TO THE EDITOR:
B-cell precursor acute lymphoblastic leukemia (BCP-ALL) is a rare aggressive malignancy in adults. BCP-ALL is frequently characterized by recurrent chromosomal translocations that deregulate proto-oncogenes or result in fusion genes encoding chimeric oncoproteins.1 Gene expression profiling studies have consolidated the notion that specific chromosomal aberrations delineate unique leukemia subtypes with distinctive biological and clinical features.2 Therefore, their identification is critical for optimal therapeutic management. The most clinically relevant example is the use of tyrosine kinase inhibitors in patients with the Philadelphia chromosome (Ph) or related BCR-ABL1 fusion, which is found in ∼25% to 30% of adult BCP-ALL. However, apart from Ph-positive ALL, a significant proportion of adult BCP-ALL lacks recurrent classifying chromosomal aberrations, contributing little to current treatment stratification strategies.3 During the last decade, genome-wide copy-number analyses have uncovered numerous genomic microdeletions in BCP-ALL that recurrently target tumor suppressor and lymphoid factor genes like CDKN2A, IKZF1, and PAX5.4 These alterations are the result of erroneous RAG-mediated recombination events that accumulate in transformed progenitor B cells.5,6 By contrast, point mutations are relatively infrequent in BCP-ALL as compared with other tumors, including other hematopoietic malignancies. Remarkably, both microdeletions and point mutations are usually thought to be secondary oncogenic events, whereas the biological and clinical features of BCP-ALL are frequently dictated by the primary cytogenetic aberrations.
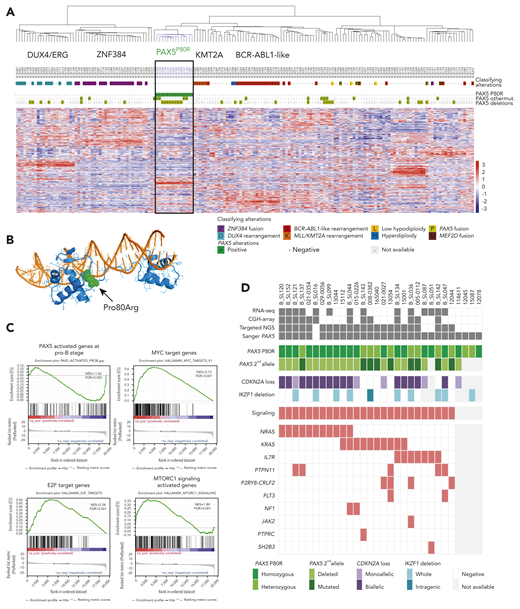
To refine oncogenic subtypes and elucidate primary genetic alterations in adult BCP-ALL, we performed transcriptome analysis by RNA-sequencing (RNA-seq) in a discovery cohort of 170 BCP-ALL cases that were virtually all negative for the common classifying cytogenetic aberrations (details of the study cohorts are provided in supplemental Figure 1, available on the Blood Web site). Unsupervised hierarchical clustering of gene-expression data identified well-defined clusters of BCP-ALL cases (Figure 1A). Four clusters corresponded to known oncogenic subtypes, namely ZNF384,7,8 ERG/DUX4,7-10 BCR-ABL1-like,11 and KMT2A/MLL, as demonstrated by relevant in-frame fusion transcripts or IGH fusions in most cases (including KMT2A fusions, which were cryptic on cytogenetics). An additional cluster comprised 14 cases, which lacked any fusion gene. Mutational analysis of RNA-seq data revealed the presence of a unique PAX5 mutation, c.239C>G, p.P80R, in all cases. PAX5 is a transcription factor from the paired box family that controls B-cell identity and development.12,13 PAX5 is also a haploinsufficient tumor suppressor gene in BCP-ALL14,15 that is targeted by various deletions and mutations in ∼30% of BCP-ALL.4,16,17 Deletions and mutations of PAX5 have been considered so far as secondary oncogenic events because they were found in various BCP-ALL subtypes and possibly in minor subclones. However, we did not identify PAX5P80R mutation in any case from other clusters nor in association with any classifying alteration, suggesting that this specific PAX5 mutation may define a distinct BCP-ALL subtype.
PAX5P80Rmutation defines a subtype of BCP-ALL with distinct transcriptional signature and genomic alterations. (A) Unsupervised hierarchical clustering of RNA-seq gene-expression data of 170 BCP-ALL cases. The color coding of genetic annotations is indicated at the bottom of panel A. (B) Structural modeling of PAX5 P80R mutation based on the crystal structure of the paired box DNA binding domain of PAX5 (PDB 1K7821 ). The Pro80 mutated to Arg (indicated in green) is located in the linker nearby the N-terminal helix. The PD is depicted in blue; the DNA helix is depicted in orange. (C) Gene Set Enrichment Analyses of genes expressed in PAX5P80R cases as compared with other cases. Gene Set Enrichment Analyses show negative enrichment of PAX5 target genes upregulated during early stages of B-cell differentiation (upper left) and positive enrichment of MYC target genes (upper right), E2F target genes (lower left), and MTORC1 signaling genes (lower right). (D) Genomic alterations of the 30 PAX5P80R cases. The color-coding is indicated at the bottom of panel D. FDR, false discovery rate; NES, normalized enrichment score.
PAX5P80Rmutation defines a subtype of BCP-ALL with distinct transcriptional signature and genomic alterations. (A) Unsupervised hierarchical clustering of RNA-seq gene-expression data of 170 BCP-ALL cases. The color coding of genetic annotations is indicated at the bottom of panel A. (B) Structural modeling of PAX5 P80R mutation based on the crystal structure of the paired box DNA binding domain of PAX5 (PDB 1K7821 ). The Pro80 mutated to Arg (indicated in green) is located in the linker nearby the N-terminal helix. The PD is depicted in blue; the DNA helix is depicted in orange. (C) Gene Set Enrichment Analyses of genes expressed in PAX5P80R cases as compared with other cases. Gene Set Enrichment Analyses show negative enrichment of PAX5 target genes upregulated during early stages of B-cell differentiation (upper left) and positive enrichment of MYC target genes (upper right), E2F target genes (lower left), and MTORC1 signaling genes (lower right). (D) Genomic alterations of the 30 PAX5P80R cases. The color-coding is indicated at the bottom of panel D. FDR, false discovery rate; NES, normalized enrichment score.
We analyzed a recurrence cohort consisting of 419 additional, unselected BCP-ALL from patients enrolled in trials conducted by the Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL)18,19 (supplemental Figure 1). By Sanger sequencing, we identified 16 additional PAX5P80R cases that, as anticipated, did not harbor any classifying cytogenetic aberration. Thus, the prevalence of the PAX5P80R subtype in the population of adult patients with newly diagnosed Ph-negative BCP-ALL enrolled in GRAALL trials was 5.3% (29 out of 551).
The Pro80 residue is included in the paired box DNA binding domain (PD) of PAX5 (Figure 1B). Structural modeling suggests that Pro80Arg mutation could potentially disrupt DNA binding by perturbing the PD N-terminal helical domain. Alternatively, it might increase DNA binding through electrostatic interactions with the DNA phosphodiester backbone, leading to an increase in nonspecific DNA binding. To gain insights into the functional consequences of PAX5P80R mutation, we analyzed the gene expression signature of PAX5P80R cases. Using gene set enrichment analysis (Figure 1C), we observed a negative enrichment of the PAX5 target genes activated during early stages of B-cell development,20 in accordance with a loss-of-function effect of the mutation. However, the transcriptional program of PAX5 was not completely abrogated despite biallelic genomic alteration, suggesting that the P80R mutant retained some function. Interestingly, pathway analysis also demonstrated significant enrichment of MYC target genes. Collectively, these data suggest that the effects of P80R mutation may involve both partial loss of function of PAX5 on its target genes and activation of an aberrant transcriptional program.
We analyzed additional genomic alterations in the 30 PAX5P80R cases (Figure 1D). The karyotype frequently displayed structural rearrangements of chromosomal arms 9p (n = 15/30) and/or 7p (n = 11/30) such as dic(9;20) and der(7;9) (supplemental Table 1). PAX5 gene is located at 9p13.2, and homozygosity of P80R was indeed observed in 19 cases. All heterozygote remaining cases with available data (n = 9/11) harbored a second PAX5 mutation (3 frameshifts, 3 missenses, 1 nonsense, and 2 splice variants) leading to PAX5 biallelic alteration. IKZF1 located at 7p12.2 was also frequently lost. Strikingly, all the PAX5P80R cases harbored mutations in signal transduction factor genes, especially in the RAS pathway. Copy-number data showed remarkably few additional focal alterations (median 1 per case, range 0-8) except focal deletions of the second CDKN2A allele located at 9p21, resulting in biallelic loss. The transcriptional signature of the PAX5P80R subtype showed significant enrichment in MYC and E2F target genes, the latter in agreement with the loss of repression by CDKN2A, and in genes activated by the mTORC1 complex, in agreement with RAS-mediated activation of mTOR signaling (Figure 1C). Altogether, the PAX5P80R subtype was characterized by a distinct transcriptional signature in relation with a unique combination of genomic alterations targeting PAX5, CDKN2A, and RAS.
Clinical features and outcome associated with the PAX5P80R BCP-ALL subtype were analyzed within a cohort of 312 patients with Ph-negative BCP-ALL enrolled in the GRAALL-2003 and GRAALL-2005 trials18,19 (supplemental Figure 1). The patients and initial disease characteristics (supplemental Table 2) were not different in the PAX5P80R group, with the exception of a lower frequency of IKZF1 intragenic deletion (0% vs 27.0%, P = .025). PAX5P80R patients had a good peripheral blood prednisone response at day 8 (100% vs 83.9%, P = .14), and they had a significantly better bone marrow blast clearance at day 15 (92.9% vs 56.5% in the remaining cohort, P = .010). At the end of induction, all PAX5P80R patients achieved complete remission (100% vs 94.3%, P = 1.0) and good molecular response, none of them having minimal residual disease (MRD) higher than 10−3 (MRD1 ≥10−3, 0% vs 31.2%, P = .036). The median follow-up for outcome analyses was 5.3 years. When censoring at allogeneic stem cell transplants, PAX5P80R patients had a significantly higher probability of 5-year event-free survival (EFS; 81.8% vs 42.7%; hazard ratio = 0.20, 95% confidence interval 0.05-0.83; P = .026) and 5-year overall survival (OS; 90.9% vs 47.0%; hazard ratio = 0.12, 95% confidence interval 0.02-0.87; P = .036) (Figure 2A-B). This difference remains without censoring at allogeneic transplantation (Figure 2C-D; supplemental Figure 2). Therefore, PAX5P80R characterizes a distinct genetic subtype with a favorable prognosis, at least in the context of a high-intensity, pediatric-inspired chemotherapy regimen.
Outcome analyses for PAX5P80Rpatients. (A) EFS and (B) OS, after censoring patients at the time of allogeneic stem cell transplantation, according to the presence of PAX5P80R. (C) EFS and (D) OS of PAX5P80R cases as compared with defined cytogenetic subtypes, without censoring patients who received allogeneic stem cell transplantation.
Outcome analyses for PAX5P80Rpatients. (A) EFS and (B) OS, after censoring patients at the time of allogeneic stem cell transplantation, according to the presence of PAX5P80R. (C) EFS and (D) OS of PAX5P80R cases as compared with defined cytogenetic subtypes, without censoring patients who received allogeneic stem cell transplantation.
In conclusion, we report here the first oncogenic subtype defined by a point mutation in ALL, which is also the first good-risk genetic subtype in adult BCP-ALL. In addition to the PAX5P80R mutation, this subtype is typically associated with inactivation of the second PAX5 allele, biallelic deletion of CDKN2A, and alterations of the RAS pathway. Systematic screening at diagnostic workup may be easily implemented in order to refine risk-adapted treatment stratification.
The online version of this article contains a data supplement.
Acknowledgments
The authors thank the patients and all the GRAALL investigators and biologists who contributed samples and data for this study. The authors thank Céline Beugnon for data collection, and Kelly Vanoukia and Agnès Pérus for their excellent technical assistance. The authors are grateful to Lawrence McIntosh for insightful exchange.
The Swiss investigators were supported by the Swiss State Secretariat for Education, Research, and Innovation, Switzerland. This work was supported by a grant from the Journée Nationale Contre la Leucémie 2014 and by a grant from Amgen.
Authorship
Contribution: M.P. performed experiments, analyzed data, and wrote the manuscript; N.B. performed statistical analyses, analyzed data, and wrote the manuscript; F.S. performed bioinformatics analyses, analyzed data, and wrote the manuscript; I.B. performed experiments and analyzed data; C.S., M.B., X.T., C.G., Y.C., T.L., E.L., and J.K. treated patients and provided clinical data; S.Q. performed bioinformatics analyses; M.L.-P. reviewed cytogenetics data; E.D., C.P., N.G., and V.A. provided samples and performed MRD analyses; V.L. managed the GRAALL organization; J.S. contributed to study design and wrote the manuscript; H.D. led the GRAALL, contributed to study design, and wrote the manuscript; E.C. designed research, analyzed data, and wrote the manuscript; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Emmanuelle Clappier, Laboratoire d’Hématologie, Hôpital Saint-Louis, 1 Avenue Claude Vellefaux, 75010 Paris, France; e-mail: emmanuelle.clappier@aphp.fr.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal