Key Points
Next-generation sequencing broadens the spectrum of germ line mutations in a cohort of patients with likely-inherited BMF.
Salient clinical features and distinct natural histories are consistently found in SAMD9L and SAMD9, MECOM/EVI1, and ERCC6L2 disorders.
Abstract
Bone marrow (BM) failure (BMF) in children and young adults is often suspected to be inherited, but in many cases diagnosis remains uncertain. We studied a cohort of 179 patients (from 173 families) with BMF of suspected inherited origin but unresolved diagnosis after medical evaluation and Fanconi anemia exclusion. All patients had cytopenias, and 12.0% presented ≥5% BM blast cells. Median age at genetic evaluation was 11 years; 20.7% of patients were aged ≤2 years and 36.9% were ≥18 years. We analyzed genomic DNA from skin fibroblasts using whole-exome sequencing, and were able to assign a causal or likely causal germ line mutation in 86 patients (48.0%), involving a total of 28 genes. These included genes in familial hematopoietic disorders (GATA2, RUNX1), telomeropathies (TERC, TERT, RTEL1), ribosome disorders (SBDS, DNAJC21, RPL5), and DNA repair deficiency (LIG4). Many patients had an atypical presentation, and the mutated gene was often not clinically suspected. We also found mutations in genes seldom reported in inherited BMF (IBMF), such as SAMD9 and SAMD9L (N = 16 of the 86 patients, 18.6%), MECOM/EVI1 (N = 6, 7.0%), and ERCC6L2 (N = 7, 8.1%), each of which was associated with a distinct natural history; SAMD9 and SAMD9L patients often experienced transient aplasia and monosomy 7, whereas MECOM patients presented early-onset severe aplastic anemia, and ERCC6L2 patients, mild pancytopenia with myelodysplasia. This study broadens the molecular and clinical portrait of IBMF syndromes and sheds light on newly recognized disease entities. Using a high-throughput sequencing screen to implement precision medicine at diagnosis can improve patient management and family counseling.
Medscape Continuing Medical Education online

In support of improving patient care, this activity has been planned and implemented by Medscape, LLC and the American Society of Hematology. Medscape, LLC is jointly accredited by the Accreditation Council for Continuing Medical Education (ACCME), the Accreditation Council for Pharmacy Education (ACPE), and the American Nurses Credentialing Center (ANCC), to provide continuing education for the healthcare team.
Medscape, LLC designates this Journal-based CME activity for a maximum of 1.00 AMA PRA Category 1 Credit(s)™. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
All other clinicians completing this activity will be issued a certificate of participation. To participate in this journal CME activity: (1) review the learning objectives and author disclosures; (2) study the education content; (3) take the post-test with a 75% minimum passing score and complete the evaluation at http://www.medscape.org/journal/blood; and (4) view/print certificate. For CME questions, see page 374.
Disclosures
Laurie Barclay, freelance writer and reviewer, Medscape, LLC, owns stock, stock options, or bonds from Alnylam, Biogen, and Pfizer. Associate Editor Jorge Cortes and the authors declare no competing financial interests.
Learning objectives
Upon completion of this activity, participants will be able to:
Compare outcomes among oncogenetic low-risk vs oncogenetic high-risk patients with childhood T-cell acute lymphoblastic leukemia (T-ALL).
Assess outcome prediction among patients with childhood T-ALL based on a combination of oncogenetic classifier, minimal residual disease, and other factors.
Determine the clinical implications of these findings regarding outcome prediction for patients with childhood T-ALL based on a combination of oncogenetic classifier, minimal residual disease, and other factors.
Release date: January 18, 2018; Expiration date: January 18, 2019
Introduction
Bone marrow (BM) failure (BMF) syndromes are a heterogeneous group of hematological disorders involving single-lineage cytopenia or pancytopenia (ie, aplastic anemia [AA]). They may either be acquired or inherited (inherited BMF [IBMF]), the most common IBMF being Fanconi anemia (FA).1-4 Overlap between BMF and myelodysplastic syndromes (MDSs) has long been recognized.5-9 Due to severe pancytopenia or disease progression to either MDSs or acute myeloid leukemia (AML), BMF patients are often considered for hematopoietic stem cell transplantation (HSCT).10 Recognizing the inherited nature of BMF is a crucial step in selecting healthy sibling donors and adapting the conditioning regimen to avoid toxicity arising from underlying genetic defects.11,12 In addition, early recognition of patients with IBMF prevents inappropriate administration of immunosuppressive therapy for acquired AA. Identifying a genetic cause also enables risk-adapted monitoring to be undertaken in patients, including cancer risk assessment and family counseling. A growing number of mutations have been described in patients with IBMF syndromes.13,14 However, most studies have focused on patients with specific suspected diseases based on a syndromic presentation (mainly FA, dyskeratosis congenita [DC], Diamond-Blackfan anemia [DBA], Shwachman-Diamond syndrome [SDS], and other chronic neutropenia [CN]) and so did not provide a wider perspective of the underlying diagnoses in other patients with IBMF.14-17
At the French Bone Marrow Failure Center laboratory, we receive samples from most pediatric and adult clinical centers in France, with the aim of diagnosing or ruling out FA and other IBMF causes. Our long-established practice has been to systematically grow skin-biopsy fibroblasts at diagnosis and use these nonhematopoietic cells, considered to be germ line, to rule out the potential confounding events that can obscure the results in blood cells.18,19 For example, spontaneous hematopoietic reversion may lead to somatic mosaicism in FA and other IBMFs.19-22 Moreover, somatic mutations associated with clonal hematopoiesis or MDS/AML should be distinguished from germ line mutations.6,9,23-26 In this study, we used an unbiased, whole-exome sequencing (WES) comprehensive analysis on fibroblast DNA from a large cohort of patients with BMF of suspected inherited origin but unresolved diagnosis after initial medical evaluation, with the aim of drawing a broad molecular and clinical portrait of this heterogeneous group of patients.
Methods
Patients and cohort
Patient samples were sent to the French Bone Marrow Failure Center laboratory at Saint-Louis Hospital (Paris, France) between February 2002 and June 2016. Written informed consent for genetic investigation, tissue banking, and research was provided by the patients or their relatives in accordance with the Declaration of Helsinki and French law. The tissue banking protocol and this study were approved by the Institutional Review Board of the University Institute of Hematology (IUH; Saint-Louis Hospital, Paris, France).
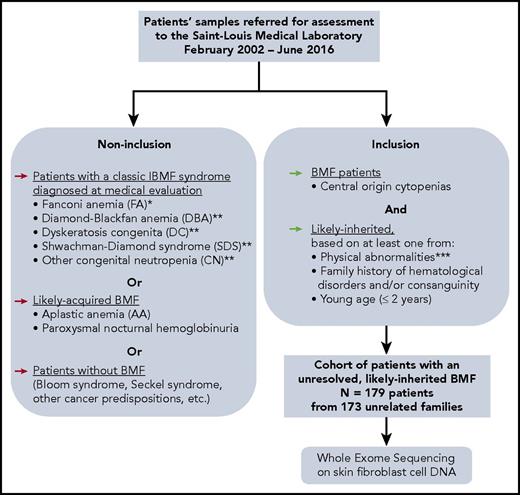
One hundred seventy-nine consecutive patients from 31 French centers (supplemental Table 1, available on the Blood Web site) with BMF of suspected inherited origin were retrospectively included in this study (Figure 1). All patients had at least 1 blood cytopenia (neutrophil count, <1.5 × 109/L; platelet count, <150 × 109/L; and/or hemoglobin [Hb] concentration <2 standard deviations [SDs] below mean, adjusted for age) of central origin confirmed by BM aspiration and/or biopsy. Results of blood tests and marrow examination were reviewed, and referring cytologists or pathologists were contacted where necessary. We characterized dysplastic BM in accordance with the revised World Health Organization (WHO) 2008 classification,27 considering blastic progression as ≥5% blast cells in the BM and moderate dyserythropoiesis as a classic sign of BMF but not as a criterion for MDSs.5
Constitution of a cohort of IBMF patients, likely inherited, with unresolved origin after initial evaluation. *FA diagnosis was performed at initial evaluation, for all patients whose samples were sent in our laboratory, as we previously described, for example, MMC sensitivity in peripheral blood and skin fibroblast cells.18,19 **Typical DBA, SDS, and other CN, and DC patients who were diagnosed up front with these syndromes based on a syndromic presentation were not included in the current “undiagnosed cases” WES study. Nevertheless, we subsequently identified additional patients with mutated DBA, SDS, or DC genes in our cohort through WES analysis. A posteriori, some of these patients had a presentation that might have been recognized as syndromic if fully evaluated. ***Physical abnormalities suggesting an inherited origin included skeletal abnormalities (thumb, jaw, or any limb malformation); neurological defects (developmental delay or intellectual disability, dyspraxia, attention deficit disorder, or abnormal MRI image); growth retardation; skin, nail, or hair abnormalities; genitourinary or renal malformations (renal agenesis, cryptorchidic testis); cardiac abnormalities (valvular heart disease, heart failure); lung abnormalities (mainly restrictive syndrome or fibrosis), liver abnormalities (cirrhosis, fibrosis). BMF, bone marrow failure; MMC, mitomycin C; WES, whole-exome sequencing.
Constitution of a cohort of IBMF patients, likely inherited, with unresolved origin after initial evaluation. *FA diagnosis was performed at initial evaluation, for all patients whose samples were sent in our laboratory, as we previously described, for example, MMC sensitivity in peripheral blood and skin fibroblast cells.18,19 **Typical DBA, SDS, and other CN, and DC patients who were diagnosed up front with these syndromes based on a syndromic presentation were not included in the current “undiagnosed cases” WES study. Nevertheless, we subsequently identified additional patients with mutated DBA, SDS, or DC genes in our cohort through WES analysis. A posteriori, some of these patients had a presentation that might have been recognized as syndromic if fully evaluated. ***Physical abnormalities suggesting an inherited origin included skeletal abnormalities (thumb, jaw, or any limb malformation); neurological defects (developmental delay or intellectual disability, dyspraxia, attention deficit disorder, or abnormal MRI image); growth retardation; skin, nail, or hair abnormalities; genitourinary or renal malformations (renal agenesis, cryptorchidic testis); cardiac abnormalities (valvular heart disease, heart failure); lung abnormalities (mainly restrictive syndrome or fibrosis), liver abnormalities (cirrhosis, fibrosis). BMF, bone marrow failure; MMC, mitomycin C; WES, whole-exome sequencing.
The inclusion criterion of suspected inherited origin was based on: at least 1 physical abnormality (growth, skeletal, neurological, genitourinary, cardiac, lung, liver, skin, nail, or hair), and/or a suggestive family history (consanguinity or family history of hematological disorder), and/or young age (≤2 years). FA patients were excluded from the study (diagnosed through mitomycin C sensitivity tests on fibroblast cells that we perform systematically on all patients at initial evaluation),18 as were excluded patients diagnosed up front based on a syndromic presentation with DBA, DC, SDS, and other CN. Overall, our selection criteria resulted in a final cohort of “unresolved, likely IBMF” patients (Figure 1).
Samples
Primary fibroblast cells were available in our repository for all included patients. Skin biopsies had been performed at medical evaluation using 3- or 4-mm square punches and sent to the laboratory the same day at room temperature in saline solution. In the laboratory, skin samples were sliced and the resulting fragments were adhered to plastic plates for 15 minutes, and then covered with medium (minimal essential medium and 20% fetal calf serum; Thermo Fisher Scientific). Individual growing fibroblast clones were picked up after 2 to 3 weeks and expanded for genomic DNA extraction and banking at low passage. Genomic DNA was extracted from fibroblast cells with the use of the QIAamp DNA Mini kit (Qiagen).
In some patients, peripheral blood nucleated cells or BM cells were also studied along with fibroblast cells to evaluate somatic evolution. For the new disease entities, especially MECOM, SAMD9, SAMD9L, and ERCC6L2 patients, parental blood DNA was obtained where possible.
WES and analysis
Whole-exome libraries were prepared using SureSelect XT Target Enrichment (Agilent Technologies). Libraries were sequenced on NextSeq500 or HiSeq1000 systems (Illumina Inc). Data analysis was performed using a homemade pipeline that included standard tools to identify point mutations, insertions, or deletions, as well as copy-number alterations (supplemental Methods; supplemental Figure 1). To securely identify IBMF-causal mutations, a flagging procedure highlighting a list of suspected and known IBMF genes was integrated as an assisting tool (supplemental Figure 1; supplemental Table 2). In addition, the broad WES data were also manually investigated for potential novel IBMF-causing genes. The pathogenic effect of the variants was assessed using guidelines from the American College of Medical Genetics and Genomics (ACMG; supplemental Tables 4 and 5).28 A systematic, integrated review was then performed for each patient in multidisciplinary roundtable sessions combining broad biological and clinical expertise to assess the relevance of detected variants to the IBMF phenotype (see supplemental Methods), resulting after iterative rounds of evaluation in the final list of disease-causal or likely causal mutations.
Sanger resequencing and primer list
Sanger-sequencing verification of variants detected by our analysis was performed on fibroblast DNA and occasionally in blood DNA as well. The complete list of primers used for verification is shown in supplemental Table 15.
Somatic alterations in SAMD9 and SAMD9L genes were sought in the peripheral blood mononucleated cells (PBMCs) or BM cells from the relevant patients.
Allelic imbalance and consanguinity assessment
Germ line allelic imbalance was detected using a homemade pipeline adapted from the ExomeAI tool29 (supplemental Methods). Consanguinity was defined using the WES data when stretches of homozygosity >3 Mb encompassing >1% of the genome, in accordance with the recommendations of the ACMG.30
Somatic allelic imbalance in chromosome 7 was assessed in SAMD9 and SAMD9L cases by comparing blood or BM DNA with fibroblast DNA.
Results
Clinical patient characteristics
Patient and disease characteristics of the 179 patients from our undiagnosed, likely IBMF cohort are summarized in Table 1. Median age when a skin biopsy was taken for genetic evaluation was 11 years; retrospectively, the median age at first hematological abnormality was 8 years. At skin biopsy, 20.7% of the patients were aged 2 years or under, and 36.9% were ≥18 years. Most patients presented with pancytopenia (68.4%), whereas the others had cytopenias in 1 or 2 lineages (18.1% and 13.6%, respectively). Evidence for dysplastic features in marrow cells (according to the 2016 revision of the WHO classification) was found in 68 of 157 cases (43.3%), including 21 of 175 patients (12%) with blastic progression (Table 1; supplemental Table 6). BM cytogenetic abnormalities were found in 45 of 161 cases (28.0%), the most frequent being monosomy 7 (supplemental Table 7; supplemental Figure 2). Physical abnormalities were found in 129 of 176 patients (73.3%): 45% with short stature, and roughly one-quarter each with skeletal/skin/nail/hair or neurological abnormalities (Table 1). The 179 patients originated from 173 distinct families. Twenty-five patients of 179 (14.0%) originated from consanguineous union (see “Methods”), whereas 50 patients of 173 (28.9%) had a known family history of hematological disorders.
Clinical characteristics of the 179 patients
| Characteristic . | Value* . |
|---|---|
| Sex, no. (%) | |
| Male | 96 (53.6) |
| Female | 83 (46.4) |
| Age at skin biopsy, no. (%), y | |
| ≤2 | 37 (20.7) |
| >2 and <18 | 76 (42.5) |
| ≥18 | 66 (36.9) |
| Age of first hematological symptoms, median (range), y | 8 (0-47) |
| Hematological characteristics | |
| Pancytopenia, no./total (%) | 121/177 (68.4) |
| Cytopenia, 1 lineage, no./total (%) | 32/177 (18.1) |
| Cytopenias, 2 lineages, no./total (%) | 24/177 (13.6) |
| Anemia, no./total (%)† | 158/178 (88.8) |
| Hb level, median (range), g/dL | 9.5 (2.5-15) |
| MCV, median (range) | 98 (68-117) |
| Thrombocytopenia, no./total (%), <150 × 109/L | 147/178 (82.6) |
| Platelet count, median (range), × 109/L | 45 (2-500) |
| Neutropenia, no./total (%), <1.5 × 109/L | 138/177 (78.0) |
| Neutrophil count, median (range), × 109/L | 0.87 (0-7) |
| BM dysplasia, no./total (%) | 68/157 (43.3) |
| BM blast cell percentage, no./total (%) | |
| <5% | 154/175 (88.0) |
| ≥5, <10%‡ | 9/175 (5.1) |
| ≥10%‡ | 12/175 (6.9) |
| Abnormal karyotype, no./total (%)§ | 45/161 (28.0) |
| Family history, no./total (%) | 66/173 (38.2) |
| Family history of hematological disorders | 50/173 (28.9) |
| Consanguinity|| | 25/179 (14.0) |
| Physical abnormalities, no./total (%) | 129/176 (73.3) |
| Growth restriction | 77/173 (44.5) |
| Skeletal malformation | 49/172 (28.5) |
| Neurological disorder | 39/172 (22.7) |
| Skin, nail, or hair abnormalities | 40/172 (23.3) |
| Genitourinary disorder | 32/171 (18.7) |
| Cardiac disorder | 15/170 (8.8) |
| Lung disorder | 8/172 (4.7) |
| Liver disorder | 8/173 (4.6) |
| Characteristic . | Value* . |
|---|---|
| Sex, no. (%) | |
| Male | 96 (53.6) |
| Female | 83 (46.4) |
| Age at skin biopsy, no. (%), y | |
| ≤2 | 37 (20.7) |
| >2 and <18 | 76 (42.5) |
| ≥18 | 66 (36.9) |
| Age of first hematological symptoms, median (range), y | 8 (0-47) |
| Hematological characteristics | |
| Pancytopenia, no./total (%) | 121/177 (68.4) |
| Cytopenia, 1 lineage, no./total (%) | 32/177 (18.1) |
| Cytopenias, 2 lineages, no./total (%) | 24/177 (13.6) |
| Anemia, no./total (%)† | 158/178 (88.8) |
| Hb level, median (range), g/dL | 9.5 (2.5-15) |
| MCV, median (range) | 98 (68-117) |
| Thrombocytopenia, no./total (%), <150 × 109/L | 147/178 (82.6) |
| Platelet count, median (range), × 109/L | 45 (2-500) |
| Neutropenia, no./total (%), <1.5 × 109/L | 138/177 (78.0) |
| Neutrophil count, median (range), × 109/L | 0.87 (0-7) |
| BM dysplasia, no./total (%) | 68/157 (43.3) |
| BM blast cell percentage, no./total (%) | |
| <5% | 154/175 (88.0) |
| ≥5, <10%‡ | 9/175 (5.1) |
| ≥10%‡ | 12/175 (6.9) |
| Abnormal karyotype, no./total (%)§ | 45/161 (28.0) |
| Family history, no./total (%) | 66/173 (38.2) |
| Family history of hematological disorders | 50/173 (28.9) |
| Consanguinity|| | 25/179 (14.0) |
| Physical abnormalities, no./total (%) | 129/176 (73.3) |
| Growth restriction | 77/173 (44.5) |
| Skeletal malformation | 49/172 (28.5) |
| Neurological disorder | 39/172 (22.7) |
| Skin, nail, or hair abnormalities | 40/172 (23.3) |
| Genitourinary disorder | 32/171 (18.7) |
| Cardiac disorder | 15/170 (8.8) |
| Lung disorder | 8/172 (4.7) |
| Liver disorder | 8/173 (4.6) |
Hb, hemoglobin; MCV, mean corpuscular volume.
May not total 179 due to missing data in some patients and percentages may not total 100 due to rounding.
Hemoglobin concentration <2 SDs below mean, adjusted for age.
Detailed in supplemental Table 6.
Detailed in supplemental Table 7.
Estimated from the WES data as detailed in “Methods.”
Analysis of gene variants
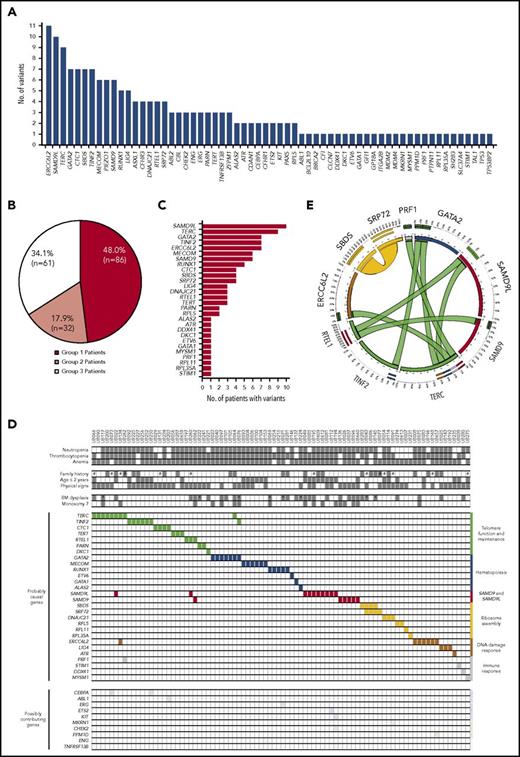
We analyzed WES data from fibroblast genomic DNA, considered here to be germ line, in the 179 patients. One hundred ninety-nine genic alterations, including nucleotide substitutions (N = 162), small insertions or deletions (N = 29), and larger chromosomal deletions (N = 8) were identified in 62 genes (Figure 2A; supplemental Tables 8 and 9). Based on comprehensive analysis combining clinical and molecular evidence (see Roundtable session description with representative examples in supplemental Methods), we provided a molecular diagnosis for 86 IBMF patients who were classified as “assigned with a causal or likely causal gene variant” (group 1; 48.0%; Figure 2B-C; supplemental Tables 8 and 10). In these group 1 patients, 40.7% of the gene variants that we regarded as causal or likely causal had been previously reported (supplemental Table 8). In 40 other patients (group 2; 22.3%), contributing gene variants were suspected but not regarded as causal (Figure 2B; supplemental Table 9), whereas no relevant variant was identified in the 53 remaining patients (group 3; 29.6%).
Gene variants in the IBMF cohort. (A) Number of gene variants identified using WES analysis in 179 patients. This collection of variants was further analyzed in comprehensive, iterative roundtable sessions that enabled us to assign each patient into 1 of 3 groups and provide final lists of genes as shown in panels B and C and supplemental Tables 8 and 9. (B) Distribution of patients into 3 groups: group 1, patients assigned with a causal or likely causal gene variant; group 2, patients with possibly contributing variants but no established molecular diagnosis; group 3, patients with no relevant genetic variant identified. (C) Distribution of the genes retained as causal or likely causal (86 patients, also see supplemental Table 8). (D) Integrated matrix of the 86 assigned patients and gene variants. The top part of the panel shows clinical, hematological criteria, and family history. #, consanguinity as screened by WES analysis; *, patients with BM blast cells ≥5% in a context of dysplasia. The remaining part of the panel shows gene mutations including co-occurring mutations. Genes are grouped by broad biological pathways. The distribution of additional, possibly contributing, variants is also shown in the bottom part. (E) Circos plot representation of co-occurring mutations in causal or likely causal BMF genes. Ribbon width is proportional to the number of co-occurrences between genes.
Gene variants in the IBMF cohort. (A) Number of gene variants identified using WES analysis in 179 patients. This collection of variants was further analyzed in comprehensive, iterative roundtable sessions that enabled us to assign each patient into 1 of 3 groups and provide final lists of genes as shown in panels B and C and supplemental Tables 8 and 9. (B) Distribution of patients into 3 groups: group 1, patients assigned with a causal or likely causal gene variant; group 2, patients with possibly contributing variants but no established molecular diagnosis; group 3, patients with no relevant genetic variant identified. (C) Distribution of the genes retained as causal or likely causal (86 patients, also see supplemental Table 8). (D) Integrated matrix of the 86 assigned patients and gene variants. The top part of the panel shows clinical, hematological criteria, and family history. #, consanguinity as screened by WES analysis; *, patients with BM blast cells ≥5% in a context of dysplasia. The remaining part of the panel shows gene mutations including co-occurring mutations. Genes are grouped by broad biological pathways. The distribution of additional, possibly contributing, variants is also shown in the bottom part. (E) Circos plot representation of co-occurring mutations in causal or likely causal BMF genes. Ribbon width is proportional to the number of co-occurrences between genes.
The 28 genes in group 1 patients are displayed in Figure 2C. SAMD9L was found mutated in 10 of the 179 patients (5.6%), followed by TERC (N = 9, 5.0%), GATA2 (N = 7, 3.9%), ERCC6L2 (N = 7, 3.9%), TINF2 (N = 7, 3.9%), MECOM/EVI1 (N = 6, 3.4%), SAMD9 (N = 6, 3.4%), and RUNX1 (N = 5, 2.8%). Of note, a tail of mutant genes was observed individually once in a single patient, some already known to be implicated in IBMF (such as DKC1 and ETV6, classifying the corresponding patients in group 1), whereas others had not been described in IBMF and so were regarded as tentative (group 2). In group 1, 60 patients had monoallelic variants (in genes such as GATA2, TERC, SAMD9L, SAMD9, and MECOM), whereas 27 had biallelic variants (in genes such as ERCC6L2, SBDS, and LIG4), suggesting autosomal-dominant and autosomal-recessive disorders, respectively (supplemental Table 8). The transmission observed was largely consistent in 24 patients who had a family history of hematological disorders (supplemental Figure 3, see family histories in the figure legend). Moreover, 8 of the 13 group 1 patients with parental consanguinity had homozygous mutations, as expected, whereas the 5 remaining patients had monoallelic mutations in an autosomal-dominant gene or compound heterozygous mutations (supplemental Figure 8; supplemental Table 8).
The assigned gene variants and various annotations of group 1 patients were displayed in an integrative matrix (Figure 2D). Interestingly, we found 2 co-occurring IBMF gene variants in 10 of the 86 group 1 patients (11.6%), involving a total of 10 of the 28 causal or likely causal genes (Figure 2D-E; supplemental Table 11). Similar findings have recently been reported in nonhematological genetic disorders, suggesting that combined mutations may modulate and enrich phenotypes.31,32
For 34 other genes, the variants were considered to be “possibly contributing,” either in isolation (defining group 2 patients; Figure 2A-B) or in association with another gene in group 1 patients (Figure 2D). More patients and case-by-case functional investigations will be required to clarify the potential role of each of these variants in the BMF phenotype.
Mutations converged toward major IBMF core biological pathways
We then clustered the causal or likely causal genes in our patient cohort according to their known function (Figure 2D; supplemental Figure 4). A first cluster comprised telomere genes (TERT, TERC, DKC1, and RTEL1), found mutated in 29 of 86 patients (33.7%). A second gene cluster, mutated in 21 patients (24.4%), comprised hematopoietic genes. These were mainly transcription factor genes involved in hematopoietic stem and progenitor cells, such as GATA2, RUNX1, and MECOM/EVI1. A third cluster included genes primarily involved in ribosome assembly, such as SBDS, SRP72, DNAJC21, and RPL5 (12 patients; 14.0%), whereas DNA damage response genes such as ERCC6L2, LIG4, and ATR were mutated in 11 patients (12.8%), and immune response genes such as DDX41 and STIM1 were mutated in 4 patients (4.7%).
Finally, we identified recurrent mutations of 2 partially homologous genes not initially included in the major IBMF core biological pathways, SAMD9L and SAMD9. These genes were adjacent at chromosomal region 7q21 and encoded 2 large proteins of ill-defined functions in the regulation of inflammatory and antiviral responses.33-35
IBMF phenotypes and syndromes
We analyzed the clinical and biological phenotypes of the 86 patients and compared them to those described in the literature and medical databases in association with the corresponding genes (supplemental Table 12).
Forty-nine of the 86 group 1 patients (57.0%) had mutated genes well known to be associated with IBMF and/or MDS/AML. These included DC, DBA, and SDS gene mutations. Many of these patients presented without the prototypical (syndromic) clinical presentation (supplemental Table 12). For example, among the 29 patients with telomere gene mutation, only 18 actually had mouth, nail, or skin lesions typical of DC, whereas only 1 in 4 patients with ribosome gene mutations presented with a typical phenotype of DBA (supplemental Table 12). These “atypical” presentations were most probably enriched by our study design (Figure 1).
In 29 patients (33.7% of group 1), mutations in genes seldom or not primarily reported hitherto in IBMF were found, including MECOM (N = 6), ERCC6L2 (N = 7), SAMD9L (N = 10), and SAMD9 (N = 6).
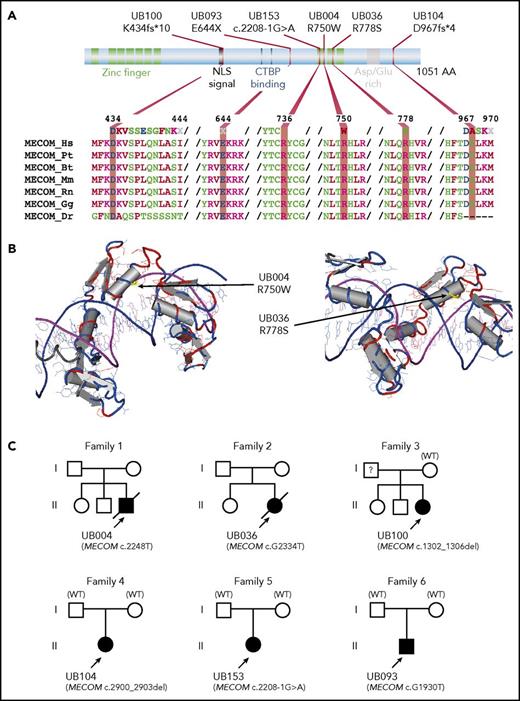
Inactivating mutations in MECOM (whose product is the EVI1 transcription factor) have been previously reported in 3 patients with radioulnar synostosis with amegakaryocytic thrombocytopenia (RUSAT) syndrome.36 In our cohort, we identified 6 unrelated, mutated patients. Where parental samples were available (N = 3, both paternity and maternity confirmed), the child mutation was de novo (Figure 3). Strikingly, all patients presented with severe neonatal AA without BM dysplasia, and all patients received HSCT before the age of 3. Only 1 presented an ulnar syndrome (all patients having undergone skeletal imaging) (Table 2). Clinical phenotypes often included cardiac abnormalities (found in 3 patients in the current cohort and leading to death in 2).
Predicted amino acid changes and family trees of patients with MECOM mutations. (A) Graphical representation of MECOM/EVI1 predicted protein with its major functional domains (top part) including zinc finger in green, nuclear localization signal (NLS) and asparagine/glutamine (Asp/Glu)-rich region in gray, and C-terminal binding protein (CTBP) binding in blue. The bottom part shows amino acid alignments using ClustalW, representing conservation among different species. (B) Three-dimensional (3D) model of the localization of amino acid substitutions (yellow part, indicated by an arrow), highlighting the proximity of these amino acids with DNA (purple/blue double helix). The structure representations originate from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB; www.rcsb.org/pdb) after BLAST alignment of MECOM variant proteins. (C) Family trees for the 6 patients with MECOM mutations. All mutations were de novo where parental DNA was available and both paternity and maternity were confirmed (families 4 to 6). In family 3, MECOM mutation was not identified in mother DNA, whereas paternal DNA was not available. See clinical details in Table 2. Bt, Bos taurus; Dr, Danio rerio; Gg, Gallus gallus; Hs, Homo sapiens; Mm, Mus musculus; Pt, Pan troglodytes; Rn, Rattus norvegicus; WT, wild-type.
Predicted amino acid changes and family trees of patients with MECOM mutations. (A) Graphical representation of MECOM/EVI1 predicted protein with its major functional domains (top part) including zinc finger in green, nuclear localization signal (NLS) and asparagine/glutamine (Asp/Glu)-rich region in gray, and C-terminal binding protein (CTBP) binding in blue. The bottom part shows amino acid alignments using ClustalW, representing conservation among different species. (B) Three-dimensional (3D) model of the localization of amino acid substitutions (yellow part, indicated by an arrow), highlighting the proximity of these amino acids with DNA (purple/blue double helix). The structure representations originate from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB; www.rcsb.org/pdb) after BLAST alignment of MECOM variant proteins. (C) Family trees for the 6 patients with MECOM mutations. All mutations were de novo where parental DNA was available and both paternity and maternity were confirmed (families 4 to 6). In family 3, MECOM mutation was not identified in mother DNA, whereas paternal DNA was not available. See clinical details in Table 2. Bt, Bos taurus; Dr, Danio rerio; Gg, Gallus gallus; Hs, Homo sapiens; Mm, Mus musculus; Pt, Pan troglodytes; Rn, Rattus norvegicus; WT, wild-type.
Clinical and hematological characteristics of patients with MECOM mutations
| Patient ID . | UB004 . | UB036 . | UB093 . | UB100 . | UB104 . | UB153 . |
|---|---|---|---|---|---|---|
| Nucleic acid change | c.C2248T | c.G2334T | c.G1930T | c.1302_1306del | c.2900_2903del | c.2208-1G>A |
| Amino acid change | p.R750W | p.R778S | p.E644X | p.K434fs | p.D967fs | — |
| Sex | M | F | M | F | F | F |
| Age | 18 mo | 6 mo | 3 mo | Neonatal | 9 mo | 9 mo |
| Family history | Simplex | Simplex | Simplex | Simplex | Simplex | Simplex |
| Hb, g/dL | 5.7 | 6.0 | 6.0 | 9.2 | 5.5 | 8.0 |
| Platelets, × 109/L | 1 | 10 | 10 | 62 | 36 | 10 |
| ANC, × 109/L | 0.35 | 0.06 | 0 | 0 | 0.4 | 0 |
| BM | Hypocellular | Hypocellular | Hypocellular | Hypocellular | Hypocellular | Hypocellular |
| BM karyotype | 46,XY | 46,XX | 46,XY | 46,XX | Trisomy 8 | 46,XX |
| Skeletal abnormality | Radioulnar synostosis | Thumb abnormalities | Clubfoot | No | No | Thumb abnormalities |
| Cardiac abnormality | Tetralogy of Fallot | Myocardial atrophy | Pulmonary stenosis | No | No | No |
| Other | — | — | Facial dysmorphia | — | — | Renal hypoplasia |
| Age at HSCT | 3 y | 6 mo | 15 mo | 9 mo | 18 mo | 3 y |
| Outcome | Died 3 mo after HSCT from a cardiac complication during severe infection | Died at 14 y from a cardiac complication during influenza infection | No major complication 9 y after HSCT (10-y old) | No major complication 1 y after HSCT (2-y old) | No major complication 8 mo after HSCT (2-y old) | No major complication 3 y after a second HSCT (6-y old) |
| Patient ID . | UB004 . | UB036 . | UB093 . | UB100 . | UB104 . | UB153 . |
|---|---|---|---|---|---|---|
| Nucleic acid change | c.C2248T | c.G2334T | c.G1930T | c.1302_1306del | c.2900_2903del | c.2208-1G>A |
| Amino acid change | p.R750W | p.R778S | p.E644X | p.K434fs | p.D967fs | — |
| Sex | M | F | M | F | F | F |
| Age | 18 mo | 6 mo | 3 mo | Neonatal | 9 mo | 9 mo |
| Family history | Simplex | Simplex | Simplex | Simplex | Simplex | Simplex |
| Hb, g/dL | 5.7 | 6.0 | 6.0 | 9.2 | 5.5 | 8.0 |
| Platelets, × 109/L | 1 | 10 | 10 | 62 | 36 | 10 |
| ANC, × 109/L | 0.35 | 0.06 | 0 | 0 | 0.4 | 0 |
| BM | Hypocellular | Hypocellular | Hypocellular | Hypocellular | Hypocellular | Hypocellular |
| BM karyotype | 46,XY | 46,XX | 46,XY | 46,XX | Trisomy 8 | 46,XX |
| Skeletal abnormality | Radioulnar synostosis | Thumb abnormalities | Clubfoot | No | No | Thumb abnormalities |
| Cardiac abnormality | Tetralogy of Fallot | Myocardial atrophy | Pulmonary stenosis | No | No | No |
| Other | — | — | Facial dysmorphia | — | — | Renal hypoplasia |
| Age at HSCT | 3 y | 6 mo | 15 mo | 9 mo | 18 mo | 3 y |
| Outcome | Died 3 mo after HSCT from a cardiac complication during severe infection | Died at 14 y from a cardiac complication during influenza infection | No major complication 9 y after HSCT (10-y old) | No major complication 1 y after HSCT (2-y old) | No major complication 8 mo after HSCT (2-y old) | No major complication 3 y after a second HSCT (6-y old) |
—, Not applicable; ANC, absolute neutrophil count; F, Female; Hb, hemoglobin concentration; HSCT, hematopoietic stem cell transplantation; ID, identifier; M, male.
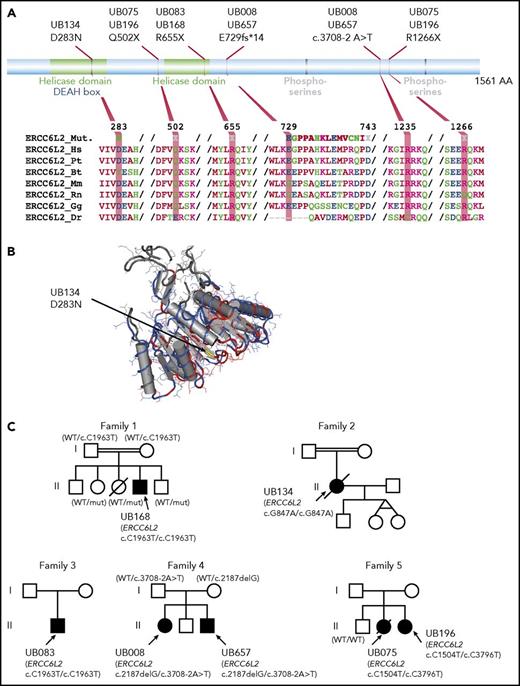
ERCC6L2 belongs to a helicase gene family. Homozygous-truncating mutations have been reported in 3 patients with BMF, learning disabilities, and microcephaly.37,38 In our cohort, biallelic ERCC6L2 mutations were found in 7 patients (5 families; median age, 13 years) (Figure 4; Table 3). Marrow was mostly hypoplastic, whereas dysplastic features were found in 2 patients, both with monosomy 7. Only 1 presented additional neurological symptoms (ie, learning difficulties, developmental delay, and vascular abnormalities in the right frontal lobe on magnetic resonance imaging [MRI]).
Predicted amino acid changes and family trees of patients with ERCC6L2 mutations. (A) Graphical representation of ERCC6L2 protein. Functional domains (top part) such as helicase domains are depicted in green, phosphoserines in gray, and DEAH boxes that denote Asp-Glu-Ala-His box in blue. The bottom part shows amino acid alignments using ClustalW, representing conservation among different species. (B) A 3D model of the ERCC6L2 amino acid substitution (yellow part, indicated by an arrow) highlighting the location of this amino acid in a conserved helicase domain. (C) Family trees for the 7 patients with ERCC6L2 biallelic mutations (5 families). Where parental DNA was available and both paternity and maternity were confirmed (family 1 and 6), each parent bore 1 allele. Patients from consanguineous union in families 1 and 2 had homozygous ERCC6L2 mutations. Of note, families 1 and 3 with identical variants were unrelated.
Predicted amino acid changes and family trees of patients with ERCC6L2 mutations. (A) Graphical representation of ERCC6L2 protein. Functional domains (top part) such as helicase domains are depicted in green, phosphoserines in gray, and DEAH boxes that denote Asp-Glu-Ala-His box in blue. The bottom part shows amino acid alignments using ClustalW, representing conservation among different species. (B) A 3D model of the ERCC6L2 amino acid substitution (yellow part, indicated by an arrow) highlighting the location of this amino acid in a conserved helicase domain. (C) Family trees for the 7 patients with ERCC6L2 biallelic mutations (5 families). Where parental DNA was available and both paternity and maternity were confirmed (family 1 and 6), each parent bore 1 allele. Patients from consanguineous union in families 1 and 2 had homozygous ERCC6L2 mutations. Of note, families 1 and 3 with identical variants were unrelated.
Clinical and hematological characteristics of patients with ERCC6L2 mutations
| Patient ID . | UB657 . | UB008 . | UB075 . | UB196 . | UB083 . | UB134 . | UB168 . |
|---|---|---|---|---|---|---|---|
| Nucleic acid change | c.2187delG c.3708-2A>T | c.2187delG c.3708-2A>T | c.C1504T c.C3796T | c.C1504T c.C3796T | c.C1963T c.C1963T | c.G847A c.G847A | c.C1963T c.C1963T |
| Amino acid change | p.E729fs — | p.E729fs — | p.Q502X p.R1266X | p.Q502X p.R1266X | p.R655X p.R655X | p.D283N p.D283N | p.R655X p.R655X |
| Other causal mutation | — | — | — | — | — | TERC | — |
| Sex | M | F | F | F | M | F | M |
| Age, y | 7 | 13 | 22 | 18 | 2 | 22 | 13 |
| Family history | Brother of UB008 | Sister of UB657 | Sister of UB196 | Sister of UB075 | N/A | Simplex | Consanguinity, brother with intellectual disability |
| Hb, g/dL | 11.4 | <12 | 11.9 | 12.9 | 10.9 | 10.7 | 9.0 |
| Platelets, × 109/L | 64 | <150 | 107 | 101 | 48 | 38 | 4 |
| ANC, × 109/L | <1.5 | <1.5 | 0.4 | 1.6 | 1.0 | 0.1 | 0.7 |
| BM | Hypocellular | Hypocellular | Dysplasia | N/A | Hypocellular | Hypocellular dysplasia | Hypocellular |
| BM karyotype | 46,XX | 46,XX | Monosomy 7 | N/A | 46,XY | 46,XX | 46,XY |
| Microcephaly | No | No | No | No | No | No | Yes |
| Neurological defect | No | No | No | No | No | No | Learning difficulties, intellectual disability, vascular abnormalities in the right frontal lobe (MRI) |
| Other | — | — | — | — | Facial dysmorphia | — | Bilateral pyeloureteral junction abnormalities |
| Age at HSCT, y | 14 | 13 | 22 | — | — | — | — |
| Outcome | No significant complication after HSCT (15-y old) | No significant complication after HSCT (27-y old) | Died at 24 y, of EBV lymphoma post-HSCT | Thrombo-cytopenia and neutropenia (26-y old) | Mild thrombo-cytopenia (15-y old) | Died at 43 y, after AML with −7, hypomethylating agent failure | Stable, macrocytosis without anemia, no neurological signs (21-y old) |
| Patient ID . | UB657 . | UB008 . | UB075 . | UB196 . | UB083 . | UB134 . | UB168 . |
|---|---|---|---|---|---|---|---|
| Nucleic acid change | c.2187delG c.3708-2A>T | c.2187delG c.3708-2A>T | c.C1504T c.C3796T | c.C1504T c.C3796T | c.C1963T c.C1963T | c.G847A c.G847A | c.C1963T c.C1963T |
| Amino acid change | p.E729fs — | p.E729fs — | p.Q502X p.R1266X | p.Q502X p.R1266X | p.R655X p.R655X | p.D283N p.D283N | p.R655X p.R655X |
| Other causal mutation | — | — | — | — | — | TERC | — |
| Sex | M | F | F | F | M | F | M |
| Age, y | 7 | 13 | 22 | 18 | 2 | 22 | 13 |
| Family history | Brother of UB008 | Sister of UB657 | Sister of UB196 | Sister of UB075 | N/A | Simplex | Consanguinity, brother with intellectual disability |
| Hb, g/dL | 11.4 | <12 | 11.9 | 12.9 | 10.9 | 10.7 | 9.0 |
| Platelets, × 109/L | 64 | <150 | 107 | 101 | 48 | 38 | 4 |
| ANC, × 109/L | <1.5 | <1.5 | 0.4 | 1.6 | 1.0 | 0.1 | 0.7 |
| BM | Hypocellular | Hypocellular | Dysplasia | N/A | Hypocellular | Hypocellular dysplasia | Hypocellular |
| BM karyotype | 46,XX | 46,XX | Monosomy 7 | N/A | 46,XY | 46,XX | 46,XY |
| Microcephaly | No | No | No | No | No | No | Yes |
| Neurological defect | No | No | No | No | No | No | Learning difficulties, intellectual disability, vascular abnormalities in the right frontal lobe (MRI) |
| Other | — | — | — | — | Facial dysmorphia | — | Bilateral pyeloureteral junction abnormalities |
| Age at HSCT, y | 14 | 13 | 22 | — | — | — | — |
| Outcome | No significant complication after HSCT (15-y old) | No significant complication after HSCT (27-y old) | Died at 24 y, of EBV lymphoma post-HSCT | Thrombo-cytopenia and neutropenia (26-y old) | Mild thrombo-cytopenia (15-y old) | Died at 43 y, after AML with −7, hypomethylating agent failure | Stable, macrocytosis without anemia, no neurological signs (21-y old) |
—, Not applicable; EBV, Epstein-Barr virus; MRI, magnetic resonance imaging; N/A, not available. Other abbreviations are explained in Table 2.
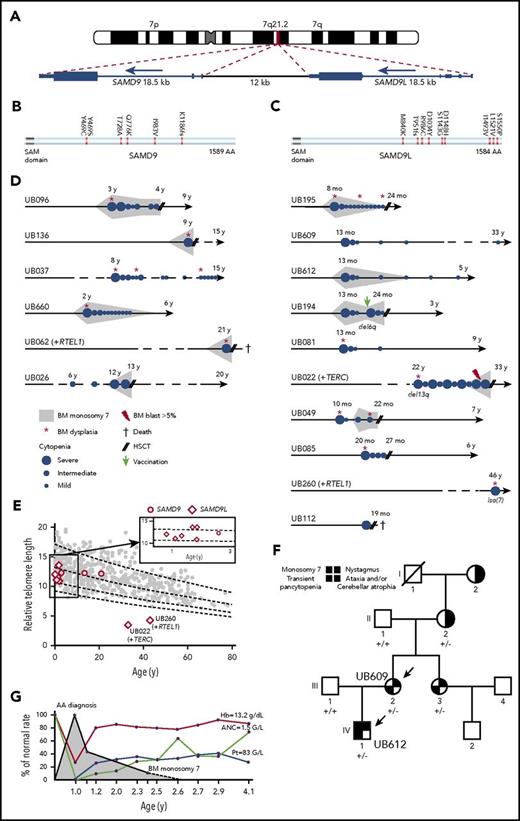
The SAMD9L gene was found mutated in 10 patients from 9 unrelated families, representing the most frequently mutated gene in our IBMF cohort (Figures 2C-D and 5; supplemental Figure 5). Activating mutations have been reported recently in the rare autosomal-dominant Ataxia-Pancytopenia syndrome, with patients from 4 families presenting with hematological and neurological symptoms.39,40 Patients from our cohort also presented with severe BMF, and monosomy 7 was found in 5 of 10 patients (Figure 5; supplemental Figure 5; Table 4). Only 2 patients had significant neurological abnormalities (hydrocephaly and arachnoid cyst on MRI), whereas 2 in the same family had nystagmus without any other severe neurological signs. Three patients had recurrent infections in infancy, with documented immunoglobulin deficiency in 2 (Table 4).
Characteristics in patients with SAMD9L and SAMD9 mutations. (A) G-banding ideogram of human chromosome 7 with genomic location of the tandem SAMD9 and SAMD9L genes. (B-C) Graphical representation of SAMD9L and SAMD9 proteins, respectively. Amino acid changes are indicated; see additional details in supplemental Figures 6 and 7. (D) Schematic of hematological history from birth to latest follow-up in the 16 patients with SAMD9 or SAMD9L mutations; see family trees in supplemental Figures 6 and 7, clinical characteristics in Table 4 and somatic evolution in supplemental Table 12. Median age of the SAMD9L and SAMD9 mutated patients at diagnosis was 13 months and 7 years, respectively (range, 8 months to 46 years, and 2 to 21 years, respectively). (E) Telomere length in PBMCs in 8 SAMD9L and 3 SAMD9 mutated patients, measured by flow cytometry fluorescence in situ hybridization. The vertical axis shows the relative telomere length determined in comparison with a control tetraploid cell line per percentage, whereas the horizontal axis displays age. Only 2 patients had short telomeres, and they also had a TERC and a RTEL1 mutation, respectively. (F) Family tree for patients UB609 and UB612 (arrows). The germ line SAMD9L mutation status is shown for the family members; +/−, heterozygous c.C2956T; +/+, no mutation. Patient UB612 (IV-1) was diagnosed with severe BMF and monosomy 7 without BM dysplasia at the age of 13 months. Transplantation was planned but eventually cancelled because his blood cell counts improved spontaneously. This patient is now 5 years old, without residual cytopenia or monosomy 7. His mother, patient UB609 (III-2), experienced a rather similar story 32 years previously; when she was 13 months old she was diagnosed with severe AA and BM transplantation was planned, which was also cancelled due to spontaneous improvement. Although they were not included in our IBMF cohort, we managed to investigate more patients in this family. The mother’s sister (III-3) also had early transitory pancytopenia, and both sisters, now aged 33 and 37 years, are doing well. Interestingly, the grandmother (II-2) had a cerebellar atrophy with ataxia, a nystagmus and the SAMD9L mutation found in her oral swab, whereas her own mother (I-2) also had the same neurological phenotype but could not be genetically explored. (G) Blood cell counts over time for patient UB612. The red curve indicates the hemoglobin concentration (Hb); light brown, the absolute neutrophil count (ANC); whereas the blue line indicates platelet counts (Pt). The gray area indicates the detection of monosomy 7 on marrow karyotyping.
Characteristics in patients with SAMD9L and SAMD9 mutations. (A) G-banding ideogram of human chromosome 7 with genomic location of the tandem SAMD9 and SAMD9L genes. (B-C) Graphical representation of SAMD9L and SAMD9 proteins, respectively. Amino acid changes are indicated; see additional details in supplemental Figures 6 and 7. (D) Schematic of hematological history from birth to latest follow-up in the 16 patients with SAMD9 or SAMD9L mutations; see family trees in supplemental Figures 6 and 7, clinical characteristics in Table 4 and somatic evolution in supplemental Table 12. Median age of the SAMD9L and SAMD9 mutated patients at diagnosis was 13 months and 7 years, respectively (range, 8 months to 46 years, and 2 to 21 years, respectively). (E) Telomere length in PBMCs in 8 SAMD9L and 3 SAMD9 mutated patients, measured by flow cytometry fluorescence in situ hybridization. The vertical axis shows the relative telomere length determined in comparison with a control tetraploid cell line per percentage, whereas the horizontal axis displays age. Only 2 patients had short telomeres, and they also had a TERC and a RTEL1 mutation, respectively. (F) Family tree for patients UB609 and UB612 (arrows). The germ line SAMD9L mutation status is shown for the family members; +/−, heterozygous c.C2956T; +/+, no mutation. Patient UB612 (IV-1) was diagnosed with severe BMF and monosomy 7 without BM dysplasia at the age of 13 months. Transplantation was planned but eventually cancelled because his blood cell counts improved spontaneously. This patient is now 5 years old, without residual cytopenia or monosomy 7. His mother, patient UB609 (III-2), experienced a rather similar story 32 years previously; when she was 13 months old she was diagnosed with severe AA and BM transplantation was planned, which was also cancelled due to spontaneous improvement. Although they were not included in our IBMF cohort, we managed to investigate more patients in this family. The mother’s sister (III-3) also had early transitory pancytopenia, and both sisters, now aged 33 and 37 years, are doing well. Interestingly, the grandmother (II-2) had a cerebellar atrophy with ataxia, a nystagmus and the SAMD9L mutation found in her oral swab, whereas her own mother (I-2) also had the same neurological phenotype but could not be genetically explored. (G) Blood cell counts over time for patient UB612. The red curve indicates the hemoglobin concentration (Hb); light brown, the absolute neutrophil count (ANC); whereas the blue line indicates platelet counts (Pt). The gray area indicates the detection of monosomy 7 on marrow karyotyping.
Clinical and hematological characteristics of patients with SAMD9L and SAMD9 mutations
| ID (UB) . | Mutation . | Sex/age . | Family history . | Hb, g/dL Plt, × 109/L ANC, × 109/L . | BM . | BM cytogenetic . | Short TLM . | GR . | Immune deficiency . | Neurological defect . | Genitourinary abnormality . | Other . | Age at BMT . | Outcome . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 195 | SAMD9L* | M/8 mo | Consanguinity | 6.5 | Hypocellular dysplasia | 45,XY, −7[7/15] | No | No | No | No | No | — | 24 mo | Alive, 2 months after HSCT, GVHD |
| 5 | ||||||||||||||
| 0.06 | ||||||||||||||
| 609 | SAMD9L | F/13 mo | Yes (see Figure 3E) | <6 | N/A | ND | ND | No | No | Nystagmus | No | — | — | Mild thrombocytopenia (33-y old) |
| <2 | ||||||||||||||
| <1 | ||||||||||||||
| 612 | SAMD9L | M/13 mo | Yes (see Figure 3E) | 6 | Hypocellular | 45,XY, −7[16/28] | No | No | Transient Ig deficiency (IgG < 3 g) | Nystagmus | No | — | — | BM FISH −7[8%] (2-y old) |
| 10 | Mild cytopenias (5-y old) | |||||||||||||
| 0.25 | ||||||||||||||
| 194 | SAMD9L | F/13 mo | Simplex | 6 | Hypocellular | 45,XX, −7 | No | No | No | No | No | — | 25 mo | Spontaneous blood cell count improvement; relapse after vaccination; clonal progression with del6q; then HSCT |
| 17 | ||||||||||||||
| 0.27 | ||||||||||||||
| 081 | SAMD9L | M/13 mo | Uncle with Hodgkin disease | 9.5 | Hypocellular dysplasia | Normal, no −7 by FISH | ND | No | No | Hydrocephaly, arachnoid cyst on MRI | No | — | — | Complete regression of cytopenias (9-y old) |
| 3 | ||||||||||||||
| 0.25 | ||||||||||||||
| 022 | SAMD9L TERC | M/27 y | Father with transient thrombo-cytopenia. Grandfather died of AML | 13 | Hypocellular dysplasia | Del13q, then −7 | Yes | No | Recurrent infection in infancy | No | No | Hepatitis, pulmonary fibrosis | 33 y | RAEB1 with del13q and −7 (33-y old); CR after 1 cycle of azacytidine; HSCT; pulmonary fibrosis (37-y old) |
| 79 | ||||||||||||||
| 1.68 | ||||||||||||||
| 049 | SAMD9L | F/10 mo | Brother with cytopenias | 11 | Hypocellular dysplasia | 45,XX, −7[17/20] | No | No | No | No | No | — | 24 mo | Alive 5 years after HSCT |
| <100 | ||||||||||||||
| 0.72 | ||||||||||||||
| 085 | SAMD9L | F/20 mo | Mother and uncle with mild intellectual disability | 8.2 | Dysplasia | Normal | No | Yes | Ig deficiency (IgG, 3 g) | Mild intellectual disability, hydrocephaly, bilateral white substance changes and arachnoid cyst on MRI | No | Asthma, coxa valga | 27 mo | Neurological defects with unsteady gait (6-y old), cerebellar hypoplasia and abnormal white matter signal and arachnoid cyst on MRI. Growth defect and parenteral nutrition |
| 20 | ||||||||||||||
| 1.7 | ||||||||||||||
| 260 | SAMD9L RTEL1 | M/46 y | — | 10.9 | Dysplasia | 46,XY, der(7)[6]/ 46,XY,ish add(7)[14] | Yes | No | — | No | No | Tongue cancer, cirrhosis | — | Lost sight |
| 19 | ||||||||||||||
| <1 | ||||||||||||||
| 112 | SAMD9L | F/19 mo | Simplex | 8.1 | N/A | Normal no −7 on FISH | No | Yes | N/A | No | No | — | 19 mo | Died in intensive care unit during severe infection (21-mo old) |
| 5 | ||||||||||||||
| 0.12 | ||||||||||||||
| 096 | SAMD9 | F/3 y | Simplex | 11.9 | Hypocellular dysplasia | 45,XX, −7[12/20] | No | Yes | Severe infections | Abnormal white matter signal on MRI | No | Hyper-thyroidism | 4 y | No GVHD, persistent hyperthyroidism (9-y old) |
| 240 | ||||||||||||||
| 0 | ||||||||||||||
| 136 | SAMD9 | M/9 y | Simplex | 12 | Dysplasia | −7 | ND | Yes | No | No | No | — | 9 y | No major complication after HSCT (15-y old) |
| 15 | ||||||||||||||
| 0.5 | ||||||||||||||
| 037 | SAMD9 | F/8 y | Simplex | 10.2 | Dysplasia | Normal | ND | Yes | Ig deficiency (IgM, IgG2, IgG4), recurrent infections | No | No | — | — | Thrombocytopenia with dysplasia in BM, normal karyotype, immunodeficiency (15-y old) |
| 14 | ||||||||||||||
| 0.94 | ||||||||||||||
| 660 | SAMD9 | M/24 mo | Simplex | 11.4 | Dysplasia | 45,XY, −7[14/20] | ND | Yes | No | Language delay | Hypospadias, testicular dysgenesis | Diarrhea | — | Complete regression of cytopenias and of −7, persists at 6-y old |
| 19 | ||||||||||||||
| 0.7 | ||||||||||||||
| 062 | SAMD9 RTEL1 | F/M/21 y | Simplex | 12.1 | Hypocellular dysplasia | 45,XY, −7 | No | No | Recurrent severe infections | No | Hypospadias, cryptorchidism, sexual ambiguity, renal hypoplasia | Diarrhea | 21 y | HSCT, died (22-y old) |
| 88 | ||||||||||||||
| 0.5 | ||||||||||||||
| 026 | SAMD9 | F/6 y | Simplex | 11.5 | Hypocellular | 45,XX, −7 | No | Yes | IgG deficiency, recurrent infections | No | No | — | 13 y | No major complication after HSCT (20-y old) |
| 102 | ||||||||||||||
| 1.0 |
| ID (UB) . | Mutation . | Sex/age . | Family history . | Hb, g/dL Plt, × 109/L ANC, × 109/L . | BM . | BM cytogenetic . | Short TLM . | GR . | Immune deficiency . | Neurological defect . | Genitourinary abnormality . | Other . | Age at BMT . | Outcome . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 195 | SAMD9L* | M/8 mo | Consanguinity | 6.5 | Hypocellular dysplasia | 45,XY, −7[7/15] | No | No | No | No | No | — | 24 mo | Alive, 2 months after HSCT, GVHD |
| 5 | ||||||||||||||
| 0.06 | ||||||||||||||
| 609 | SAMD9L | F/13 mo | Yes (see Figure 3E) | <6 | N/A | ND | ND | No | No | Nystagmus | No | — | — | Mild thrombocytopenia (33-y old) |
| <2 | ||||||||||||||
| <1 | ||||||||||||||
| 612 | SAMD9L | M/13 mo | Yes (see Figure 3E) | 6 | Hypocellular | 45,XY, −7[16/28] | No | No | Transient Ig deficiency (IgG < 3 g) | Nystagmus | No | — | — | BM FISH −7[8%] (2-y old) |
| 10 | Mild cytopenias (5-y old) | |||||||||||||
| 0.25 | ||||||||||||||
| 194 | SAMD9L | F/13 mo | Simplex | 6 | Hypocellular | 45,XX, −7 | No | No | No | No | No | — | 25 mo | Spontaneous blood cell count improvement; relapse after vaccination; clonal progression with del6q; then HSCT |
| 17 | ||||||||||||||
| 0.27 | ||||||||||||||
| 081 | SAMD9L | M/13 mo | Uncle with Hodgkin disease | 9.5 | Hypocellular dysplasia | Normal, no −7 by FISH | ND | No | No | Hydrocephaly, arachnoid cyst on MRI | No | — | — | Complete regression of cytopenias (9-y old) |
| 3 | ||||||||||||||
| 0.25 | ||||||||||||||
| 022 | SAMD9L TERC | M/27 y | Father with transient thrombo-cytopenia. Grandfather died of AML | 13 | Hypocellular dysplasia | Del13q, then −7 | Yes | No | Recurrent infection in infancy | No | No | Hepatitis, pulmonary fibrosis | 33 y | RAEB1 with del13q and −7 (33-y old); CR after 1 cycle of azacytidine; HSCT; pulmonary fibrosis (37-y old) |
| 79 | ||||||||||||||
| 1.68 | ||||||||||||||
| 049 | SAMD9L | F/10 mo | Brother with cytopenias | 11 | Hypocellular dysplasia | 45,XX, −7[17/20] | No | No | No | No | No | — | 24 mo | Alive 5 years after HSCT |
| <100 | ||||||||||||||
| 0.72 | ||||||||||||||
| 085 | SAMD9L | F/20 mo | Mother and uncle with mild intellectual disability | 8.2 | Dysplasia | Normal | No | Yes | Ig deficiency (IgG, 3 g) | Mild intellectual disability, hydrocephaly, bilateral white substance changes and arachnoid cyst on MRI | No | Asthma, coxa valga | 27 mo | Neurological defects with unsteady gait (6-y old), cerebellar hypoplasia and abnormal white matter signal and arachnoid cyst on MRI. Growth defect and parenteral nutrition |
| 20 | ||||||||||||||
| 1.7 | ||||||||||||||
| 260 | SAMD9L RTEL1 | M/46 y | — | 10.9 | Dysplasia | 46,XY, der(7)[6]/ 46,XY,ish add(7)[14] | Yes | No | — | No | No | Tongue cancer, cirrhosis | — | Lost sight |
| 19 | ||||||||||||||
| <1 | ||||||||||||||
| 112 | SAMD9L | F/19 mo | Simplex | 8.1 | N/A | Normal no −7 on FISH | No | Yes | N/A | No | No | — | 19 mo | Died in intensive care unit during severe infection (21-mo old) |
| 5 | ||||||||||||||
| 0.12 | ||||||||||||||
| 096 | SAMD9 | F/3 y | Simplex | 11.9 | Hypocellular dysplasia | 45,XX, −7[12/20] | No | Yes | Severe infections | Abnormal white matter signal on MRI | No | Hyper-thyroidism | 4 y | No GVHD, persistent hyperthyroidism (9-y old) |
| 240 | ||||||||||||||
| 0 | ||||||||||||||
| 136 | SAMD9 | M/9 y | Simplex | 12 | Dysplasia | −7 | ND | Yes | No | No | No | — | 9 y | No major complication after HSCT (15-y old) |
| 15 | ||||||||||||||
| 0.5 | ||||||||||||||
| 037 | SAMD9 | F/8 y | Simplex | 10.2 | Dysplasia | Normal | ND | Yes | Ig deficiency (IgM, IgG2, IgG4), recurrent infections | No | No | — | — | Thrombocytopenia with dysplasia in BM, normal karyotype, immunodeficiency (15-y old) |
| 14 | ||||||||||||||
| 0.94 | ||||||||||||||
| 660 | SAMD9 | M/24 mo | Simplex | 11.4 | Dysplasia | 45,XY, −7[14/20] | ND | Yes | No | Language delay | Hypospadias, testicular dysgenesis | Diarrhea | — | Complete regression of cytopenias and of −7, persists at 6-y old |
| 19 | ||||||||||||||
| 0.7 | ||||||||||||||
| 062 | SAMD9 RTEL1 | F/M/21 y | Simplex | 12.1 | Hypocellular dysplasia | 45,XY, −7 | No | No | Recurrent severe infections | No | Hypospadias, cryptorchidism, sexual ambiguity, renal hypoplasia | Diarrhea | 21 y | HSCT, died (22-y old) |
| 88 | ||||||||||||||
| 0.5 | ||||||||||||||
| 026 | SAMD9 | F/6 y | Simplex | 11.5 | Hypocellular | 45,XX, −7 | No | Yes | IgG deficiency, recurrent infections | No | No | — | 13 y | No major complication after HSCT (20-y old) |
| 102 | ||||||||||||||
| 1.0 |
—, Not applicable; BMT, BM transplantation; CR, complete remission; FISH, fluorescence in situ hybridization; GR, growth restriction; GVHD, graft-versus-host disease; Ig, immunoglobulin; K, karyotype; N/A, not available; ND, not done; Plt, platelet count; TLM, telomere. Other abbreviations are explained in Tables 1-3.
SAMD9L homozygous mutation.
Mutations in SAMD9 were found in 6 unrelated patients (Figure 5; supplemental Figure 6). Recently, a rare multiorgan syndrome called MIRAGE (myelodysplasia, infection, restriction of growth, adrenal hypoplasia, genital phenotypes, and enteropathy) has been associated with SAMD9 germ line mutations.41 In our cohort, the 6 patients presented with mild BMF, including multilineage dysplasia and monosomy 7 in 5 patients (Figure 5D; Table 4), whereas only 1 presented a portrait of MIRAGE syndrome. Four patients presented recurrent infections, with documented immunoglobulin deficiency in 2.
Strikingly, 11 of the 13 patients with SAMD9 and SAMD9L mutations who were not transplanted up front (ie, within 6 months) showed spontaneous improvement in blood cell counts within months, and even possible monosomy 7 disappearance (5 patients), with a median follow-up of 4 years (7 months to 32 years; censored at HSCT or relapse of cytopenia) (Figure 5; Table 4). Strikingly, HSCT that was planned for 5 of these patients was eventually cancelled and patients are still alive without consistent cytopenia with a follow-up of 4 to 32 years. In this respect, 2 related patients included in this cohort were highly informative based on their family history (Figure 5F; see family history in the legend of the figure). This recurrent clinical feature of spontaneous improvement in SAMD9/SAMD9L patients suggested that hematopoietic somatic mosaicism might lead to attenuated BMF.40,42 We therefore sequenced blood and/or marrow mononuclear cells obtained after spontaneous improvement in 12 patients from our cohort with germ line SAMD9 or SAMD9L mutations, and found somatic lesions such as acquired 7q uniparental disomy (UPD) or additional SAMD9 or SAMD9L cis mutations in 6 of them, confirming the spontaneous genetic correction hypothesis (supplemental Table 13; supplemental Figure 7). Of note, variant allele frequency (VAF) of the germ line–mutated allele in the blood at diagnosis may be low due to monosomy 7 or UPD, such as in patient UB085, whose VAF was 10% in peripheral blood cells by contrast with fibroblast DNA.
Discussion
Although an expert center can definitely rule out an FA diagnosis in patients using clastogenic agent sensitivity tests and fibroblast cells, many patients with BMF of suspected genetic origin remain undiagnosed after initial etiological workup. Using thorough clinical/biological analysis combined with WES of nonhematopoietic cells, we provided a molecular diagnosis for 48.0% of our cohort of undiagnosed patients with BMF of suspected genetic origin. Skin fibroblast DNA secured the germ line nature of the identified mutations, beyond potential hematopoietic reversion with somatic mosaicism, clonal hematopoiesis, or clonal progression that commonly occur throughout the lives of patients with IBMF.6,7,9,19-23 Fibroblast DNA use was especially helpful in SAMD9/SAMD9L patients in our study. Moreover, the comprehensive use of an unbiased, genome-wide analysis in a large cohort enabled us to extend the repertoire of the common genes whose mutations can confer IBMF, that is, beyond conventional IBMF genes and prototypical phenotypes. We found recurrent involvement of genes not usually mentioned at BMF diagnosis, such as SAMD9/SAMD9L, ERCC6L2, MECOM, and LIG4. Moreover, many patients with a mutation in a conventional IBMF gene, such as in a DC or DBA gene, had not been diagnosed at initial evaluation, either because the specific genetic test was not readily available or because the clinical diagnosis was cryptic or had not been suspected by the referring physician at this time.
Genes either associated or suspected to be associated with IBMF syndromes have been screened elsewhere.15-17 Notably, these studies were performed in blood cells (in which revertant cells can expand), included miscellaneous hematological disorders, and did not address SAMD9, SAMD9L, MECOM, or ERCC6L2 mutations, which we found to be prominent in our IBMF cohort. Nevertheless, we failed to obtain a molecular diagnosis for more than half of our patients. Reasons for this may include regions of low-coverage sequencing, mutations in noncoding regions (including introns and regulatory sequences), small/intermediate-size copy-number abnormalities as well as cryptic structural variants, for which WES is ineffective. Some patients may also have multifactorial genetics or acquired BMF combined with physical abnormalities.
Although no frequent novel IBMF gene was identified, many patients had gene variants that we considered to be “possibly contributing” (classifying patients in group 2), but which were not regarded as causal in our current study. More cases and further investigations will be required to investigate these and other IBMF candidates. Notably, genes such as TP53 and BRCA2 have previously been associated with MDS/AML, but whether they can actually cause AA will have to be clarified (see discussion in supplemental Methods, “Roundtable sessions”). In this respect, the data made available from this study will serve as a resource for further investigations of IBMF syndromes by the research community.
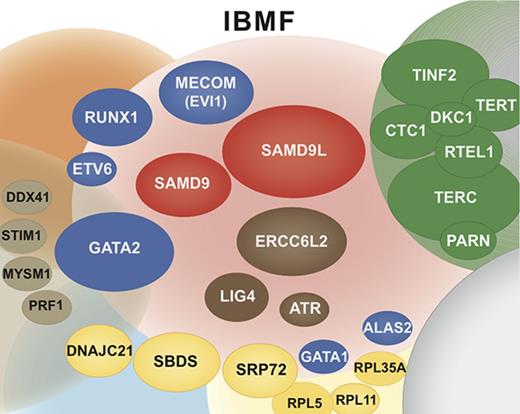
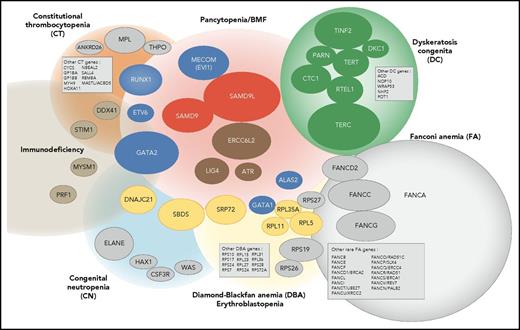
Overall, our study confirms that IBMF syndromes are highly heterogeneous. The clinical phenotypes are variable, often mild, do not always fully conform to classic syndromes, and largely overlap from 1 syndrome to the other; their family history can be absent. Moreover, new clinical entities have to be integrated into an overall molecular portrait of IBMF diseases, along with classic syndromes such as FA and DC (Figure 6). In this broader context, predicting an individual genotype based on clinical and biological data are even more challenging and not always reliable. Adding to the complexity, a proportion of IBMF patients may carry 2 mutated genes. Therefore, a broad approach encompassing the various IBMF genes, including their regulatory and intronic regions, using extended gene panels and possibly WES, will be useful in obtaining an accurate genetic diagnosis at medical evaluation. Consequently, we have now implemented systematic screening at BMF diagnosis with an updated list of genes that can confidently be considered to be IBMF causal. Regarding the DNA source, skin biopsies remain invaluable in many clinical situations to overcome potential confounding hematopoietic reversion or somatic mutations, even though they are more demanding for the patient and time-consuming in the laboratory.
Integrated portrait of IBMF diseases. Genetically defined entities are shown. The genes found mutated in our current IBMF cohort are shown as colored ellipses, the sizes of which correspond to patient frequency; these cases are integrated in an updated general portrait of IBMF diseases.
Integrated portrait of IBMF diseases. Genetically defined entities are shown. The genes found mutated in our current IBMF cohort are shown as colored ellipses, the sizes of which correspond to patient frequency; these cases are integrated in an updated general portrait of IBMF diseases.
Our work sheds light on and extends the clinical spectrum of several seldom-reported IBMF entities that are associated with salient and recurrent features. First, de novo mutations in MECOM defined a unique disease with severe AA occurring within the first months of life that requires HSCT. Second, ERCC6L2 biallelic mutations defined a mild pancytopenia with possible MDS. Third, SAMD9 and SAMD9L mutations defined hematological entities that appear to be a prominent cause of IBMF. These patients typically experienced early-onset severe BMF, with frequent dysplastic features and monosomy 7. Strikingly, the hematological situation improved spontaneously in many patients upon the acquisition of somatic lesions leading to the replacement of the mutated allele by the wild-type allele. This once again demonstrates the interest of working on nonhematopoietic cells, that is, skin fibroblast cells, to prevent genetic misdiagnosis due to low-mutation allele frequency in blood cells. Moreover, it is possible that patients such as these, even those with dysplasia and monosomy 7, may benefit from a careful watch-and-wait policy rather than up-front HSCT, a practice-changing view of IBMF/hypoplastic MDS with monosomy 7 in young children. This approach would need confirmation by prospective studies in patients with SAMD9/SAMD9L mutations.
In conclusion, our study highlights the relevance of a translational genomics approach requiring close collaboration between families, physicians, biologists, bioinformatics specialists, and scientists to accurately diagnose IBMF. Precision-medicine approaches such as the use of next-generation sequencing at diagnosis can improve family counseling and patient management, including decisions for invasive and costly procedures such as HSCT. It also raises practical and ethical issues for families that benefit from an evaluation in an expert center.43 Beyond medical issues, our work should also prompt various basic and clinical research initiatives, whereas our clinically and biologically annotated cohort with nonhematopoietic DNA constitutes a foundation for further investigations on IBMF/MDS pathogenesis, classification, risk prediction, and follow-up.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and their families, the Association Laurette Fugain, and the Association Saint-Louis Contre la Leucémie for their support, the physicians and nurses from French pediatrics, hematology, oncology, and genetics centers for taking care of the patients, along with the Société Française d’Hématologie and the Société d’Hématologie et d’Immunologie Pédiatrique. The authors thank Niclas Setterblad, Antonio Alberdi, and Julien Pelé at the Genomic Platform, Institut Universitaire d’Hématologie, Paris, France.
This work was supported by the European Research Council Consolidator grant no. 311660, the Agence Nationale pour la Recherche “Saint-Louis Institute Program” no. ANR-10-IBHU-0002, the Agence Nationale de la Recherche (ANR) program Alliance Parisienne des Instituts de Recherche en Cancérologie (PACRI), and the Cancéropole Île-de-France (IDF). M.S. was supported by the Alliance Nationale pour les Sciences de la Vie et de la Santé (AVIESAN)–Institut National du Cancer (INCa) Program “Formation à la Recherche Translationnelle.” Saint-Louis/Robert Debré Hospital was supported by the French Government (Direction de l’Hospitalisation et de l’Organisation des Soins) as a Centre de Référence Maladies Rares “Aplasies Médullaires Constitutionnelles,” filière Maladies Rares Immuno-Hématologiques (MARIH; R.P.d.L., T.L., J.-H.D., A.B., G.S., and J.S.).
Authorship
Contribution: O.B., M.S., G.S. and J.S. designed the studies; O.B., M.S., S.Q., E. Lainey, L.H., E.C., N.V., M.D.C., J.M.-P., W.C., A.R., L.D.J., and J.S. performed research and analyzed data; M.S., T.L., R.P.d.L., J.-H.D., F.S.d.F., E. Lengline, R.I., N. Boissel, L.A., P.F., S.M., C.S., M.M., C.D., N. Blin, B.B., I.P., M.H., S.B., A.P., G.L., G.M., Y.B., A.B., and G.S. collected and analyzed clinical data; O.B., M.S., G.S., and J.S. wrote the manuscript; and all authors approved the manuscript and the submission.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jean Soulier, Saint-Louis Hospital, APHP, and University Paris Diderot, Bat. Jean Bernard U944 INSERM, 1 Av Claude Vellefaux, 75010 Paris, France; e-mail: jean.soulier@sls.aphp.fr; and Gérard Socié, Hematology/Transplantation APHP, Saint-Louis Hospital, University Paris Diderot, and INSERM UMR 1160, 1 Av Claude Vellefaux, 75010 Paris, France; e-mail: gerard.socie@paris7.jussieu.fr.
References
Author notes
O.B. and M.S. contributed equally to this article.