Abstract
Acute lymphoblastic leukemia (ALL) is the commonest childhood tumor and remains a leading cause of cancer death in the young. In the last decade, microarray and sequencing analysis of large ALL cohorts has revolutionized our understanding of the genetic basis of this disease. These studies have identified new ALL subtypes, each characterized by constellations of structural and sequence alterations that perturb key cellular pathways, including lymphoid development, cell-cycle regulation, and tumor suppression; cytokine receptor, kinase, and Ras signaling; and chromatin modifications. Several of these pathways, particularly kinase-activating lesions and epigenetic alterations, are logical targets for new precision medicine therapies. Genomic profiling has also identified important interactions between inherited genetic variants that influence the risk of leukemia development and the somatic genetic alterations that are required to establish the leukemic clone. Moreover, sequential sequencing studies at diagnosis, remission, and relapse have provided important insights into the relationship among genetic variants, clonal heterogeneity, and the risk of relapse. Ongoing studies are extending our understanding of coding and noncoding genetic alterations in B-progenitor and T-lineage ALL and using these insights to inform the development of faithful experimental models to test the efficacy of new treatment approaches.
Introduction
Acute lymphoblastic leukemia (ALL) is the most common childhood cancer.1 Five-year survival rates now exceed 85% in children, but survival following relapse is poor.2 ALL comprises multiple entities with distinct genetic alterations, clinical features, and treatment response. Over the last decade, studies utilizing microarray analysis of gene expression, DNA copy-number alterations, and next-generation sequencing have provided major insights into the pathogenesis and clinical behavior of ALL.3-22 Most ALL genomes harbor sequence and structural DNA alterations involving coding genes, as well as alterations of noncoding elements such as noncoding RNAs23 and enhancer elements.24,25
Here, we consider results from these studies in several categories: (1) identification of new subtypes of ALL that lack recurring gross chromosomal alterations; (2) characterization of the constellations of genetic alterations that define each ALL subtype; (3) the relationship between genetic alterations, clonal heterogeneity, and relapse; (4) identification of inherited genetic variants and mutations linked to ALL susceptibility and outcome; (5) and translating new discoveries to improved diagnostic, prognostic, and precision medicine approaches.
Genetic subtypes of ALL
Gross chromosomal alterations are a hallmark of ALL (Table 1).26 High hyperdiploidy (>50 chromosomes) occurs in 25% to 30% of childhood B-cell precursor ALL (B-ALL) and is associated with favorable outcome. Hypodiploidy with <44 chromosomes is uncommon (2% to 3% of cases) and is associated with poor outcome.27,28
Key subtypes of ALL
| Subtype . | Prevalence (%)* . | Comment . |
|---|---|---|
| B-cell precursor ALL | ||
| Hyperdiploidy with >50 chromosomes | 20-30 | Excellent prognosis |
| Hypodiploidy with <44 chromosomes | 2-3 | Poor prognosis; high frequency of Ras pathway and Ikaros gene family mutations |
| t(12;21)(p13;q22) translocation encoding ETV6-RUNX1 | 15-25 | Excellent prognosis |
| t(1;19)(q23;p13) translocation encoding TCF3-PBX1 | 2-6 | Increased incidence in African-Americans; generally excellent prognosis; association with CNS relapse |
| t(9;22)(q34;q11.2) translocation encoding BCR-ABL1 | 2-4 | Historically poor outcome; improved with addition of imatinib and/or dasatinib to intensive chemotherapy |
| Ph-like ALL | 10-15 | Multiple kinase-activating lesions; associated with older age, elevated white blood cell count, and IKZF1 alteration; potentially amenable to TKI therapy |
| t(4;11)(q21;q23) translocation encoding MLL-AF4 fusion | 1-2 | Common in infant ALL (especially age <6 mo); poor prognosis |
| t(8;14)(q24;q32), t(2;8)(q12;q24), t(2;8)(q12;q24) encoding; MYC rearrangement | 2 | Favorable prognosis with short-term high-dose chemotherapy |
| CRLF2 rearrangement (IGH-CRLF2; P2RY8-CRLF2) | 5-7 | Common in Down syndrome–associated and Ph-like ALL (∼50% each); associated with IKZF1 deletion and/or mutation and JAK1/2 mutation and poor prognosis in non–Down syndrome–associated ALL |
| ERG-dysregulated ALL | ∼7 | Distinct gene expression profile; the majority have focal ERG deletions and favorable outcome despite IKZF1 alterations |
| PAX5 rearrangement | ∼2 | Multiple partners, commonly from dic(7;9), dic(9;12), and dic(9;20) |
| iAMP21 | ∼2 | Complex structural alterations of chromosome 21; rarely associated with a constitutional Robertsonian translocation rob(15;21)(q10;q10)c; poor prognosis |
| T-lineage ALL | ||
| t(1;7)(p32;q35) and t(1;14)(p32;q11) translocations and interstitial 1p32 deletion; TAL1 dysregulation | 15-18 | Generally favorable outcome |
| t(11;14)(p15;q11) translocation and 5′ LMO2 deletion; LMO2 dysregulation | 10 | Generally favorable outcome |
| t(10;14)(q24;q11) and t(7;10)(q35;q24) translocations; TLX1 [HOX11] dysregulation | 7 | Good prognosis |
| t(5;14)(q35;q32) translocation; TLX3 dysregulation | 20 | Commonly fused to BCL11B, also a target of deletion and/or mutation; poor prognosis |
| t(10;11)(p13;q14) translocation; PICALM-MLLT10 [CALM-AF10] | 10 | May have poor outcome |
| MLL-MLLT1 [MLL-ENL] | 2-3 | Superior prognosis to other types of MLL-rearranged leukemia |
| 9q34 amplification encoding NUP214-ABL1 | 6 | Potentially amenable to TKIs, also identified in high-risk B-ALL; other kinase fusions identified in T-ALL include EML1-ABL1, ETV6-JAK2, and ETV6-ABL1 |
| t(7;9)(q34;q34) translocation | <1 | Rearrangement of NOTCH1; also sequence mutations in >50% T-ALL |
| Early T-cell precursor ALL | 10-15 | Immature immunophenotype; expression of myeloid and/or stem cell markers; historically poor outcome, although improved in recent studies; genetically heterogeneous with mutations in hematopoietic regulators, cytokine, and Ras signaling, and epigenetic modifiers |
| Subtype . | Prevalence (%)* . | Comment . |
|---|---|---|
| B-cell precursor ALL | ||
| Hyperdiploidy with >50 chromosomes | 20-30 | Excellent prognosis |
| Hypodiploidy with <44 chromosomes | 2-3 | Poor prognosis; high frequency of Ras pathway and Ikaros gene family mutations |
| t(12;21)(p13;q22) translocation encoding ETV6-RUNX1 | 15-25 | Excellent prognosis |
| t(1;19)(q23;p13) translocation encoding TCF3-PBX1 | 2-6 | Increased incidence in African-Americans; generally excellent prognosis; association with CNS relapse |
| t(9;22)(q34;q11.2) translocation encoding BCR-ABL1 | 2-4 | Historically poor outcome; improved with addition of imatinib and/or dasatinib to intensive chemotherapy |
| Ph-like ALL | 10-15 | Multiple kinase-activating lesions; associated with older age, elevated white blood cell count, and IKZF1 alteration; potentially amenable to TKI therapy |
| t(4;11)(q21;q23) translocation encoding MLL-AF4 fusion | 1-2 | Common in infant ALL (especially age <6 mo); poor prognosis |
| t(8;14)(q24;q32), t(2;8)(q12;q24), t(2;8)(q12;q24) encoding; MYC rearrangement | 2 | Favorable prognosis with short-term high-dose chemotherapy |
| CRLF2 rearrangement (IGH-CRLF2; P2RY8-CRLF2) | 5-7 | Common in Down syndrome–associated and Ph-like ALL (∼50% each); associated with IKZF1 deletion and/or mutation and JAK1/2 mutation and poor prognosis in non–Down syndrome–associated ALL |
| ERG-dysregulated ALL | ∼7 | Distinct gene expression profile; the majority have focal ERG deletions and favorable outcome despite IKZF1 alterations |
| PAX5 rearrangement | ∼2 | Multiple partners, commonly from dic(7;9), dic(9;12), and dic(9;20) |
| iAMP21 | ∼2 | Complex structural alterations of chromosome 21; rarely associated with a constitutional Robertsonian translocation rob(15;21)(q10;q10)c; poor prognosis |
| T-lineage ALL | ||
| t(1;7)(p32;q35) and t(1;14)(p32;q11) translocations and interstitial 1p32 deletion; TAL1 dysregulation | 15-18 | Generally favorable outcome |
| t(11;14)(p15;q11) translocation and 5′ LMO2 deletion; LMO2 dysregulation | 10 | Generally favorable outcome |
| t(10;14)(q24;q11) and t(7;10)(q35;q24) translocations; TLX1 [HOX11] dysregulation | 7 | Good prognosis |
| t(5;14)(q35;q32) translocation; TLX3 dysregulation | 20 | Commonly fused to BCL11B, also a target of deletion and/or mutation; poor prognosis |
| t(10;11)(p13;q14) translocation; PICALM-MLLT10 [CALM-AF10] | 10 | May have poor outcome |
| MLL-MLLT1 [MLL-ENL] | 2-3 | Superior prognosis to other types of MLL-rearranged leukemia |
| 9q34 amplification encoding NUP214-ABL1 | 6 | Potentially amenable to TKIs, also identified in high-risk B-ALL; other kinase fusions identified in T-ALL include EML1-ABL1, ETV6-JAK2, and ETV6-ABL1 |
| t(7;9)(q34;q34) translocation | <1 | Rearrangement of NOTCH1; also sequence mutations in >50% T-ALL |
| Early T-cell precursor ALL | 10-15 | Immature immunophenotype; expression of myeloid and/or stem cell markers; historically poor outcome, although improved in recent studies; genetically heterogeneous with mutations in hematopoietic regulators, cytokine, and Ras signaling, and epigenetic modifiers |
CNS, central nervous system.
Frequencies refer to childhood ALL.
Chromosomal rearrangements creating chimeric fusion genes commonly involve hematopoietic transcription factors, epigenetic modifiers, cytokine receptors, and tyrosine kinases.26 Common rearrangements in B-lineage ALL are the t(12;21)(p13;q22) encoding ETV6-RUNX1 (TEL-AML1), t(1;19)(q23;p13) encoding TCF3-PBX1 (E2A-PBX1), t(9;22)(q34;q11.2) resulting in formation of the “Philadelphia” chromosome (Ph) encoding BCR-ABL1, rearrangements of MLL (KMT2A) at 11q23 to a range of fusion partners, and rearrangement of the cytokine receptor gene CRLF2 at the pseudoautosomal region 1 (PAR1) at Xp22.3/Yp11.3.6,29 Approximately 20% of childhood B-ALL cases lack one of these alterations and have alternative sentinel genetic lesions, including deregulation of the ETS-family transcription factor ERG, or one of a diverse range of alterations that drive kinase signaling in Ph-like ALL. T-ALL is characterized by activating mutations of NOTCH1 and rearrangements of transcription factors TLX1 (HOX11), TLX3 (HOX11L2), LYL1, TAL1, and MLL (Table 1).
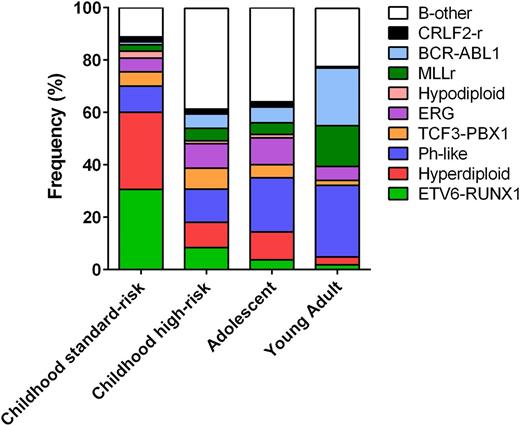
The prevalence of these subtypes varies with age (Figure 1). High hyperdiploidy and ETV6-RUNX1 are each present in 25% to 30% of childhood ALL cases but occur in <3% of young adults (age 21-39 years). Conversely, BCR-ABL1–positive ALL comprises 2% to 5% of childhood vs one-quarter of adult ALL. Ph-like ALL rises in prevalence from 10% of children with standard-risk ALL to over 25% of young adults.7,22,30
Prevalence of ALL subtypes across age groups. The prevalence of ALL subtypes varies between children with standard-risk ALL (age 1-9 years, white blood cell count <50 × 109/L), children with high-risk ALL (age 10-15 years and/or white blood cell count >50 × 109/L), adolescents (age 16-20 years), and young adults with ALL (age 21-39 years). B-other, B-cell ALL with other subtypes.
Prevalence of ALL subtypes across age groups. The prevalence of ALL subtypes varies between children with standard-risk ALL (age 1-9 years, white blood cell count <50 × 109/L), children with high-risk ALL (age 10-15 years and/or white blood cell count >50 × 109/L), adolescents (age 16-20 years), and young adults with ALL (age 21-39 years). B-other, B-cell ALL with other subtypes.
Frequently mutated pathways in ALL
Most ALL cases harbor multiple somatic genetic alterations in addition to gross chromosomal alterations (Table 2 and Figure 2).3,4,7 Chromosomal rearrangements and aneuploidy are early events in leukemogenesis, with DNA copy-number alterations and sequence mutations acquired subsequently. This is supported by several observations: (1) monochorionic twins concordant for an ALL chimeric fusion but discordant for secondary genetic alterations,31 (2) the universal presence of translocations or aneuploidy in ALL cells from a given patient or patient-derived xenograft but variability in submicroscopic genetic alterations,32,33 (3) genomic studies of matched diagnosis and relapse samples that typically show concordance for translocations in each patient at each time point but evolution in additional genetic changes,34 and (4) sequencing studies showing that many ALL cases harbor multiple subclonal sequence mutations in individual genes but constancy of chromosomal rearrangements.35
Key genetic alterations in ALL
| Gene . | Alteration . | Frequency . | Pathway and consequences of alteration . | Clinical relevance . | Reference . |
|---|---|---|---|---|---|
| PAX5 | Focal deletions, translocations, and sequence mutations | 30% of B-ALL | Transcription factor required for B-lymphoid development; mutations impair DNA binding and transcriptional activation | Important in leukemogenesis, but not associated with adverse outcome | 3-5 |
| IKZF1 | Focal deletions or sequence mutations | 15% of all pediatric B-ALL cases, including 70%-80% of BCR-ABL1-positive ALL and one-third of high-risk BCR-ABL1-negative B-ALL | Transcription factor required for development of HSC to lymphoid precursor; deletions and mutations result in loss of function or dominant-negative isoforms | Associated with poor outcome | 4, 5, 7, 51, 102 |
| JAK1/2 | Pseudokinase and kinase domain mutations | 18%-35% of DS ALL and 10.7% high-risk BCR-ABL1-negative ALL | Constitutive JAK-STAT activation | Potential for targeting with TKIs that inhibit JAK1/2 | 8, 39, 43 |
| CRLF2 | Rearrangement as IGH-CRLF2 or P2RY8-CRLF2 resulting in overexpression | 5%-16% of pediatric and adult B-ALL and >50% DS ALL; 50% of Ph-like ALL | Associated with mutant JAK in up to 50% of cases; CRLF2 mutations and JAK mutations are cotransforming in cell lines/F3 cells and result in constitutive STAT activation; in high-risk B-ALL, associated with IKZF1 alterations, JAK mutations, and poor outcome | Detected by flow cytometry or molecular assays; potential for targeting with JAK inhibitors | 6, 29, 41, 42, 44 |
| Kinase-activating alterations in Ph-like ALL | Rearrangements of 13 cytokine receptors and tyrosine kinases; sequence mutations of IL7R and FLT; deletions of SH2B3 | 10% childhood B-ALL, up to 30% ALL in adult ALL; associated with Ph-like gene expression profile | Activation of kinase signaling pathways and amenable to TKI therapy | Associated with high-risk features and increased risk of relapse; anecdotal reports of response to TKI therapy | 14, 22, 30, 54 |
| NOTCH1 | Mutations, rarely deletions | >60% T-ALL | Activation of NOTCH1 signaling; mutations in FBXW7 and PTEN also influence NOTCH1 signaling and prognosis | Key pathogenic lesion in T-ALL; direct targeting limited by toxicity; variable associations with outcome | 62 |
| CREBBP | Focal deletion and sequence mutations | 19% of relapsed ALL; also mutated in non-Hodgkin lymphoma | Mutations result in impaired histone acetylation and transcriptional regulation | Mutations selected for at relapse and associated with glucocorticoid resistance | 11, 35 |
| NT5C2 | Focal mutations | Up to 20% relapsed mutation | Mutations in gain of function | Mutations selected at relapse and associated with resistance to thiopurines | 17, 84 |
| TP53 | Deletions and focal mutations | ∼90% of LH ALL, otherwise uncommon in predominant clone at diagnosis | Frequently germline in LH ALL; confer resistance to therapy | Hallmark of LH ALL at diagnosis; associated with disease relapse | 15, 83 |
| Gene . | Alteration . | Frequency . | Pathway and consequences of alteration . | Clinical relevance . | Reference . |
|---|---|---|---|---|---|
| PAX5 | Focal deletions, translocations, and sequence mutations | 30% of B-ALL | Transcription factor required for B-lymphoid development; mutations impair DNA binding and transcriptional activation | Important in leukemogenesis, but not associated with adverse outcome | 3-5 |
| IKZF1 | Focal deletions or sequence mutations | 15% of all pediatric B-ALL cases, including 70%-80% of BCR-ABL1-positive ALL and one-third of high-risk BCR-ABL1-negative B-ALL | Transcription factor required for development of HSC to lymphoid precursor; deletions and mutations result in loss of function or dominant-negative isoforms | Associated with poor outcome | 4, 5, 7, 51, 102 |
| JAK1/2 | Pseudokinase and kinase domain mutations | 18%-35% of DS ALL and 10.7% high-risk BCR-ABL1-negative ALL | Constitutive JAK-STAT activation | Potential for targeting with TKIs that inhibit JAK1/2 | 8, 39, 43 |
| CRLF2 | Rearrangement as IGH-CRLF2 or P2RY8-CRLF2 resulting in overexpression | 5%-16% of pediatric and adult B-ALL and >50% DS ALL; 50% of Ph-like ALL | Associated with mutant JAK in up to 50% of cases; CRLF2 mutations and JAK mutations are cotransforming in cell lines/F3 cells and result in constitutive STAT activation; in high-risk B-ALL, associated with IKZF1 alterations, JAK mutations, and poor outcome | Detected by flow cytometry or molecular assays; potential for targeting with JAK inhibitors | 6, 29, 41, 42, 44 |
| Kinase-activating alterations in Ph-like ALL | Rearrangements of 13 cytokine receptors and tyrosine kinases; sequence mutations of IL7R and FLT; deletions of SH2B3 | 10% childhood B-ALL, up to 30% ALL in adult ALL; associated with Ph-like gene expression profile | Activation of kinase signaling pathways and amenable to TKI therapy | Associated with high-risk features and increased risk of relapse; anecdotal reports of response to TKI therapy | 14, 22, 30, 54 |
| NOTCH1 | Mutations, rarely deletions | >60% T-ALL | Activation of NOTCH1 signaling; mutations in FBXW7 and PTEN also influence NOTCH1 signaling and prognosis | Key pathogenic lesion in T-ALL; direct targeting limited by toxicity; variable associations with outcome | 62 |
| CREBBP | Focal deletion and sequence mutations | 19% of relapsed ALL; also mutated in non-Hodgkin lymphoma | Mutations result in impaired histone acetylation and transcriptional regulation | Mutations selected for at relapse and associated with glucocorticoid resistance | 11, 35 |
| NT5C2 | Focal mutations | Up to 20% relapsed mutation | Mutations in gain of function | Mutations selected at relapse and associated with resistance to thiopurines | 17, 84 |
| TP53 | Deletions and focal mutations | ∼90% of LH ALL, otherwise uncommon in predominant clone at diagnosis | Frequently germline in LH ALL; confer resistance to therapy | Hallmark of LH ALL at diagnosis; associated with disease relapse | 15, 83 |
DS, Down syndrome; HSC, hematopoietic stem cell; NR, none reported.
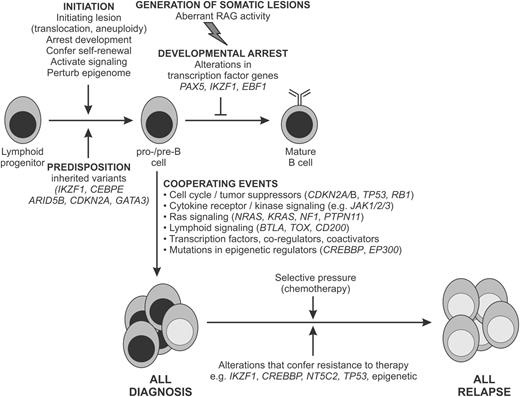
Schema for the genetic pathogenesis of B-ALL. Key inherited and somatic genomic variants and their relationship to the development of ALL and treatment failure are shown. Representative alterations are shown.
Schema for the genetic pathogenesis of B-ALL. Key inherited and somatic genomic variants and their relationship to the development of ALL and treatment failure are shown. Representative alterations are shown.
The frequency of somatic mutations in ALL is low (<20 nonsilent mutations per case),13,15,18,22 with exceptions being occasional relapse cases with hypermutator phenotypes.35 Indeed, MLL-rearranged infant ALL has one of the lowest mutational frequencies in human cancer.36 The most common type of structural genetic alterations in ALL are focal deletions arising from aberrant activity of the recombinase activating genes.5,20
Genes encoding transcriptional regulators of lymphoid development are among the most frequently mutated genes, particularly in B-ALL. These include PAX5, IKZF1, and EBF1 that encode DNA-binding transcription factors required for lymphoid development. Most common are somatic deletions or sequence mutations of PAX5 and IKZF1, and less common are translocations of PAX5 and deletions of EBF1.4 Each results in loss of function or expression of dominant-negative alleles and impaired lymphoid maturation that contributes to leukemogenesis.37,38
Additional commonly altered pathways in ALL include tumor suppression and cell-cycle regulation (TP53, RB1, and CDKN2A/CDKN2A); cytokine receptor, tyrosine kinase, and Ras signaling (ABL1, ABL2, CRLF2, CSF1R, EPOR, FLT3, IL2RB, IL7R, JAK1/2/3, NTRK3, and PDGFRB), Ras signaling (NF1, KRAS, NRAS, and PTPN11), and epigenetic modification (EZH2, CREBBP, SETD2, MLL2 [KMT2D], and NSD2 [WHSC1]).4,7,39 The type of alteration (chromosomal rearrangement, deletion/amplification, or sequence mutation) and gene involved varies between subtypes.39 For example, IKZF1 alterations are a hallmark of BCR-ABL1–positive and Ph-like ALL5 ; in contrast, other members of the IKAROS transcription factor family, IKZF2 and IKZF3, are selectively mutated in hypodiploid ALL.15 Several key genetic alterations are predictive of outcome, including the association of IKZF1 alterations with treatment failure in both BCR-ABL1–positive and Ph-like ALL.7,40
Alterations driving kinase signaling and Ph-like ALL
Ph-like ALL cases lack BCR-ABL1, exhibit a gene expression profile similar to BCR-ABL1–positive ALL, harbor alterations of B lymphoid transcription factor genes, and have poor outcome.7,30 The prevalence of Ph-like ALL increases with age and varies according to ethnicity, in part because CRLF2 alterations are associated with Hispanic ethnicity and native American genetic ancestry.41
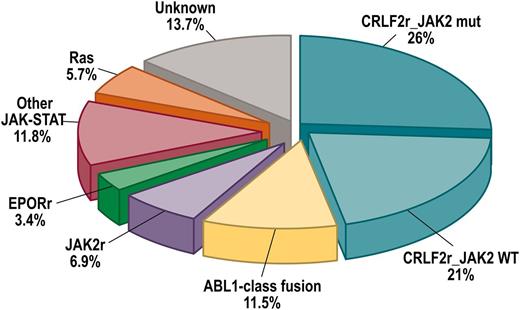
Ph-like ALL is characterized by a diverse range of genetic alterations that dysregulate cytokine receptor and tyrosine kinase signaling (Table 3 and Figure 3).14,22 Analysis of over 1700 cases of B-ALL identified several types of kinase alteration in Ph-like ALL: rearrangements of CRLF2 (47% of cases), rearrangements of ABL-class tyrosine kinase genes (12%), rearrangements of JAK2 (7%) and EPOR (10%), mutations activating Janus kinase and signal transducer and activator of transcription (JAK-STAT) signaling (11%) and Ras (6%), and less common kinase alterations (NTRK3 and PTK2B).
Kinase rearrangements and therapeutic targets in Ph-like ALL
| Kinase . | Potential TKI . | Number of gene partners . | Number of cases . | 5′ genes . |
|---|---|---|---|---|
| ABL1 | Dasatinib | 6 | 14 | ETV6, NUP214, RCSD1, RANBP2, SNX2, and ZMIZ1 |
| ABL2 | Dasatinib | 3 | 7 | PAG1, RCSD1, and ZC3HAV1 |
| CSF1R | Dasatinib | 1 | 4 | SSBP2 |
| PDGFRB | Dasatinib | 4 | 11 | EBF1, SSBP2, TNIP1, and ZEB2 |
| CRLF2 | JAK2 inhibitor | 2 | 30 | IGH and P2RY8 |
| JAK2 | JAK2 inhibitor | 10 | 19 | ATF7IP, BCR, EBF1, ETV6, PAX5, PPFIBP1, SSBP2, STRN3, TERF2, and TPR |
| EPOR | JAK2 inhibitor | 2 | 9 | IGH and IGK |
| DGKH | Unknown | 1 | 1 | ZFAND3 |
| IL2RB | JAK1/JAK3 inhibitor | 1 | 1 | MYH9 |
| NTRK3 | Crizotinib | 1 | 1 | ETV6 |
| PTK2B | FAK inhibitor | 2 | 1 | KDM6A and STAG2 |
| TSLP | JAK2 inhibitor | 1 | 1 | IQGAP2 |
| TYK2 | TYK2 inhibitor | 1 | 1 | MYB |
| Kinase . | Potential TKI . | Number of gene partners . | Number of cases . | 5′ genes . |
|---|---|---|---|---|
| ABL1 | Dasatinib | 6 | 14 | ETV6, NUP214, RCSD1, RANBP2, SNX2, and ZMIZ1 |
| ABL2 | Dasatinib | 3 | 7 | PAG1, RCSD1, and ZC3HAV1 |
| CSF1R | Dasatinib | 1 | 4 | SSBP2 |
| PDGFRB | Dasatinib | 4 | 11 | EBF1, SSBP2, TNIP1, and ZEB2 |
| CRLF2 | JAK2 inhibitor | 2 | 30 | IGH and P2RY8 |
| JAK2 | JAK2 inhibitor | 10 | 19 | ATF7IP, BCR, EBF1, ETV6, PAX5, PPFIBP1, SSBP2, STRN3, TERF2, and TPR |
| EPOR | JAK2 inhibitor | 2 | 9 | IGH and IGK |
| DGKH | Unknown | 1 | 1 | ZFAND3 |
| IL2RB | JAK1/JAK3 inhibitor | 1 | 1 | MYH9 |
| NTRK3 | Crizotinib | 1 | 1 | ETV6 |
| PTK2B | FAK inhibitor | 2 | 1 | KDM6A and STAG2 |
| TSLP | JAK2 inhibitor | 1 | 1 | IQGAP2 |
| TYK2 | TYK2 inhibitor | 1 | 1 | MYB |
Data are from Roberts et al.22
Frequency of subtypes of Ph-like ALL. Combined prevalence of Ph-like ALL subtypes in children, adolescents, and young adults including CRFL2-rearranged JAK2 mutant (CRLF2r_JAK2 mut) and CRFL2-rearranged JAK2 wild-type (CRFL2r_JAK2 WT), ABL1-class rearrangements (ABL1, ABL2, CSF1R, and PDGFRB), JAK2 and EPOR rearrangements and other mutations in JAK-STAT signaling (FLT3, IL7R, SH2B3, JAK1/3, TYK2, IL2RB, and TSLP), Ras mutations (KRAS, NRAS, NF1, PTPN11, and BRAF), and unknown alterations. Data from Roberts et al.22 HR, high-risk.
Frequency of subtypes of Ph-like ALL. Combined prevalence of Ph-like ALL subtypes in children, adolescents, and young adults including CRFL2-rearranged JAK2 mutant (CRLF2r_JAK2 mut) and CRFL2-rearranged JAK2 wild-type (CRFL2r_JAK2 WT), ABL1-class rearrangements (ABL1, ABL2, CSF1R, and PDGFRB), JAK2 and EPOR rearrangements and other mutations in JAK-STAT signaling (FLT3, IL7R, SH2B3, JAK1/3, TYK2, IL2RB, and TSLP), Ras mutations (KRAS, NRAS, NF1, PTPN11, and BRAF), and unknown alterations. Data from Roberts et al.22 HR, high-risk.
ABL-class rearrangements encode fusion genes involving ABL1, ABL2 (ARG), CSF1R (encoding the macrophage colony stimulating factor receptor), and PDGFRB.14,22 Multiple fusion partners have been identified, each involving the kinase as the downstream partner and preserving an intact kinase domain (Figure 4). JAK2 is rearranged to at least 14 different partner genes in Ph-like ALL. EPOR rearrangements include reciprocal or cryptic translocations with immunoglobulin and other loci (eg, IGH and IGK) that deregulate receptor expression and also truncate EPOR, causing increased JAK-STAT signaling.14,22
ABL1-class rearrangements in Ph-like ALL. The figure shows each ALL sample as a column, each kinase rearrangement as a green box, and the diverse range of fusion partners in blue. On the right, representative schema of fusion proteins are shown, showing preservation of the kinase domain in the C terminus of the protein and fusion partners uniformly in the 5′ region of the protein, with domains that mediate overexpression of the kinase, cellular mislocalization, and dimerization of the fusion protein. TKD, tyrosine kinase domain.
ABL1-class rearrangements in Ph-like ALL. The figure shows each ALL sample as a column, each kinase rearrangement as a green box, and the diverse range of fusion partners in blue. On the right, representative schema of fusion proteins are shown, showing preservation of the kinase domain in the C terminus of the protein and fusion partners uniformly in the 5′ region of the protein, with domains that mediate overexpression of the kinase, cellular mislocalization, and dimerization of the fusion protein. TKD, tyrosine kinase domain.
CRLF2 encodes cytokine receptor like factor 2, also known as the thymic stromal-derived lymphopoietin receptor (TSLPR) that forms a heterodimeric receptor with the interleukin-7 receptor α chain (IL7Rα) for thymic stromal lymphopoietin (TSLP). CRLF2 is rearranged to the immunoglobulin heavy chain locus (IGH-CRLF2) or by a deletion upstream of CRLF2 that results in expression of P2RY8-CRLF2.6,29,41 CRLF2 rearrangements are most common in Ph-like and Down syndrome–associated ALL and are age dependent, with P2RY8-CRLF2 associated with young age and IGH-CRLF2 with older age and Hispanic ancestry.6,29 Less common are activating CRLF2 point mutations (eg, p.Phe232Cys).42 The majority of CRLF2-rearranged cases have additional alterations driving JAK-STAT or Ras signaling, particularly activating JAK1 or JAK2 mutations.6,29,43 The most frequent site of mutation is p.Arg683 in the JAK2 pseudokinase domain, but mutations also occur in the pseudokinase domain of JAK1 and the kinase domains of JAK1 and JAK2.8 Other mutations observed in CRLF2-rearranged cases include IL7R sequence mutations, SH2B3 deletions, TSLP rearrangements, and Ras mutations.8,22
In most studies, CRLF2 rearrangements are associated with poor prognosis, particularly in cases with concomitant IKZF1 alteration.41,44 CRLF2-rearranged cells exhibit activated JAK-STAT, phosphatidylinositol 3-kinase (PI3K)/mechanistic target of rapamycin (mTOR), and BCL-2 signaling, and therapies targeting these pathways alone or in combination have shown efficacy in preclinical models.45,46
Other alterations that activate kinase signaling in Ph-like ALL include those activating IL7R, FLT3, IL2RB (interleukin 2 receptor subunit β), JAK1 or JAK3, or loss-of-function deletions/mutations of SH2B3 (LNK), a negative regulator of JAK-STAT signaling.47 IL7R mutations are also observed in T-cell precursor ALL (T-ALL) and B-ALL.13,48,49 These mutations are often in the transmembrane domain and cause constitutive JAK-STAT signaling that may be abrogated with JAK inhibitors.13,50
A minority of Ph-like cases have mutations activating Ras signaling (NRAS, KRAS, PTPN11, and NF1),22 although these are not exclusively observed in Ph-like ALL. Several kinases are infrequently rearranged in Ph-like ALL, including NTRK3 and TYK2.22
Ph-like ALL is associated with high-risk clinical features, a poor response to induction chemotherapy, elevated minimal residual disease (MRD) levels, and/or poor survival.7,14,22,30,51,52 Several observations indicate that tyrosine kinase inhibitors (TKIs) may be effective in Ph-like ALL. Kinase fusions and sequence mutations confer cytokine-independent proliferation in mouse pre–B-cell lines that is abrogated by tyrosine kinase inhibitors (TKIs), ABL1 inhibitors such as imatinib and dasatinib for ABL-class fusions, and JAK2 inhibitors for JAK-STAT–activating alterations.14,22,53 Human Ph-like leukemic cells show similar pathway activation and sensitivity to TKI in vitro and in xenograft models. There are several reports of Ph-like ALL with ABL-class rearrangements poorly responsive to chemotherapy with profound TKI responses,22,54 which have led to prospective precision medicine trials of TKI therapy in Ph-like ALL.
Hypodiploid ALL
Hypodiploid B-ALL (<44 chromosomes) is associated with poor outcome.27,28 Several subgroups of hypodiploid ALL are recognized. The most common are near-haploid (NH, 24-31 chromosomes) and low-hypodiploid (LH, 32-39 chromosomes) ALL, which have distinct transcriptional profiles and submicroscopic genetic alterations.15 High-hypodiploid (40-43 chromosomes) ALL is rare, and near-diploid ALL (44 or 45 chromosomes) is a distinct entity frequently characterized by ETV6-RUNX1 fusion or rearrangements forming dicentric chromosomes.
NH ALL is characterized by Ras-activating mutations, most commonly focal deletions of NF1, but also NRAS, KRAS, and PTPN11 mutations, and inactivation of IKZF3.15 LH ALL is characterized by biallelic alteration of TP53, deletions of CDKN2A/B and RB1, and inactivation of IKZF2.15 TP53 mutations are germline in half of childhood LH ALL, making this a manifestation of Li-Fraumeni syndrome requiring clinical TP53 mutation testing. Germline mutations in other oncogenes are also observed in hypodiploid ALL, including NRAS and PTPN11.15 Hypodiploid ALL leukemic cells exhibit activation of PI3K/mTOR and mitogen-activated protein kinase kinase/extracellular signal-regulated kinase (MEK-ERK) signaling that is sensitive to monotherapy with PI3K inhibitors, but not mitogen-activated protein kinase kinase inhibitors, as well as BCL2 inhibitors, and the potential for these agents in this high-risk ALL subset is currently being tested in preclinical studies.
ERG-deregulated ALL
Approximately 7% of childhood and adult B-ALL cases have a distinct gene expression profile but lack a unifying chromosomal rearrangement.55 Most have a focal deletion of ERG,56,57 which encodes an ETS family transcription factor also rearranged in prostate cancer.58 These cases often express an aberrant ERG protein. IKZF1 alterations are relatively common in ERG-altered ALL but, in contrast to BCR-ABL1–positive and Ph-like ALL, are not associated with inferior outcome.57,59
Intrachromosomal amplification of chromosome 21
Intrachromosomal amplification of chromosome 21 (iAMP21), defined as gain of at least 3 copies of part of chromosome 21 incorporating RUNX1, occurs in ∼2% of B-ALL and is associated with older age at diagnosis and poor outcome.60 The structural alterations identified in iAMP21 are usually complex with large regions of amplification and commonly flanking deletions.21 iAMP21 is usually observed in cases lacking a recurring chromosomal translocation, but its risk is increased several-thousand-fold in persons with a constitutional Robertsonian t(15;21).21
T-lineage ALL
T-ALL is also characterized by translocations that deregulate transcription factors, commonly by rearrangement to T-cell antigen receptor loci, and recurring sequence mutations and DNA copy-number alterations that disrupt developmental, signaling, and tumor suppressor pathways, including BCL11B,61 NOTCH1,62 FBXW7,63 MYB,64 PTEN,65 RB1, and additional genes whose role in the pathogenesis of T-ALL remains to be elucidated, including PHF610 and WT1.9,66
Early T-cell precursor (ETP) ALL is a recently described subtype of high-risk ALL defined by reduced expression of T-cell markers (CD1a, CD8, and CD5) and aberrant expression of myeloid or stem cell markers.67 Many studies reported poor outcome of ETP ALL,67,68 but this is mitigated by contemporary risk-adapted therapy.69,70
ETP ALL is genetically heterogeneous, with mutation of multiple cellular pathways including (1) hematopoietic and lymphoid development (RUNX1, IKZF1, ETV6, GATA3, and EP300); (2) Ras, cytokine receptor, and kinase signaling (NRAS, IL7R, KRAS, JAK1, JAK3, NF1, PTPN11, and SH2B3); and (3) loss-of-function mutations targeting epigenetic regulators.12,13,71,72 These commonly involve the polycomb repressor complex 2 (PRC2; EZH2, SUZ12, and EED), which mediates histone 3 lysine 27 (H3K27) trimethylation and repression of gene transcription, and SETD2, which encodes a H3K36 trimethylase that marks actively transcribed genes.
The gene expression profile of ETP ALL is similar to that of hematopoietic stem cells, suggesting that ETP ALL may be one of a spectrum of neoplasms of hematopoietic stem cells and progenitors that retain multilineage potential, including “near” ETP ALL (with normal CD5 expression) and mixed-phenotype acute leukemia. Existing data indicate that these forms of leukemia harbor mutations targeting similar genes and pathways.73 The involvement of JAK-STAT and PRC2 pathways in ETP ALL suggests that JAK inhibition and/or chromatin-modifying agents may be therapeutically useful.49,50,74
Recent studies have provided additional insights into the biology of T-ALL. Exome sequencing identified recurrent mutations in ribosomal proteins and CNOT3, which encodes part of a transcriptional regulatory complex.18 Mutations and chimeric fusions involving kinases observed in B-ALL have also been identified in T-ALL including ABL1,75 PTK2B (FAK), and JAK2,19 suggesting a subset of T-ALL may also be amenable to targeted therapy with TKIs.
Noncoding genomic alterations are also important in T-ALL.23,24 A recent study identified novel long noncoding RNAs in T-ALL, including several regulated by NOTCH1.23 One long noncoding RNA was examined in detail and found to regulate oncogenic signaling through the insulin-like growth factor-1 receptor.23
Intriguingly, a noncoding region 1.5 Mb distal to MYC is commonly duplicated in T-ALL, is controlled by NOTCH1, and regulates expression of MYC.24 Targeted disruption of this region in a mouse model demonstrated that this locus was important for leukemogenesis and provides an explanation for the previously observed regulation of MYC expression by NOTCH1 in T-ALL.76
Another mechanism of leukemogenesis in T-ALL is mutations upstream of the T-ALL oncogene TAL1, which generate a binding site for the MYB transcription factor, thereby recruiting a protein complex including TAL1 and the H3K27 acetylator CREBBP, resulting in formation of an oncogenic “superenhancer” region characterized by high levels of H3K27 acetylation.25 These examples underscore the importance of careful integrated analysis of coding and noncoding genomic, transcriptomic, and epigenomic features in ALL.
Epigenetic alterations in ALL
Multiple studies have identified mutations in genes encoding epigenetic regulators and mutations in noncoding regions of the genome that are modified by such regulators. Furthermore, mutations in epigenetic modifiers such as CREBBP and SETD2 are enriched at relapse.11,35,77
Alterations in cytosine methylation are also important in leukemogenesis. Leukemia cells typically exhibit global hypomethylation, with selective hypermethylation of CpG-rich regions in gene promoters. Promoter hypermethylation is associated with gene silencing and hypomethylation with transcriptional activation. Genome-wide profiling of cytosine methylation, gene expression, and DNA copy-number alterations has shown recently that ALL subtypes have distinct methylation and gene expression signatures and that methylation is an important determinant of ALL gene expression. Methylation perturbations may be a consequence of the founding genetic alterations and/or cooperate with structural genetic changes to promote leukemogenesis.78,79
Mutations in genes encoding epigenetic regulators and chromatin-modifying proteins have been identified in most subtypes of ALL. In addition to PRC2 mutations in ETP ALL,13,71 notable examples are mutations of WHSC1 (NSD2) in ETV6-RUNX1 ALL,80 and mutation of CREBBP (a H3K18, H3K27, and nonhistone acetyl transferase), SETD2 (a H3K36 trimethylase), KDM6A, and MLL2 in relapsed and hypodiploid ALL.11,15,35,77 Consequently, there is interest in testing the efficacy of drugs that modulate histone modifications in ALL, including inhibitors of bromodomain readers,81 histone demethylases,82 and histone deacetylases.
Genetic heterogeneity, clonal evolution, and relapse in ALL
Most diagnostic ALL samples exhibit clonal diversity, and this diversity evolves over time. Analysis of paired diagnosis/relapse specimens has shown that most cases exhibit substantial genomic changes during disease progression, with acquisition of new deletions and mutations, and loss of diagnosis-specific lesions.34 Founding chromosomal translocations are almost usually conserved from diagnosis to relapse, along with a proportion of DNA copy-number alterations and point mutations in most cases. Many deletions or mutations that emerge in the predominant relapse clone may be detected at low levels at diagnosis using sensitive technology.35 Together, these findings indicate that the predominant clones observed at diagnosis and relapse clones arise from a common “ancestral” or “preleukemic” clone that harbors some of the genetic alterations required for leukemogenesis but then evolves down multiple paths. Treatment dramatically influences this evolution and clonal composition by suppressing or eradicating one or more of the predominant clones at diagnosis and facilitating the emergence of subclones that harbor and/or acquire mutations that confer resistance to therapy.
Genome sequencing, coupled with deep coverage of mutated sites, has enabled this clonal structure and temporal progression to be dissected with precision.35 First, many relapse-acquired lesions involve genes regulating B-cell development (IKZF1), tumor suppression (TP53),83 Ras signaling, chromatin modification (CREBBP and SETD2),77 and drug metabolism (NT5C2).17,35,84 Second, relapse-acquired alterations may induce a more stem cell–like state (eg, IKZF1) or directly confer resistance to individual chemotherapeutic agents (eg, CREBBP and NT5C2). CREBBP mutations result in resistance to glucocorticoids,11 and mutations in the 5′-nucleotidase gene NT5C2 result in resistance to nucleoside analogs.84 Third, many cases have multiple subclonal mutations involving the same pathway or gene at diagnosis. Initial chemotherapy results in suppression or elimination of all but one usually minor clone that survives to acquire additional mutations that confer resistance to therapy and propagate relapse.35 Finally, many mutations present in the predominant clone at relapse may be detected at early time points in therapy. This is important given the increasing application of next-generation sequencing approaches to MRD detection.85
Inherited genetic variation and ALL risk and outcome
In the last few years, there have been growing data supporting an important role of common inherited variants and rare deleterious mutations in risk of developing ALL.
Genome-wide association studies using microarrays to genotype millions of single-nucleotide polymorphisms in cases and ethnically matched controls have identified associations between polymorphisms in genes including IKZF1, ARID5B, CEBPE, CDKN2A, and GATA3 and the risk of developing ALL.86-90 Associations with specific ALL subtypes and relapse have also been identified.91-95 Several genes such as IKZF1, CEBPE, and GATA3 encode transcription factors that are also targets of somatic genetic alteration in ALL.96 CDKN2A/B encode the INK4/ARF family of tumor suppressors and cell-cycle regulators; this locus is commonly deleted in B- and T-ALL. In addition, ARID5B polymorphisms are associated with ALL risk and outcome in specific ethnic groups,92 and GATA3 variants are associated with Ph-like ALL.94
Deleterious germline mutations have been identified in familial and sporadic ALL. In addition to the germline TP53 and Ras mutations observed in LH ALL,15 additional hypodiploid cases harbor germline mutations involving genes mediating DNA repair that are likely to be pathogenic.15 Down syndrome is associated with an increased risk of AML and ALL, and the rare constitutional Robertsonian translocation rob(15;21)(q10;q10)c is associated with a markedly increased risk of ALL with iAMP21.21 Familial ALL is uncommon, but such kindreds are highly informative. Two studies have reported kindreds with autosomal-dominant ALL, in which affected individuals harbored a novel germline PAX5 mutation, p.Gly183Ser, that attenuated the transcriptional activity of PAX5.97 Leukemic cells exhibited loss of the nonmutated PAX5 allele by deletion of chromosome 9p, suggesting that germline heterozygosity of this variant is tolerated but that severe attenuation of PAX5 activity is required for leukemogenesis.97 Recent reports have described several kindreds with deleterious inherited mutations in the ETS domain of ETV6,98,99 a common target of mutation and rearrangement in leukemia.
Prospects for precision medicine in ALL
One can envisage several opportunities for incorporating genomic discoveries into ALL management, from initial diagnosis and risk stratification to the delivery of targeted therapy. Sequencing will be increasingly used for molecular diagnosis and will likely supplant current approaches that require multiple complex cytogenetic and molecular tests that are unable to identify all clinically relevant genomic alterations. Notable examples include ERG-deregulated and Ph-like ALL, which are either not evident on conventional cytogenetic analysis or are highly genetically heterogeneous. Diagnostic sequencing approaches must be capable of identifying sequence alterations, DNA copy-number alterations, and structural rearrangements in ALL. Thus, it is likely that genome-wide sequencing approaches, with their attendant challenges in turnaround time and interpretation, will be increasingly adopted as a diagnostic approach.
Second, sequencing will identify subtypes and specific genetic alterations with prognostic importance, and the potential role of these in risk stratification must be clarified. Current risk-stratification algorithms consider a limited range of genetic information (eg, aneuploidy and selected rearrangements) with age, initial white blood cell count, central nervous system involvement, and MRD response.100 Such algorithms have improved survival by optimizing use of intensive chemotherapy regimens and by limiting the use of prophylactic cranial irradiation.101 Genomic features such as IKZF1 alterations, CRLF2 rearrangement, iAMP21, and Ph-like ALL are associated with poor outcome in many studies,14,22 but their ability to refine prognosis in studies incorporating intensive therapy and MRD measurement has varied or has been difficult to measure due to complexity of testing or small sample sizes.52,102 Also, the prognostic importance of specific alterations is subtype dependent, with interplay between molecular lesions. For example, IKZF1 alterations are associated with poor outcome in B-ALL, with the exception of ERG-altered ALL. Thus, comprehensive genomic studies in large cohorts treated with current therapies, including treatment modifications based upon MRD response, are required.
Third, sequencing will identify targets or pathways to guide implementation of novel therapies, either in frontline studies or following relapse, particularly for high-risk ALL subtypes with poor outcomes amenable to treatment with currently approved agents.103 The paradigm for precision medicine in leukemia is the use of ABL-class TKIs in chronic myeloid leukemia and combination therapy with TKIs and chemotherapy in Ph+ ALL.104,105 Although important questions remain in Ph+ ALL (including the optimal TKI, the optimal chemotherapy backbone, the length of TKI therapy, the role of allogeneic hematopoietic stem cell transplantation in first remission, and the role of TKIs after hematopoietic stem cell transplantation), any patient diagnosed with Ph+ ALL in the United States or western Europe will receive a TKI as part of therapy. There is consequently enthusiasm to extend TKI therapy to Ph-like ALL, but it is critical to determine whether kinase-activating alterations are similar drivers to BCR-ABL1; that is, to show that they arise early in disease development, that they are present in all subclones at all stages of disease, and that the leukemia is dependent upon continued activity of the mutant kinase.
Subsets of Ph-like ALL, including those with ABL-class, JAK2, and EPOR rearrangements, appear to fulfill the above requirements. ABL-class fusions phenocopy BCR-ABL1 in both transforming function and TKI sensitivity in experimental models, and there are anecdotal reports of remarkable short-term responses to ABL-class TKIs in refractory Ph-like ALL. Thus, clinical trials using these agents are being developed by multiple groups including the Children’s Oncology Group (COG) and St. Jude Children’s Research Hospital. Given this, it is essential to consider how Ph-like ALL is identified and the complex underlying genomic landscape dissected. One may focus on selective testing for subsets of alterations, screening for the Ph-like gene expression profile, and/or genome sequencing approaches. We emphasize that the most important goal is to identify underlying kinase-activating lesions (eg, ABL-class fusions) rather than the gene expression profile that defines Ph-like ALL. This may be accomplished by focused molecular/fluorescence in situ hybridization assays for known fusions and rearrangements, coupled with selective genome sequencing for enigmatic cases (the approach adopted for the large cohorts studied by the COG), or sequencing of all patients at the time of diagnosis. To establish TKI efficacy, the ideal approach would be to conduct a randomized trial of chemotherapy with or without TKIs. This is likely not feasible, and establishing benefit will rely more upon trials that use historical control comparisons, as the COG did with imatinib in Ph+ ALL.104
It will be even more challenging to extend precision medicine approaches to other genetically defined subsets of ALL, as few recapitulate the key features of BCR-ABL1 ALL. A number of pathways are recurrently mutated in B- and T-ALL, including NOTCH1 in T-ALL, epigenetic modifiers in both B- and T-ALL, transcription factors involved in B-cell differentiation, the Ras pathway, and others. Moreover, it is likely that combinatorial therapy with drugs targeting multiple “nodes” in perturbed signaling pathways will be effective.46 Although there are promising new drugs and preliminary preclinical data in support of these approaches, data justifying use in front-line ALL therapy are lacking. It will be critical to conduct clinical trials that are designed to clearly establish whether or not new therapies improve outcome.
Finally, sequencing is likely to have an important role in monitoring response to therapy, by deeply sequencing antigen receptor loci85 or other targets of genetic alteration, particularly those known to facilitate resistance to therapy (for example, IKZF1, CREBBP, and NT5C2).106 Sequencing of antigen receptor loci has shown promise as an alternative to flow cytometric or polymerase chain reaction–based assays. An important challenge for future studies is deploying technology to sensitively detect mutations and structural changes and to determine how such information may be used to change therapy when a mutation that confers resistance to a specific agent emerges.
Conclusions and future directions
The ALL genome remains a work in progress. The number of comprehensively sequenced ALL genomes is small, particularly in older individuals. In existing data sets, the number of frequently mutated genes is typically low, with a “long tail” of infrequently mutated genes. Thus, we need to sequence many more cases broadly to systematically and comprehensively identify all relevant coding and noncoding alterations. This will be important to more completely understand the key pathways driving leukemia development and to inform the interpretation of clinical sequencing efforts. We will likely also see much greater use of single-cell sequencing technologies to profile structural alterations, transcriptional profiles, and epigenomic marks. This will be especially useful to gain a fuller understanding of the role of clonal composition and dynamics in ALL. An important challenge is experimental validation of the functional effects of the identified mutations. There have been few faithful models of ALL, in part due to the poor understanding of the polygenic basis of this disease. We are now at a new dawn where we have the genomic information to build the right models of leukemia and to use these to test new therapies to improve patient outcome.
Acknowledgments
The authors thank their colleagues at St. Jude Children’s Research Hospital, the Children’s Oncology Group, and the National Cancer Institute Therapeutically Applicable Research to Generate Effective Treatments (TARGET) consortium (https://ocg.cancer.gov/programs/target). Several of the studies described were supported by the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital, the National Cancer Institute of the US National Institutes of Health, Alex’s Lemonade Stand Foundation, the American Association for Cancer Research, the American Society of Hematology, the Henry Schueler 41&9 Foundation, the Leukemia and Lymphoma Society. The National Health and Medical Research Council (Australia), the Pew Charitable Trusts, Stand Up To Cancer, and the St. Baldrick’s Foundation. The authors apologize to authors of the many excellent manuscripts that could not be cited due to space considerations.
Authorship
Contribution: S.P.H. and C.G.M. reviewed the literature and wrote the manuscript.
Conflict-of-interest disclosure: S.P.H. and C.G.M. are inventors on patent 8 568 964 “Identification of novel subgroups of high-risk pediatric precursor-B cell acute lymphoblastic leukemia, outcome correlations and diagnostic and therapeutic methods related to same.”
Correspondence: Stephen P. Hunger, Center for Childhood Cancer Research, Children’s Hospital of Philadelphia, Colket Translational Research Building, Room 3060, 3501 Civic Center Blvd, Philadelphia, PA 19104; e-mail: hungers@email.chop.edu; and Charles G. Mullighan, St. Jude Children’s Research Hospital, 262 Danny Thomas Pl, Mail Stop 342, Memphis, TN 38105, e-mail: charles.mullighan@stjude.org.