune, Talaris Therapeutics, and TScan Therapeutics. D.N. received personal fee from Pharmacyclics, served as consultant to the American Society of Hematology Research Collaborative and has stock ownership in Madrigal Pharmaceuticals. K.J.L. reports equity in Standard BioTools Inc. F.S.H. reports grants and personal fees from Bristol-Myers Squibb; personal fees from Merck; grants and personal fees from Novartis; and personal fees from Surface, Compass Therapeutics, Apricity, Bicara, Pieris Pharmacutical, Checkpoint Therapeutics, Genentech/Roche, Bioentre, Gossamer, Iovance, Catalym, Immunocore, Amgen, Kairos, Rheos, Zumutor, Corner Therapeuitcs, Curis, and Astra Zeneca; outside the submitted work; In addition, F.S.H. has a patent Methods for Treating MICA-Related Disorders (#20100111973) with royalties paid; a patent Tumor antigens and uses thereof (#7250291) issued; a patent Angiopoietin-2 Biomarkers Predictive of Anti-immune checkpoint response (#20170248603) pending; a patent Compositions and Methods for Identification, Assessment, Prevention, and Treatment of Melanoma using PD-L1 Isoforms (#20160340407) pending; a patent Therapeutic peptides (#20160046716) pending; a patent Therapeutic Peptides (#20140004112) pending; a patent Therapeutic Peptides (#20170022275) pending; a patent Therapeutic Peptides (#20170008962) pending; a patent THERAPEUTIC PEPTIDES Therapeutic Peptides Patent number: 9402905 issued; a patent METHODS OF USING PEMBROLIZUMAB AND TREBANANIB pending; a patent Vaccine compositions and methods for restoring NKG2D pathway function against cancers Patent number: 10279021 issued; a patent antibodies that bind to major histocompatibility complex class I polypeptide-related sequence A Patent number:10106611 issued; and a patent ANTI-GALECTIN ANTIBODY BIOMARKERS PREDICTIVE OF ANTI-IMMUNE CHECKPOINT AND ANTI-ANGIOGENESIS RESPONSES Publication number: 20170343552 pending. M.S.D. has received consulting fees from AbbVie, Adaptive Biosciences, Aptitude Health, Ascentage Pharma, AstraZeneca, BeiGene, Bio Ascend, BMS, Curio Science, Eli Lilly, Genentech, Janssen, Merck, Ono Pharmaceuticals, Research to Practice, Secura Bio, TG Therapeutics, and Takeda and research support from AstraZeneca, Ascentage Pharma, Genentech, MEI Pharma, Novartis, Surface Oncology, and TG Therapeutics. F.M. is a cofounder of and has equity in Harbinger Health, has equity in Zephyr AI, and serves as a consultant for Harbinger Health, Zephyr AI, and Red Cell Partners. F.M. declares that none of these relationships are directly or indirectly related to the content of this manuscript. R.J.S. serves on the Board of Directors for Be the Match/National Marrow Donor Program and DSMB for Juno Therapeutics, Celgene USA, and BMS; reports personal fees from Vor Biopharma, Smart Immune, Daiichi Sankyo Inc, Neovii, Bluesphere Bio, Cugene, and Jasper. J.S.G. reports serving on steering committee and receiving personal fees from AbbVie, Astellas, and Takeda and institutional research funds from AbbVie, Genentech, Prelude, and AstraZeneca. C.J.W. holds equity in BioNTech, Inc and receives research support from Pharmacyclics. The remaining authors declare no competing financial interests.
Correspondence: Catherine J. Wu, Dana-Farber Cancer Institute and Harvard Medical School, Division of Stem Cell Transplantation and Cellular Therapies, Broad Institute of Harvard and MIT, 450 Brookline Ave, Boston, MA 02215; e-mail: cwu@partners.org.
References
Author notes
∗R.J.S., J.S.G., and C.J.W. are joint senior authors.
Whole exome sequencing, bulk RNA sequencing, single cell RNA/TCR and CITE sequencing will be made available in NCBI’s Database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap) under accession number phs003015.v1.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement”in accordance with 18 USC section 1734.

![Study overview and predictive markers of response to decitabine and ipilimumab treatment. (A) The ETCTN/CTEP 10026 study consisted of a cohort of relapsed AML after HSCT (arm A) and transplant-naïve AML/MDS (arm B) with morphologic disease. Following baseline assessment at study entry (Screening), a priming cycle of decitabine monotherapy (Lead-in) was administered. Subsequent cycles of therapy were given as a combination of decitabine and ipilimumab (C1, C2, …). Biopsies were also acquired at end of treatment. The swimmer plots indicate clinical course and duration of treatment for responders (blue) and nonresponders (red). Performed assays are indicated. (B) Median variant allele frequency of recurrent somatic mutations in bone marrow aspirates at screening grouped by response status across both arms (response [n = 16] and non-response [n = 25] according to study protocol) and study arm. Statistical testing using 2-sided t-test. (C) Changes of HSC score (van Galen et al31) throughout the study. Blue indicates responders; red indicates nonresponders. Statistical testing using Wilcoxon rank-sum test. (D,E) Differential gene-expression analysis of bone marrow core biopsies between responders (n = 6) and NRs (n = 18) across both arms according to study protocol (D) and gene set enrichment analysis (E) FDR, false discovery rate.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/15/10.1182_blood.2022018246/6/m_blood_bld-2022-018246-gr1.jpeg?Expires=1769104712&Signature=l--egVyZygdkomMMmKpnKDl-tJxrFwpsoFd0r1ryOT1Rc1igwpE24BS~y8zCGxWBEHlx7QJHLxVqhVrNKxRvr-Ra6CurYRpPdBvcP62D7FxjWiKCU7kFihhvVWK48PIi39PF~4S4Le~5FSvT-CncuKxhhoLINIGQJyLbrdFxFj34qKW51rZX5HZfuVGVpuMziU7V2MRluWDB9r2bpX9jPq6Ocy4udzVfQXr6WXl9~Y7K~33JG3XZXwdFIUtSvC4wt-DO3IYOThpGnESF7ehNSWFeyYsfO9-8soSLyuVndsWWStXqdJo1wEDZjdqBt1yYopvZuuOWNz~uM2Am2jhapQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal