Abstract
Introduction
Myeloid leukemia are heterogeneous groups of diseases composed of acute myeloid leukemia (AML), myelodysplastic syndrome (MDS), chronic myelomonocytic leukemia (CMML), myeloproliferative neoplasms (MPN) and others. While these leukemias have distinct pathological and clinical manifestations, MDS, CMML, and MPN commonly share the risk of AML transformation. Clinical similarity and difference among the myeloid leukemia might be the reflection of underlying homogeneous and heterogeneous genetic basis of the diseases. We analyzed mutation data on a large cohort of pan-myeloid leukemia cases and tested whether mutation data can predict the clinical phenotype and the outcomes using machine-learning (ML) algorithms.
Methods
Bone marrow samples from 868 patients (pts) with myeloid leukemias (AML: N = 505, MDS/CMML: N = 255, and MPN: N = 108) were analyzed by the targeted capture sequencing of 295 cancer genes (N = 638, median 561x) or whole exome sequencing (N = 230, median 200x). The cohort was divided in the ratio of 4:1 to create a training set and validation set. Using a training set, we generated a ML-based model that predicts clinical phenotype based on the somatic mutation data. Performance of the model was assessed in the validation set based on the parameters including accuracy, precision, recall and area under receiver operating characteristic curve. In addition to the internal validation, we tested our ML-based model in 279 MDS/CMML pts data from the Cleveland Clinic (Makishima et al. Nature Genetics 2017).
Results
DNA sequencing detected 2,311 high-confidence somatic mutations (1,372 SNVs and 939 indels) in 87 genes in 769 (89%) pts (median 3 [IQR 2-4] mutations/pt in AML, 2 [IQR 1-4] in MDS/CMML, and 2 [IQR 1-3] in MPN). Most commonly shared mutations across all myeloid leukemias were ASXL1 (frequency in AML/MDS CMML/MPN: 16%/30%/24%), TET2 (17%/26%/16%), SRSF2 (14%/21%/7%), DNMT3A (26%/8%/6%), and RUNX1 (14%/15%/5%), which is consistent with the often preleukemic nature of these mutations. Mutations in NPM1, FLT3, KIT, and MYC were almost exclusively detected in AML, whereas mutations in CALR and MPL were specific to MPN.
Accuracy of phenotype prediction by ML algorithm relying solely on mutation data was 88% in AML, 63% in MDS/CMML and 85% in MPN. Mutations in FLT3 (co-efficient: 2.2289), NPM1 (1.58176), IDH2 (1.43436), GATA2 (1.38649), IDH1 (1.34405), CEBPA (1.2631), WT1 (1.2577), DNMT3A (1.1073), and KIT (1.07877) had the strong power in predicting AML, whereas mutations in SF3B1 (1.32368) and CUX1 (1.00176) had the strong power in predicting MDS/CMML. JAK2 (3.42899), CALR (2.6211) and MPL (2.17668) mutations had the strong power in predicting MPN.
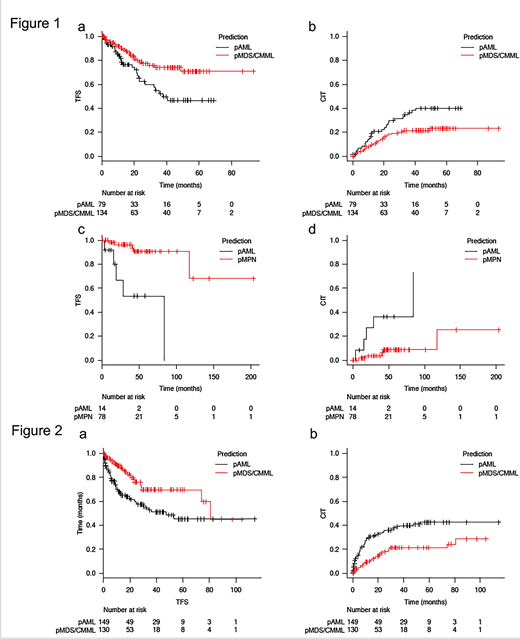
MDS/CMML or MPN cases that were misclassified (predicted) as AML (pAML) by the ML algorithm likely had a genetic similarity with AML, which might predict their risk to AML transformation. Indeed, in the internal validation set, MDS/CMML pts who were predicted as AML (MDS/CMML-pAML) by the ML algorithm had significantly worse transformation free survival (TFS) and cumulative incidence of transformation (CIT) compared to MDS/CMML pts who were predicted as MDS/CMML (MDS/CMML-pMDS/CMML) (2-year TFS: 63% [95% CI: 48-74] in MDS/CMML-pAML vs. 79% [95% CI: 69-86] in MDS/CMML-pMDS/CMML, p = 0.01, 2-year CIT: 30% [95% CI: 20-41] in MDS/CMML-pAML vs. 18% [95% CI: 12-26] in MDS/CMML-pMDS/CMML, p = 0.02), Figure 1 a-b). The similar association was also seen in MPN where MPN pts who were predicted as AML by the ML algorithm (MPN-pAML) had significantly worse TFS and CIT compared to MPN pts who were predicted as MPN (MPN-pMPN) (2-year TFS: 67% [95% CI: 27-88] in MPN-pAML vs. 96% [95% CI: 86-99] in MPN-pMPN, p < 0.01, 2-year CIT: 27% [95% CI: 6-55] in MPN-pAML vs. 4% [95% CI: 1-11] in MPN-pMPN, p < 0.01, Figure 1 c-d). We further validated our ML-based algorithm in the external cohort of 279 MDS/CMML pts from the Cleveland Clinic. (2-year TFS: 60% [95% CI: 50-69] in MDS/CMML-pAML vs. 76% [95% CI: 65-84] in MDS/CMML-pMDS/CMML, p < 0.01, 2-year CIT: 34% [95% CI: 26-42] in MDS/CMML-pAML vs. 18% [95% CI: 11-26] in MDS/CMML-pMDS/CMML, p < 0.01, Figure 2 a-b).
Conclusion
Morphology agnostic, mutation based classification of myeloid leukemia identified MDS/CMML and MPN cases that have high risk of AML transformation. These data support the idea to supplement the conventional morphological and clinical classification by the molecular alterations.
Kadia:Abbvie: Consultancy; Amgen: Consultancy, Research Funding; Jazz: Consultancy, Research Funding; Jazz: Consultancy, Research Funding; Takeda: Consultancy; Celgene: Research Funding; BMS: Research Funding; Abbvie: Consultancy; Pfizer: Consultancy, Research Funding; Takeda: Consultancy; Novartis: Consultancy; Celgene: Research Funding; Amgen: Consultancy, Research Funding; Novartis: Consultancy; Pfizer: Consultancy, Research Funding; BMS: Research Funding. Daver:Incyte: Consultancy; Incyte: Research Funding; Kiromic: Research Funding; Pfizer: Consultancy; Otsuka: Consultancy; BMS: Research Funding; Alexion: Consultancy; Novartis: Consultancy; Karyopharm: Consultancy; Daiichi-Sankyo: Research Funding; Novartis: Research Funding; ImmunoGen: Consultancy; Sunesis: Consultancy; Pfizer: Research Funding; Sunesis: Research Funding; Karyopharm: Research Funding; ARIAD: Research Funding. Pemmaraju:SagerStrong Foundation: Research Funding; Affymetrix: Research Funding; plexxikon: Research Funding; daiichi sankyo: Research Funding; samus: Research Funding; celgene: Consultancy, Honoraria; abbvie: Research Funding; cellectis: Research Funding; stemline: Consultancy, Honoraria, Research Funding; novartis: Research Funding. DiNardo:Abbvie: Honoraria; Celgene: Honoraria; Bayer: Honoraria; Agios: Consultancy; Karyopharm: Honoraria; Medimmune: Honoraria. Ravandi:Jazz: Honoraria; Sunesis: Honoraria; Xencor: Research Funding; Sunesis: Honoraria; Amgen: Honoraria, Research Funding, Speakers Bureau; Orsenix: Honoraria; Bristol-Myers Squibb: Research Funding; Seattle Genetics: Research Funding; Abbvie: Research Funding; Xencor: Research Funding; Orsenix: Honoraria; Astellas Pharmaceuticals: Consultancy, Honoraria; Macrogenix: Honoraria, Research Funding; Jazz: Honoraria; Abbvie: Research Funding; Macrogenix: Honoraria, Research Funding; Amgen: Honoraria, Research Funding, Speakers Bureau; Astellas Pharmaceuticals: Consultancy, Honoraria; Seattle Genetics: Research Funding; Bristol-Myers Squibb: Research Funding. Verstovsek:Incyte: Consultancy; Italfarmaco: Membership on an entity's Board of Directors or advisory committees; Celgene: Membership on an entity's Board of Directors or advisory committees; Novartis: Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau. Cortes:Pfizer: Consultancy, Research Funding; Arog: Research Funding; Daiichi Sankyo: Consultancy, Research Funding; Novartis: Consultancy, Research Funding; Astellas Pharma: Consultancy, Research Funding. Maciejewski:Apellis Pharmaceuticals: Consultancy; Ra Pharmaceuticals, Inc: Consultancy; Alexion Pharmaceuticals, Inc.: Consultancy, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Alexion Pharmaceuticals, Inc.: Consultancy, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Apellis Pharmaceuticals: Consultancy; Ra Pharmaceuticals, Inc: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal