Key Points
STAT3 gain-of-function mutations regulate a conserved core of transcriptional targets.
The transcriptional regulator SBNO2 is induced by hyperactive STAT3 and selectively required in STAT3-dependent hematopoietic malignancies.
Abstract
Gain-of-function mutations in the signal transducer and activator of transcription 3 (STAT3) gene are recurrently identified in patients with large granular lymphocytic leukemia (LGLL) and in some cases of natural killer (NK)/T-cell and adult T-cell leukemia/lymphoma. To understand the consequences and molecular mechanisms contributing to disease development and oncogenic transformation, we developed murine hematopoietic stem and progenitor cell models that express mutated STAT3Y640F. These cells show accelerated proliferation and enhanced self-renewal potential. We integrated gene expression analyses and chromatin occupancy profiling of STAT3Y640F-transformed cells with data from patients with T-LGLL. This approach uncovered a conserved set of direct transcriptional targets of STAT3Y640F. Among these, strawberry notch homolog 2 (SBNO2) represents an essential transcriptional target, which was identified by a comparative genome-wide CRISPR/Cas9-based loss-of-function screen. The STAT3-SBNO2 axis is also present in NK-cell leukemia, T-cell non-Hodgkin lymphoma, and NPM-ALK-rearranged T-cell anaplastic large cell lymphoma (T-ALCL), which are driven by STAT3-hyperactivation/mutation. In patients with NPM-ALK+ T-ALCL, high SBNO2 expression correlates with shorter relapse-free and overall survival. Our findings identify SBNO2 as a potential therapeutic intervention site for STAT3-driven hematopoietic malignancies.
Introduction
Signal transducer and activator of transcription 3 (STAT3) is part of the Janus kinase (JAK)/STAT signaling pathway that plays important roles in cellular processes including survival, growth, inflammation, and differentiation.1-3 Under physiological conditions, canonical STAT3 signaling is tightly regulated through receptor stimulation upon ligand binding (eg, interleukin 6 [IL-6] family cytokines or epidermal growth factor [EGF]). Upon activation, STAT3 proteins are tyrosine-phosphorylated (pY705STAT3) by JAK, dimerize, and translocate to the nucleus where they regulate transcription.1,2 Noncanonical pathways involving STAT proteins have also been reported. As such, STAT3 regulates the activity of the electron transport chain in mitochondria, and effects of unphosphorylated STAT3 as regulator of transcription have been described.4 Pathophysiological conditions including cancer are accompanied by deregulated JAK-STAT signaling. Hyperactivated STAT3 is frequently associated with a poor prognosis.1 STAT3 hyperactivation is either induced by activating mutations in the protein itself (eg, Y640F5), upstream signaling nodes (eg, in JAK26), excessive production of receptor ligands (eg, IL-67), inactivation of negative regulators (eg, suppressor of cytokine signaling [SOCS] 31,8), or direct activation though oncogenes (eg, anaplastic lymphoma kinase [ALK] fusion proteins9).
Recurrent STAT3 gain-of-function mutations have been identified in patients with hematopoietic malignancies. These include large granular lymphocytic leukemia (LGLL), adult T-cell lymphoma/leukemia, and natural killer (NK)/NKT-cell lymphoma/leukemia.5,10 The mutations described to date are located predominantly within the Src homology 2 (SH2) domain and are proposed to enhance STAT3 activation through stabilization of dimerization. STAT3 SH2 mutations are present in 40% of patients with LGLL.11 A small fraction (3%) of STAT3 mutations is found outside the SH2 domain, with to date unclear functions.12 LGLL usually represents a chronic indolent disease that originates from either T cells or NK cells and leads to autoimmune phenotypes including rheumatoid arthritis or neutropenia.11,13 The occurrence of STAT3 mutations in patients with LGLL is associated with more severe symptoms and reduced overall survival.14 Despite these clear prognostic effects, the functional consequences of STAT3 mutations are enigmatic. Recent evidence suggests that STAT3 mutations result in global epigenetic reprogramming via DNA hypermethylation in LGLL cells.15 Of note, in adult T-cell leukemia/lymphoma, STAT3 SH2 mutations are associated with more indolent disease.16
In this study, we explored the effects of mutant STAT3 on the transcriptional regulation in hematopoietic progenitor cells. The analysis of direct transcriptional targets of STAT3Y640F and the integration of genome-wide CRISPR/Cas9-based loss-of-function (LOF) data in combination with patient data, enabled us to identify strawberry notch homolog 2 (SBNO2) as a critical target of mutant STAT3. SBNO2 was selectively required for STAT3-driven hematopoietic malignancies and might open a novel therapeutic window of opportunity.
Methods
Workflow of genome-wide LOF CRISPR/Cas9 screens in HPC7 cells
The design and construction of the murine genome-wide Vienna single-guide RNA (sgRNA) library used, as well as the experimental workflow of LOF screening has been described previously.17,18 Here, Cas9-expressing single-cell clones of HPC7 cells expressing STAT3Y640F or empty vector were generated and infected with a sgRNA library at a low multiplicity of infection (MOI) (≤10%). sgRNA library–positive cells were monitored regularly by staining for Thy1.1 (APC anti-rat CD90/mouse CD90.1 clone: 53-2.1, eBioscience; at 1:200 final concentration). Four days after transduction, infected cells were selected using G418 (InVivoGen) (1 mg/mL), which was kept in the medium until the end of the screen. End point samples were harvested after the same amount of cell duplications (4) and cell pellets corresponding to at least a 500-fold library representation were harvested. Next-generation sequencing (NGS) libraries of screen end point samples and sgRNA plasmid pools were prepared as previously described17,18 and sequenced on a HiSeq 2500 platform (Illumina) using SR50 chemistry at the next-generation sequencing facility at Vienna BioCenter Core Facilities.
Gene expression analyses of patient samples with CD8+ T-cell LGLL (T-LGLL)
The study was undertaken in accordance with the principles of the Declaration of Helsinki and was approved by the ethics committees in the Helsinki University Central Hospital (Helsinki, Finland). All patients and healthy control participants gave written informed consent and their clinical details have been described previously.19
Gene expression analyses of patient samples with NK cell leukemia/T-cell non-Hodgkin lymphoma (T-NHL)
Primary samples were obtained from patients after informed consent; DNA and RNA was isolated from total leukocytes and whole-genome sequencing and RNA sequencing was performed as previously described20 at the MLL Munich Leukemia Laboratory.
All other methods are described in detail in supplemental Methods, available on the Blood website.
Results
STAT3 mutations enhance proliferation and self-renewal
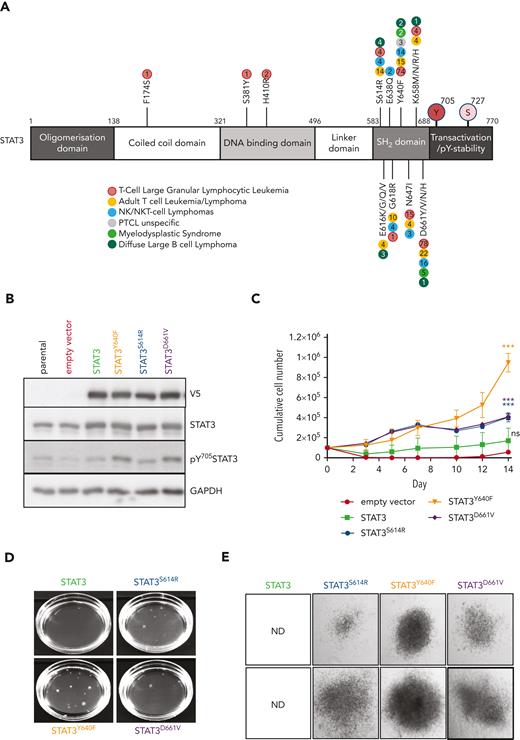
To understand the impact of the most common somatic STAT3 mutations occurring in patients with hematological malignancies (Figure 1A), we studied their effect in murine hematopoietic progenitor cells. HPC721 or HPCLSK cells22 were transduced with retroviral vectors encoding wild-type (WT) or mutant STAT323 (S614R, Y640F, or D661V) linked to a C-terminal V5 tag. The empty vector encoding dsRED only was used as control. The expression of the STAT3 mutations resulted in elevated levels of pY705STAT3, indicative of increased transcriptional activity (Figure 1B; supplemental Figure 1A). Cells expressing mutated STAT3, in particular STAT3Y640F, displayed significantly increased proliferation, colony formation potential, and size (Figure 1C-E; supplemental Figure 1B-C). Cells with mutated STAT3 retained the ability to form colonies upon serial replating (Figure 1F). We focused all further attempts on STAT3Y640F because it induced the most pronounced effects in vitro. IL-6 is a prominent mediator of inflammation and strongly activates STAT3;24 hence, we used this cytokine to study the effects of STAT3Y640F upon cytokine stimulation. Murine progenitor cells expressing STAT3Y640F reacted with significantly enhanced pY705STAT3 levels, indicative of hypersensitivity (supplemental Figure 1D). Similarly, dephosphorylation of pY705STAT3 upon IL-6 withdrawal was delayed in cells harboring STAT3Y640F (Figure 1G). In contrast, serine phosphorylation at STAT3S727 was induced in both STAT3 and STAT3Y640F to the same extent (supplemental Figure 1E). Our data led us to conclude that hematopoietic progenitor cells react to STAT3 gain-of-function mutations in vitro with increased proliferation, enhanced self-renewal capacity, and hypersensitivity to cytokine stimulation.
STAT3 mutations enhance proliferation and self-renewal. (A) Schematic overview of most common somatic STAT3 mutations.12,23 (B) Immunoblotting of pY705STAT3, STAT3, V5, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from HPC7 cells expressing empty vector, STAT3, or STAT3 mutants (Y640F, S614R, and D661V). (C) Growth curves of HPC7 cells expressing either empty vector, STAT3, or STAT3 mutants (Y640F, S614R, and D661V) (mean ± standard deviation [SD], n ≥ 3). (D) Representative image of colony formation assay (CFA) of HPCLSK expressing either empty vector, STAT3, or STAT3 mutants (Y640F, S614R, and D661V) (mean ± SD, n ≥ 3. (E) Representative images of colonies 10 days after plating derived from HPCLSK CFA assay, at original magnification ×4. (F) Serial replating CFA from HPC7 expressing either empty vector, STAT3, or STAT3 mutants (Y640F, S614R, and D661V) (mean ± SD, n ≥ 3). (G) Immunoblot of HPC7 cells expressing either empty vector, STAT3, or STAT3Y640 that were cytokine starved for 3 hours and then stimulated with IL-6 for 20 minutes (100 ng/mL), then washed with phosphate-buffered saline (PBS) and plated in cytokine-free media. Cells were harvested at the indicated time points and pY705STAT3, STAT3, V5, and GAPDH expression analyzed. Unpaired, 2-tailed Student t test was used in panel C, and 1-way analysis of variance was used in panel F for P value determination. ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ND, not determined; PTCL, peripheral T-cell lymphoma; stim, stimulation.
STAT3 mutations enhance proliferation and self-renewal. (A) Schematic overview of most common somatic STAT3 mutations.12,23 (B) Immunoblotting of pY705STAT3, STAT3, V5, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from HPC7 cells expressing empty vector, STAT3, or STAT3 mutants (Y640F, S614R, and D661V). (C) Growth curves of HPC7 cells expressing either empty vector, STAT3, or STAT3 mutants (Y640F, S614R, and D661V) (mean ± standard deviation [SD], n ≥ 3). (D) Representative image of colony formation assay (CFA) of HPCLSK expressing either empty vector, STAT3, or STAT3 mutants (Y640F, S614R, and D661V) (mean ± SD, n ≥ 3. (E) Representative images of colonies 10 days after plating derived from HPCLSK CFA assay, at original magnification ×4. (F) Serial replating CFA from HPC7 expressing either empty vector, STAT3, or STAT3 mutants (Y640F, S614R, and D661V) (mean ± SD, n ≥ 3). (G) Immunoblot of HPC7 cells expressing either empty vector, STAT3, or STAT3Y640 that were cytokine starved for 3 hours and then stimulated with IL-6 for 20 minutes (100 ng/mL), then washed with phosphate-buffered saline (PBS) and plated in cytokine-free media. Cells were harvested at the indicated time points and pY705STAT3, STAT3, V5, and GAPDH expression analyzed. Unpaired, 2-tailed Student t test was used in panel C, and 1-way analysis of variance was used in panel F for P value determination. ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ND, not determined; PTCL, peripheral T-cell lymphoma; stim, stimulation.
Genome-wide CRISPR/Cas9-based LOF screen identifies genetic dependencies of STAT3Y640F-driven cells
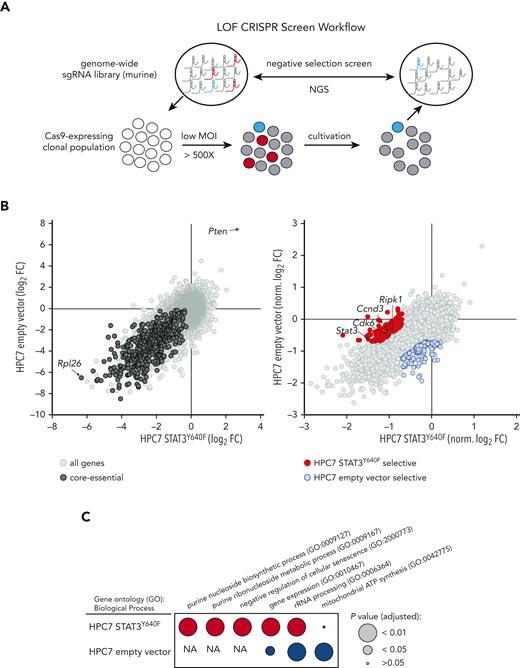
To identify genetic dependencies associated with mutant STAT3Y640F-driven cells, we performed comparative genome-wide CRISPR/Cas9-based LOF screens (Figure 2A) in clonal Cas9-HPC7 cells expressing either STAT3Y640F or the empty vector. Single-cell clones were tested for Cas9 functionality by performing competitive proliferation assays using sgRNA targeting Myb (positive control) or Rosa26 (negative control) (supplemental Figure 2A-C). Functional clones of each genotype were subsequently transduced with the second-generation, genome-wide, murine Vienna sgRNA library17,18 at low MOI while maintaining at least a 500× whole-library representation. After the same number of cell duplications, we compared the sgRNA abundances of each screen end point with those of the initial plasmid pool. The high quality of the screen was verified by the fact that targeting of core-essential genes displayed strong negative selection (eg, Rpl26) whereas common tumor suppressor genes (eg, Pten) were enriched (Figure 2B left panel; supplemental Figure 2D; supplemental Table 1).
Genome-wide CRISPR/Cas9-based LOF screen identifies genetic dependencies of STAT3Y640F-driven cells. (A) Workflow of a genome-wide LOF CRISPR/Cas9 screen. Clonal Cas9-expressing HPC7 cells (empty vector or STAT3Y640F) were transduced with the Vienna sgRNA library at low multiplicity of infection (MOI) to ensure single integration. After equal population duplications, cells were harvested and genomic DNA extracted. After library preparation cells were subjected to next-generation sequencing (NGS) to compare sgRNA abundances. (B) Comparative analysis of 2 CRISPR-based LOF screens in empty vector vs STAT3Y640F-driven HPC7 progenitor cells. Gene effects were depicted with respect to effect size of defined core-essential and nonessential genes. Depletion of core-essential genes (left) and normalized STAT3Y640F- and empty vector–specific dependencies (right) have been depicted. Dependencies defined as less than –0.7 normLog2FC compared with the plasmid pool and a difference of at least 0.5 normLog2FC. (C) Comparison of gene ontology analyses (Biological Process 2021) (https://maayanlab.cloud/Enrichr/25-27) of selective genetic dependencies of STAT3Y640F (red) and selective dependencies of empty vector screen (blue). FC, fold change; NA, not applicable; sgRNA, single guide RNA.
Genome-wide CRISPR/Cas9-based LOF screen identifies genetic dependencies of STAT3Y640F-driven cells. (A) Workflow of a genome-wide LOF CRISPR/Cas9 screen. Clonal Cas9-expressing HPC7 cells (empty vector or STAT3Y640F) were transduced with the Vienna sgRNA library at low multiplicity of infection (MOI) to ensure single integration. After equal population duplications, cells were harvested and genomic DNA extracted. After library preparation cells were subjected to next-generation sequencing (NGS) to compare sgRNA abundances. (B) Comparative analysis of 2 CRISPR-based LOF screens in empty vector vs STAT3Y640F-driven HPC7 progenitor cells. Gene effects were depicted with respect to effect size of defined core-essential and nonessential genes. Depletion of core-essential genes (left) and normalized STAT3Y640F- and empty vector–specific dependencies (right) have been depicted. Dependencies defined as less than –0.7 normLog2FC compared with the plasmid pool and a difference of at least 0.5 normLog2FC. (C) Comparison of gene ontology analyses (Biological Process 2021) (https://maayanlab.cloud/Enrichr/25-27) of selective genetic dependencies of STAT3Y640F (red) and selective dependencies of empty vector screen (blue). FC, fold change; NA, not applicable; sgRNA, single guide RNA.
To maximize comparability between data sets, we normalized gene effects to a proposed set of core-essential and nonessential genes28 (Figure 2B right panel). Detailed comparisons (less than –0.7 normLog2FC vs plasmid pool and >0.5 normLog2FC difference to empty vector) allowed us to identify and define 129 genes that were selectively required in STAT3Y640F cells. Among these genes, we found Stat3, a positive control for our screen, as well as other genes required for proliferation such as Cdk6 or Ccnd3(Figure 2B right panel). Other previously described canonical STAT3 target genes such as Socs3 or Pim11 did not show a particular selectivity for STAT3Y640F-expressing cells (supplemental Figure 2E; supplemental Table 1). Gene ontology analysis of STAT3Y640F-specific genetic dependencies revealed an enrichment in purine metabolism and negative regulation of cellular senescence, a common feature of cancer cells.29,30 In contrast, analysis of empty vector–specific genetic dependencies revealed 155 genes that showed an enrichment in mitochondrial respiratory chain complex I assembly, mitochondrial electron transport, and chromatin-mediated maintenance of transcription, all expected pathways in WT STAT3 cells.31 (Figure 2B right panel; Figure 2C; supplemental Table 1).
STAT3Y640F enhances DNA binding and transcriptional activity
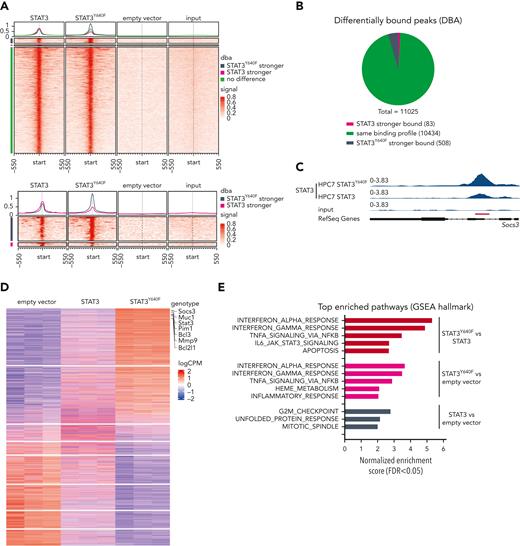
Chromatin immunoprecipitation sequencing (ChIP-seq) in the progenitor cell line HPC7 expressing STAT3,STAT3Y640F, or empty vector as control was used to understand whether mutant STAT3 alters chromatin occupancy. Our aim was to understand which of the 129 genes required in mutant cells are direct STAT3 targets. To optimally compare all ChIP-seq data, we used the C-terminal V5 tag that does not interfere with nuclear localization and DNA binding (supplemental Figure 3A-B). The vast majority (10 434; 95%) of regions were bound by WT STAT3 and STAT3Y640F at comparable intensities. We did not detect any distinct pattern of DNA binding sites of STAT3Y640F compared with WT STAT3 (Figure 3A-B). At 508 regions (4%, associated with 470 genes), we detected a stronger binding of STAT3Y640Fcompared with WT STAT3, including the prototypical STAT3 target Socs3 (Figure 3C; supplemental Table 2). In total, 83 regions (1%, corresponding to 78 genes) were bound stronger by WT STAT3 (Figure 3A-B; supplemental Table 2). Annotation of differentially and equally bound genomic regions revealed similar proportions of binding at promoters and intragenic and intergenic regions (supplemental Figure 3C). In summary, STAT3Y640F binds chromatin at the same sites as WT STAT3 with a small subset of regions showing altered binding intensities.
STAT3Y640Fenhances DNA binding and transcriptional activity. (A) Density heat map visualizing signal strength around peak summits across union consensus binding sites in STAT3, STAT3Y640F, empty vector, and input ChIP-seq samples. (B) Results from differential binding analysis of STAT3 and STAT3Y640F-V5–tagged ChIP-seq analysis depicted as a pie chart. (C) Representative illustration of ChIP-seq tracks showing the binding profiles of STAT3 and STAT3Y640F at the Socs3 promoter region. (D) Transcriptome analysis: heat map of genes differentially expressed (FDR < 0.05) in any of the 3 differential expression analyses performed (STAT3 vs empty vector, STAT3Y640F vs empty vector, STAT3Y640F vs STAT3). Canonical STAT3 target genes are highlighted. (E) Significant hallmark gene sets (FDR < 0.05) from gene set enrichment analysis (GSEA) of 3 differential expression analyses shown in panel D. DBA, differential binding analysis. FDR, false discovery rate.
STAT3Y640Fenhances DNA binding and transcriptional activity. (A) Density heat map visualizing signal strength around peak summits across union consensus binding sites in STAT3, STAT3Y640F, empty vector, and input ChIP-seq samples. (B) Results from differential binding analysis of STAT3 and STAT3Y640F-V5–tagged ChIP-seq analysis depicted as a pie chart. (C) Representative illustration of ChIP-seq tracks showing the binding profiles of STAT3 and STAT3Y640F at the Socs3 promoter region. (D) Transcriptome analysis: heat map of genes differentially expressed (FDR < 0.05) in any of the 3 differential expression analyses performed (STAT3 vs empty vector, STAT3Y640F vs empty vector, STAT3Y640F vs STAT3). Canonical STAT3 target genes are highlighted. (E) Significant hallmark gene sets (FDR < 0.05) from gene set enrichment analysis (GSEA) of 3 differential expression analyses shown in panel D. DBA, differential binding analysis. FDR, false discovery rate.
Transcriptome analysis in HPC7 cells expressing STAT3, STAT3Y640F, or empty vector completed our studies and was performed to understand whether the slightly diverse chromatin binding of STAT3Y640F would result in altered transcription. Reassuringly, we found classical STAT3 target genes (including Socs3, Pim1, and Bcl2l1)1 among the upregulated genes (Figure 3D). In total, we found 2157 genes upregulated by the STAT3Y640F mutant (STAT3Y640F vs empty vector; and STAT3Y640F vs STAT3) (supplemental Table 3). Of note, we identified a unique gene signature that was only present in STAT3Y640F mutant cells and that included 1108 upregulated and 992 downregulated genes (false discovery rate [FDR] < 0.05). Fewer genes were exclusively affected by WT STAT3 (684 genes upregulated, 639 genes downregulated; FDR < 0.05) (supplemental Figure 3D; supplemental Table 3). Gene set enrichment analyses (GSEA) revealed that STAT3Y640F-regulated genes were implicated in the immune response (interferon response, tumor necrosis factor α [TNFα] signaling via NF-κB, inflammatory response, and IL-6_JAK_STAT3 signaling). In contrast, STAT3-regulated gene sets were enriched in cell cycle signaling (G2M checkpoint) and mitotic spindle assembly (Figure 3E). The analysis was confirmed in HPCLSK cells recapitulating the STAT3Y640F gene expression patterns (supplemental Figure 3E-F; supplemental Table 3).
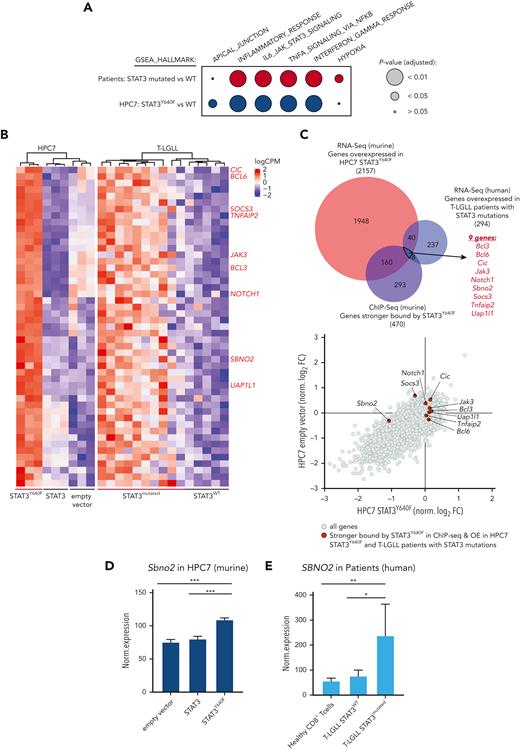
SBNO2 is an essential direct transcriptional target of mutated STAT3
The human relevance of our study was addressed by using RNA sequencing data derived from CD8+ T cells from patients with T-LGLL with either WT or mutant STAT3.15,19 In line with the murine model systems, gene expression profiles of STAT3-mutated samples from patients with T-LGLL showed an enrichment of pathways involved in inflammatory response (including TNFα, interferon alpha/gamma signaling, and IL-6_JAK_STAT3_signaling) (Figure 4A; supplemental Figure 4A; supplemental Table 4). By overlapping human and murine data sets, we identified 49 genes whose expression was increased in both human and murine STAT3-mutated cells (Figure 4B-C; supplemental Table 4). Of these 49 genes, 9 showed an increased binding of STAT3Y640F on chromatin: Jak3, notch receptor 1 (Notch1), B-cell lymphoma 3 (Bcl3), Bcl6, Tnfα-induced protein 2 (Tnfaip2), uridine diphosphate-N-acetylglucosamine pyrophosphorylase-1-like-1 (Uap1l1), Capicua (Cic), Sbno2, and Socs3 (Figure 4B-C, highlighted in red). To understand which of these genes represent potential dependencies of STAT3 mutant cells, we integrated these data with the outcome of the CRISPR LOF screen. We explored genes that fulfilled the following criteria: (1) overexpression in STAT3Y640F-driven murine hematopoietic progenitor cells, (2) increased binding by STAT3Y640F compared with WT STAT3, (3) overexpression in patients with T-LGLL with STAT3 mutations compared with patients with WT STAT3, and (4) selective genetic dependency in STAT3Y640F-driven murine hematopoietic progenitor cells as determined by genome-wide LOF screening (Figure 4C-D). This unbiased global analysis identified Sbno2 as a direct target and the potential selective vulnerability of cells driven by the STAT3Y640F mutation. In addition, we could also show that SBNO2 was upregulated in patients with T-LGLL with STAT3 mutations compared with healthy CD8+ T cells (Figure 4E). We complemented our STAT3-ChIP-seq data sets with previously published H3K27ac ChIP-seq data from thrombopoietin (TPO)-stimulated HPC7 cells.32 This analysis showed that the promoter region of Sbno2 (isoform 2) harbored both, increased binding by STAT3Y640F, and increased H3K27ac signal in HPC7 cells upon TPO treatment, indicating increased transcriptional activity. (supplemental Figure 4B). Similar to IL-6, TPO induces STAT3 signaling33 and thereby supports our concept of activated STAT3 binding the SBNO2 promoter region. In line with this, previous studies in murine bone marrow–derived macrophages34 and astrocytes35 showed a cytokine-induced upregulation of SBNO2 via STAT3. SBNO2 is a putative DExD/H-box containing helicase35 that acts as a transcriptional coregulator and is capable of exerting repressive and activating functions.34
SBNO2 is an essential direct transcriptional target of mutated STAT3. (A) GSEA analysis of differentially expressed genes between HPC7 cells expressing STAT3 or STAT3Y640F (blue) or primary CD8+ cells from patients with T-LGLL expressing either WT (n = 5) or mutated STAT3 (expressing: Y640F, n = 4; D661Y, n = 3; D661H, n = 1; D661V, n = 1; D661V+Y640F, n = 1) (red). (B) Heat map of commonly regulated genes (FDR < 0.05) between STAT3Y640F-expressing mouse HPC7 cell lines and samples from patients with T-LGLL. Genes that are stronger bound by STAT3Y640F are annotated in red on the right. (C) Overlap of genes that are overexpressed in murine and human STAT3-mutation–driven cells (i), are stronger bound by STAT3Y640F (top) (ii), and represent selective dependencies in STAT3Y640F-driven HPC7 cells (bottom) (iii). (D) Normalized expression of Sbno2 in HPC7 cells expressing either STAT3, STAT3Y640F, or empty vector (mean ± SD, n = 3). (E) Normalized expression of SBNO2 in healthy CD8+ T cells (n = 5) or T-LGLL cells expressing either WT (n = 5) or mutant (n = 10) STAT3 (mean ± SD). Levels of significance were calculated using unpaired t-tests in panels D-E. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. FDR, false discovery rate. GSEA Gene set enrichment analyses.
SBNO2 is an essential direct transcriptional target of mutated STAT3. (A) GSEA analysis of differentially expressed genes between HPC7 cells expressing STAT3 or STAT3Y640F (blue) or primary CD8+ cells from patients with T-LGLL expressing either WT (n = 5) or mutated STAT3 (expressing: Y640F, n = 4; D661Y, n = 3; D661H, n = 1; D661V, n = 1; D661V+Y640F, n = 1) (red). (B) Heat map of commonly regulated genes (FDR < 0.05) between STAT3Y640F-expressing mouse HPC7 cell lines and samples from patients with T-LGLL. Genes that are stronger bound by STAT3Y640F are annotated in red on the right. (C) Overlap of genes that are overexpressed in murine and human STAT3-mutation–driven cells (i), are stronger bound by STAT3Y640F (top) (ii), and represent selective dependencies in STAT3Y640F-driven HPC7 cells (bottom) (iii). (D) Normalized expression of Sbno2 in HPC7 cells expressing either STAT3, STAT3Y640F, or empty vector (mean ± SD, n = 3). (E) Normalized expression of SBNO2 in healthy CD8+ T cells (n = 5) or T-LGLL cells expressing either WT (n = 5) or mutant (n = 10) STAT3 (mean ± SD). Levels of significance were calculated using unpaired t-tests in panels D-E. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. FDR, false discovery rate. GSEA Gene set enrichment analyses.
Human NK-cell leukemia driven by STAT3 mutations depend on SBNO2
To functionally validate the relevance of SBNO2, we first performed competitive proliferation assays in the STAT3 mutant–expressing HPC7 in vitro model. CRISPR/Cas9- and RNA interference (RNAi)–mediated suppression of Stat3 or Sbno2 severely impaired proliferation of cells, showing the importance of the STAT3-SBNO2 axis in this system. Knock down of STAT3Y640F, confirmed by reduced Stat3 messenger RNA (mRNA) levels, resulted in reduced Sbno2, confirming the transcriptional regulation of the latter by activated STAT3 (supplemental Figure 5A-D).
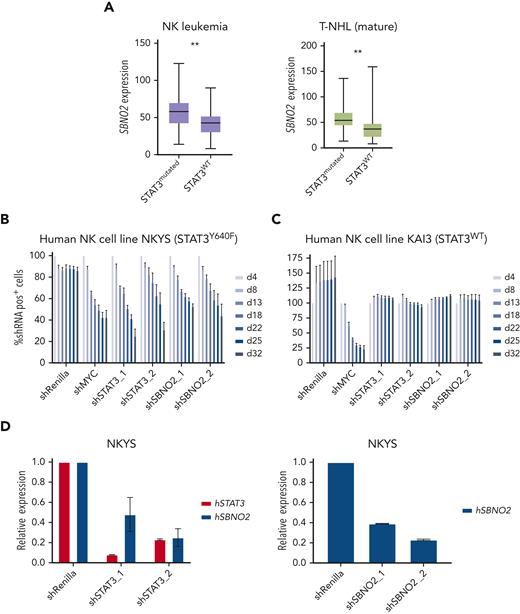
Next, we assessed whether SBNO2 overexpression is more generally associated with STAT3 mutations in human lymphoid malignancies. We therefore analyzed primary samples from patients with STAT3 mutation–driven vs WT STAT3 NK-cell leukemia (Figure 5A left panel) or T-NHL (Figure 5A right panel). SBNO2 was significantly overexpressed in both patient cohorts with mutant STAT3. To verify functional consequences, we compared human STAT3 mutation–driven NK-cell leukemia cell lines with STAT3-independent NK-cell leukemia cell lines36 (supplemental Figure 5E). Knock down of STAT3 or SBNO2 significantly impaired survival of STAT3 mutation-driven NK-cell leukemia cells (NKYS) whereas STAT3-independent cells (KAI3) remained unaffected in RNAi-based competitive proliferation assays. Similar to our findings in murine HPC7 cells, knock down of mutated STAT3 in NKYS cells also led to decreased SBNO2 expression (Figure 5B-D; supplemental Figure 5F-G). In summary, these experiments validated the presence of a STAT3-SBNO2 axis in mutant STAT3 cells and identified SBNO2 as a selective dependency in STAT3 mutation–driven leukemia.
Human NK-cell leukemia driven by STAT3 mutations depends on SBNO2 expression. (A) Normalized SBNO2 expression from patients with NK-cell leukemia (left) and from patients with T-NHL (right) with either mutated STAT3 (NK, n = 19; T-NHL, n = 34) or WT STAT3 (NK, n = 50; T-NHL, n = 56) have been depicted. Competitive proliferation assays in human NK-cell leukemia cell lines harboring either mutated STAT3 (NKYS) (B) or WT STAT3 (KAI3) (C), transduced with short hairpin RNA (shRNA) expression vectors targeting either Renilla (negative control), MYC (positive control), STAT3, or SBNO2. Relative abundance of shRNA+ cells was normalized to day 4 after transduction (mean ± SD, n = 3). (D) Quantitative polymerase chain reaction (qPCR) expression analyses of STAT3 or SBNO2 in NKYS cells 5 days after transduction with shRNA vectors targeting STAT3 or SBNO2, respectively (mean ± SD, n ≥ 3). Levels of significance were calculated using unpaired t-test in panel A. ∗∗P < .01.
Human NK-cell leukemia driven by STAT3 mutations depends on SBNO2 expression. (A) Normalized SBNO2 expression from patients with NK-cell leukemia (left) and from patients with T-NHL (right) with either mutated STAT3 (NK, n = 19; T-NHL, n = 34) or WT STAT3 (NK, n = 50; T-NHL, n = 56) have been depicted. Competitive proliferation assays in human NK-cell leukemia cell lines harboring either mutated STAT3 (NKYS) (B) or WT STAT3 (KAI3) (C), transduced with short hairpin RNA (shRNA) expression vectors targeting either Renilla (negative control), MYC (positive control), STAT3, or SBNO2. Relative abundance of shRNA+ cells was normalized to day 4 after transduction (mean ± SD, n = 3). (D) Quantitative polymerase chain reaction (qPCR) expression analyses of STAT3 or SBNO2 in NKYS cells 5 days after transduction with shRNA vectors targeting STAT3 or SBNO2, respectively (mean ± SD, n ≥ 3). Levels of significance were calculated using unpaired t-test in panel A. ∗∗P < .01.
NPM-ALK–driven hematopoietic malignancies are dependent on SBNO2 expression
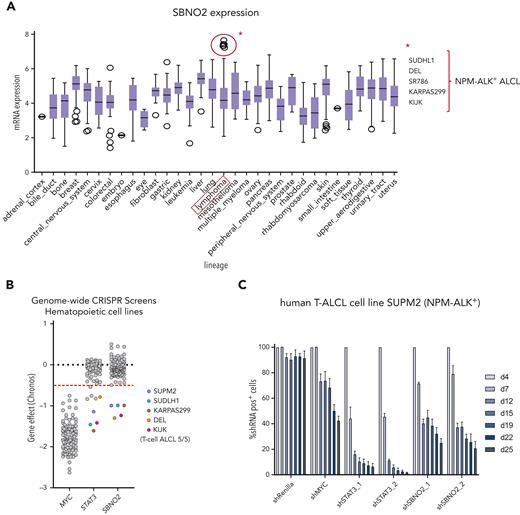
To understand the general relevance of SBNO2 in hematopoietic malignancies, we analyzed publicly available gene expression and functional genomics data sets (Broad Institute's Cancer Dependency Map-DepMap).37 To date, these data sets lack LOF data from NK-cell malignancies but allowed us to identify 5 human lymphoma cell lines with high SBNO2 expression (Figure 6A). The common denominator of this subgroup was the expression of the NPM-ALK+ fusion oncogene, which directly activates STAT3, a hallmark of NPM-ALK+ ALCL.38 The integration of publicly available reverse phase protein microarray data verified the positive association between SBNO2 and pY705STAT3 levels or SOCS3 expression (supplemental Figure 6A-B). The reanalysis of publicly available STAT3 ChIP-seq data confirmed that SBNO2 is directly regulated by STAT3. In the NPM-ALK+ ALCL cell line, SUDHL1, we found pronounced STAT3 binding at the SBNO2 promoter region (isoform 2) that was abolished upon treatment of cells with the ALK inhibitor crizotinib.39 This promoter region also harbored a strong H3K27ac signal,40 suggesting active transcription by STAT3 (supplemental Figure 6C).
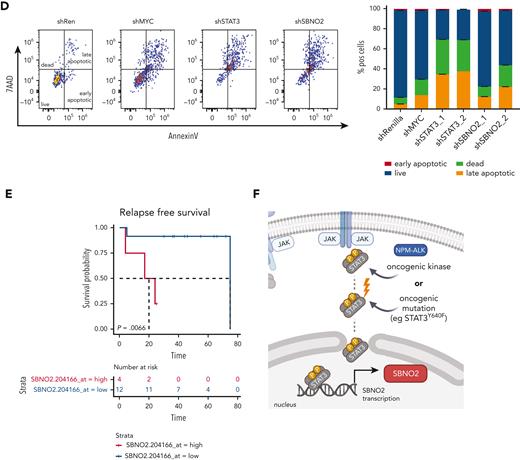
NPM-ALK–driven hematopoietic malignancies are addicted to SBNO2 expression, and SBNO2 expression is a prognostic marker in patients with NPM-ALK+ALCL. (A) SBNO2 expression across all cell lines within the cancer dependency map (DepMap).37 Cell lines with high SBNO2 expression highlighted in red. (B) Reanalysis of DepMap data from 116 CRISPR-based LOF screens done using hematopoietic cell lines. Each dot represents a distinct screen. Gene effects were normalized to FDR-adjusted median effect size of defined core-essential and nonessential genes. (C) Competitive proliferation assays performed on human NPM-ALK+ T-ALCL cells transduced with shRNA vectors targeting either Renilla (negative control), MYC (positive control), STAT3, or SBNO2, respectively. Abundance of shRNA+ cells was normalized to day 4 after transduction (mean ± SD, n = 3). (D) Flow cytometric analysis (left) and quantification of annexin/7AAD-positve apoptotic cells (right) at the end point of competition assay shown in panel C have been shown. (E) Kaplan-Meier–based survival analysis of patients with NPM-ALK+ with high vs low SBNO2 expression upon reanalysis of publicly available data sets.41 Correlation of SBNO2 expression and relapse-free time was assessed through the log-rank test in panel E. (F) Schematic illustration of the proposed model. STAT3 hyperactivation induced through either oncogenic upstream signaling (eg, via NPM-ALK) or activating STAT3 mutations (eg, STAT3Y640F) induce SBNO2 expression that is essential for cancer cell proliferation/survival.
NPM-ALK–driven hematopoietic malignancies are addicted to SBNO2 expression, and SBNO2 expression is a prognostic marker in patients with NPM-ALK+ALCL. (A) SBNO2 expression across all cell lines within the cancer dependency map (DepMap).37 Cell lines with high SBNO2 expression highlighted in red. (B) Reanalysis of DepMap data from 116 CRISPR-based LOF screens done using hematopoietic cell lines. Each dot represents a distinct screen. Gene effects were normalized to FDR-adjusted median effect size of defined core-essential and nonessential genes. (C) Competitive proliferation assays performed on human NPM-ALK+ T-ALCL cells transduced with shRNA vectors targeting either Renilla (negative control), MYC (positive control), STAT3, or SBNO2, respectively. Abundance of shRNA+ cells was normalized to day 4 after transduction (mean ± SD, n = 3). (D) Flow cytometric analysis (left) and quantification of annexin/7AAD-positve apoptotic cells (right) at the end point of competition assay shown in panel C have been shown. (E) Kaplan-Meier–based survival analysis of patients with NPM-ALK+ with high vs low SBNO2 expression upon reanalysis of publicly available data sets.41 Correlation of SBNO2 expression and relapse-free time was assessed through the log-rank test in panel E. (F) Schematic illustration of the proposed model. STAT3 hyperactivation induced through either oncogenic upstream signaling (eg, via NPM-ALK) or activating STAT3 mutations (eg, STAT3Y640F) induce SBNO2 expression that is essential for cancer cell proliferation/survival.
In nontransformed cells, STAT3 chromatin binding at the Sbno2 promoter required cytokine stimulation; STAT3 ChIP-seq of murine CD4+ T cells (ex vivo)42 revealed STAT3 binding on the Sbno2 promoter only upon IL-21 stimulation (supplemental Figure 6C). Similar to IL-6, IL-21 is a strong activator of STAT3.43 The STAT3-SBNO2 axis was also detectable when we reanalyzed 116 publicly available genome-wide LOF screens in hematopoietic cell lines (depmap).37 All cell lines that require SBNO2 expression also depended on STAT3 (Figure 6B) and, intriguingly, the analysis identified NPM-ALK–driven T-cell ALCL cell lines.
Again, RNAi-based competitive proliferation assays showed that knock down of STAT3 or SBNO2 severely impaired the proliferation and survival of NPM-ALK+ SUPM2 cells (Figure 6C-D), whereas the STAT3-independent BCR-ABL1+-driven K562 cells remained unaffected (supplemental Figure 6D). Moreover, gene expression analysis by quantitative polymerase chain reaction upon STAT3 knockdown confirmed that SBNO2 is STAT3-regulated in SUPM2 cells (supplemental Figure 6E).
SBNO2 expression is a prognostic marker in patients with NPM-ALK+ ALCL
Given the functional relevance of SBNO2 in STAT3-driven disease, we hypothesized that high SBNO2 expression is linked to adverse disease outcome in patients with NPM-ALK+ ALCL. Therefore, we analyzed a cohort of patients with NPM-ALK+ for the effect of SBNO2 expression on disease prognosis.41 Indeed, high SBNO2 expression correlated with a shorter relapse-free survival (P = .0066) (Figure 6E). When analyzing the overall survival, a similar trend was observed that did not reach statistical significance (P = .17) (supplemental Figure 6F). These data are in line with the hypothesis that aberrant STAT3 signaling, induced through mutations in STAT3 or upstream activation by the NPM-ALK fusion oncogene, leads to the rewiring of signaling and turns SBNO2 into a critical signaling factor (Figure 6F). The STAT3-SBNO2 axis thereby represents a potential new therapeutic intervention site for STAT3-driven T-/NK-cell lymphomas.
Discussion
The JAK-STAT pathway is one of the most commonly altered signaling pathways in cancer. STAT3 activation is frequently affected, and its targeted inhibition remains a primary goal for improving therapeutic strategies.44 Here, we studied STAT3 mutations that are found in hematological malignancies, particularly in patients with LGLL, and focused on the most common STAT3Y640F mutation.
We used murine hematopoietic progenitor cell lines (HPC7 and HPCLSK), which retain a high level of differentiation plasticity, to investigate the phenotypic and transcriptional features of mutated STAT3. In line with previous studies, we found that mutations within the SH2 domain of STAT3 elevate pY705STAT3 phosphorylation and promote hypersensitivity toward cytokine stimulation, exemplified through using IL-6.5STAT3 mutations increase the proliferation rate and enhance colony formation and colony size, indicative of self-renewal potential, a major features of transformed cells.45
Persistent hyperactivation of STAT3 has also been implicated as a promoter of tumorigenesis via aberrant regulation of inflammatory processes.2,46 STAT3 acts downstream of a wide range of cytokines that may elicit opposing functions. For instance, both IL-6 and IL-10 activate STAT3 but induce different cellular responses. IL-6 is primarily a proinflammatory cytokine, whereas IL-10 exerts anti-inflammatory effects.2,47 In patients with mutated STAT3 LGLL, a proinflammatory gene signature is dominant, which was reflected in the murine progenitor cell lines expressing STAT3Y640F (HPC7 and HPCLSK).13,48,49 These consistent findings validate the choice of our experimental system. Chromatin occupancy profiling of WT STAT3 and STAT3Y640F overlapped substantially, in line with a model in which STAT3 mutations do not provide gain-of-function through differential DNA binding but rather represent the consequences of prolonged and pronounced signaling leading to a shift in transcriptional patterns. This concept is further supported by the fact that in NPM-ALK–driven disease, in which STAT3 mutations are absent but the signaling pathway is hyperactivated, also depend on SBNO2 expression.
Different STAT3 mutations deregulate hundreds of genes in patient-derived T-LGLL cells and in STAT3Y640F murine hematopoietic progenitor cells. The overlap between these data sets is small but identifies a hardwired set of commonly deregulated genes. It is conceivable that some features of STAT3-mutated disease are not recapitulated in murine cells in vitro. Differences between the human disease and the murine model may be attributed to the lack of microenvironment in the murine system, and/or individual mutations in cells from human patients. In addition, it is worth noting that we covered a range of different SH2 domain mutations in our human T-LGLL data set, whereas we restricted our analysis to STAT3Y640F in our murine models. The confirmation of our findings in samples from patients with NK-leukemia or T-NHL that bear different STAT3 mutations verifies the importance of the STAT3-SBNO2 axis for a broad range of patients with mutated STAT3. In addition, we currently cannot exclude that the different STAT3 mutations in combination with individual patient–dependent gene alterations lead to distinct target gene patterns.
The stringent approach used enabled us to identify a core of 9 direct transcriptional targets (Jak3, Notch1, Bcl6, Tnfaip2, Uap1l1, Sbno2, Bcl3, Cic, and Socs3), which are preferentially occupied by mutant STAT3 and overexpressed in murine cell lines. The high STAT3 activity in STAT3Y640F-transformed cells resembles an IL-6–induced activation that boosts the expression of STAT3 targets including Socs3,1,8Notch1,50 or Bcl3.51 These genes have been associated with malignant transformation and/or inflammation. As such, Notch1 has been implicated in malignant transformation in lymphoid cells.52,53 The NF-κB pathway is frequently deregulated in inflammation and cancer54 and also deregulated by Notch1,55Bcl6,56,57 or Bcl3.58Bcl6 interferes with the development of lymphoma by repressing the tumor suppressor TP53.59Tnfaip2 and Jak3 have been attributed many functions and participate in hematopoiesis, carcinogenesis, inflammation, and immune responses.60 It is, therefore, tempting to speculate that the observed proinflammatory signature in STAT3 mutant cells results from an altered interplay between NF-κB and STAT3 target genes. The transcriptional repressor and tumor suppressor Cic acts downstream of the RAS/MAPK signaling cascade,61,62 whereas Uap1l1 is involved in glycosylation, thereby promoting cancer cell proliferation.63,64
Specific STAT3 inhibitors are currently not available. To date, the first line of treatment for patients with LGLL is the use of low-dose immunosuppressive agents (eg, methotrexate). Relapse occurs frequently but disease-related deaths are rare and predominantly caused by severe infections.13,48 Recent reports indicate that JAK inhibitors (ruxolitinib or tofacitinib) are beneficial in cases of refractory LGLL.65,66 JAK inhibitors exerts broad effects as they interfere with the entire JAK-STAT signaling pathway.67 More specific ways to block STAT3 are desired, which prompted us to set up a genome-wide LOF screening in WT STAT3 vs mutated STAT3Y640F murine hematopoietic progenitor cells. Interestingly, knock out of some prototypical STAT3 target genes failed to induce a fitness defect and did not represent a selective vulnerability of STAT3Y640F-driven cells in the genome-wide CRISPR/Cas9-based LOF screen in vitro. Among the few selective genetic dependencies of STAT3-mutation-driven cells that we identified, Sbno2 was the only gene that fulfilled all our criteria of a promising target: (1) transcriptionally controlled by STAT3, (2) selective genetic dependency of STAT3Y640F-driven cells, and (3) critical for human STAT3-mutated T-cell and NK-cell malignancies. The analysis of 116 published37 genome-wide CRISPR LOF screens confirmed the codependency between SBNO2 and STAT3. In addition, the STAT3-SBNO2 axis was present in NPM-ALK+ ALCL, which is driven by aberrant STAT3 signaling.9,38 Exceeding STAT3 pathway activation or mutations in STAT3 itself may confer dependency on SBNO2. SBNO2 and its paralog SBNO1 are only poorly characterized. In Drosophila, the sno gene is involved in endothelial growth factor receptor and Notch-related signaling and cell fate determination during development;68-70 in zebrafish, a role in brain development has been reported.71Sbno2 knockout mice are viable but display increased bone mass, because of SBNO2 regulating osteoclast fusion.72 In another study, Sbno2 knockout mice showed a slight decrease in body weight compared with WT but developed normally with no obvious signs of disease.73 In mice, Sbno2 acts as acute inflammatory response gene in astrocytes upon stimulation with IL-635 and in a follow-up study, a protective role for SBNO2 was proposed against hyper-IL-6-induced neurodegeneration.73
SBNO2 consists of a long and a short isoform, the latter being IL-6 inducible.35 Because IL-6 is an activator of STAT3 signaling,2 our findings recapitulate this report; STAT3Y640F enhances expression of the short SBNO2 isoform. Further analysis of publicly available ChIP-seq data sets confirms this concept and provides evidence that cytokines (eg,TPO32) and oncogenes (eg, NPM-ALK40), which hyperactivate STAT3, cause increased binding of STAT3 to the promoter of the short isoform of SBNO2. SBNO2 exerts context-dependent effects, it may induce proinflammatory or anti-inflammatory responses or act as oncogene or tumor suppressor, similar to STAT3.74 The role as suppressor of inflammation has been shown in macrophages, in which SBNO2, induced by IL-10–activated STAT3, inhibits NF-κB–mediated signaling.34 It is attractive to speculate that NF-κB activation disrupts the balance of how STAT3 and SBNO2 act under physiological and pathophysiological conditions. We identified SBNO2 as a selective dependency in STAT3-driven hematological malignancies. SBNO2 downregulation provokes apoptosis of NPM-ALK–rearranged T-ALCL, potentially opening novel therapeutic opportunities.
Acknowledgments
The authors thank Sabine Fajmann, Petra Kudweis, Philipp Jodl, and Isabella Mayer for excellent experimental support. The authors thank the Biomedical Sequencing Facility at the CeMM Research Center for Molecular Medicine at the Austrian Academy of Sciences and the next-generation sequencing facility at Vienna BioCenter Core Facilities, a member of the Vienna BioCenter, Vienna, Austria. The authors also thank Stephan Hutter and Wecke Walter for bioinformatics support in the analysis of primary patient samples.
This work was supported by the Austrian Science Fund (FWF) Special Research Program SFB-F6107 and the European Research Council under the European Union's Horizon 2020 research and innovation program grant agreements (#694354) (V.S.), (#636855) (F.G.), and (#647355) (S.M.). S.M. was supported by the Academy of Finland Heal-Art consortium (314442) and special COVID-19 funding (335527), ERA PerMed (JAKSTAT-TARGET consortium), Sigrid Juselius Foundation, Signe and Ane Gyllenberg Foundation, Helsinki Institute for Life Science, and Cancer Foundation Finland. B.M. was supported by the Fellinger Cancer Research Association.
Authorship
Contribution: V.S., T.B., B.M., and J.S. conceptualized the study; T.B. performed most of the experiments; T.B. and J.S. analyzed all wet laboratory experiments; R.G., T.K., T.E., and J.H. analyzed sequencing data; J.S., E.D., and S.K. performed experiments; G.H. and S.M. provided bioinformatic patient data analysis; J.Z., S.M., G.H., and F.G. were involved in experimental design and scientific discussions; T.B. and V.S. wrote the manuscript; V.S. supervised the study; and all authors revised the manuscript.
Conflict-of-interest disclosure: S.M. has received honoraria and research funding from Novartis, Pfizer, and Bristol-Myers Squibb (not related to this study). G.H. received honoraria from Novartis and Incyte (not related to this study). J.Z. is a founder, shareholder, and scientific adviser of Quantro Therapeutics GmbH (no relation to this study). J.Z. and the Zuber laboratory receive research support and funding from Boehringer Ingelheim. The remaining authors declare no competing financial interests.
Correspondence: Veronika Sexl, Institute of Pharmacology and Toxicology, University of Veterinary Medicine Vienna, Veterinaerplatz 1, 1210 Vienna, Austria; e-mail: veronika.sexl@uibk.ac.at.
References
Author notes
In-house–generated RNA sequencing data from HPC7 and HPCLSK cells as well as V5-STAT3 chromatin immunoprecipitation sequencing data from HPC7 reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE211111).
Bigwig files from STAT3 chromatin immunoprecipitation sequencing from GSE117164 and GSE102317 were used to confirm binding of STAT3 at the SBNO2 promoter. Bigwig files from GSE100835 and GSE158916 were used to show H3K27ac binding.
The RNA sequencing data from healthy patients and those with T-cell large granular lymphocyte leukemia reported in this article are available in the European Genome-phenome Archive (accession number EGAS00001005297).
The RNA sequencing data from patients with natural killer cell leukemia reported in this article are available in the European Genome-phenome Archive (accession number EGAS00001006009).
Microarray and clinical data from patients with NPM-ALK+ ALCL were downloaded from ArrayExpress using the accession code E-TABM-117. Reverse phase protein microarray data and messenger RNA expression data from DepMap were obtained from the Bioconductor depmap package (version 1.11.1; 22Q1 depmap release).
Data are available on request from the corresponding author, Veronika Sexl (veronika.sexl@uibk.ac.at).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![STAT3 mutations enhance proliferation and self-renewal. (A) Schematic overview of most common somatic STAT3 mutations.12,23 (B) Immunoblotting of pY705STAT3, STAT3, V5, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from HPC7 cells expressing empty vector, STAT3, or STAT3 mutants (Y640F, S614R, and D661V). (C) Growth curves of HPC7 cells expressing either empty vector, STAT3, or STAT3 mutants (Y640F, S614R, and D661V) (mean ± standard deviation [SD], n ≥ 3). (D) Representative image of colony formation assay (CFA) of HPCLSK expressing either empty vector, STAT3, or STAT3 mutants (Y640F, S614R, and D661V) (mean ± SD, n ≥ 3. (E) Representative images of colonies 10 days after plating derived from HPCLSK CFA assay, at original magnification ×4. (F) Serial replating CFA from HPC7 expressing either empty vector, STAT3, or STAT3 mutants (Y640F, S614R, and D661V) (mean ± SD, n ≥ 3). (G) Immunoblot of HPC7 cells expressing either empty vector, STAT3, or STAT3Y640 that were cytokine starved for 3 hours and then stimulated with IL-6 for 20 minutes (100 ng/mL), then washed with phosphate-buffered saline (PBS) and plated in cytokine-free media. Cells were harvested at the indicated time points and pY705STAT3, STAT3, V5, and GAPDH expression analyzed. Unpaired, 2-tailed Student t test was used in panel C, and 1-way analysis of variance was used in panel F for P value determination. ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. ND, not determined; PTCL, peripheral T-cell lymphoma; stim, stimulation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/15/10.1182_blood.2022018494/6/m_blood_bld-2022-018494-gr1fg.jpeg?Expires=1770127520&Signature=0ObLVeeB8A1SvHiJ9CuFWuWkPncPjRLe92Q-j6hDJRWXb6BolTNgyM5DWlu0BpKB2dYQD0fXYBVBtbgeRvHJ9~WyIRg5fZA2x4xYJ45tkjW2~eKjUr5Sv0XifxZ-T39FmDdu3BZdyJPbh9flEGI89u4JGrVM4qvZMz4AJAb2KL-v78Ry9qflpZAMei8qToautTg5sdceRsD-fzIGHqBsDgu1MAp51dkVmqkAHmFPBto~AoB3uxny2BeBp~5lmq01A8xgdt01C5z9TwRyNcVdwK4xmKcBHMIkiLUrupyl~aT6WG9ZVFrNb6JVTGFTWSXi2dr9Lv6tAC53LOV2W0PKXw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal