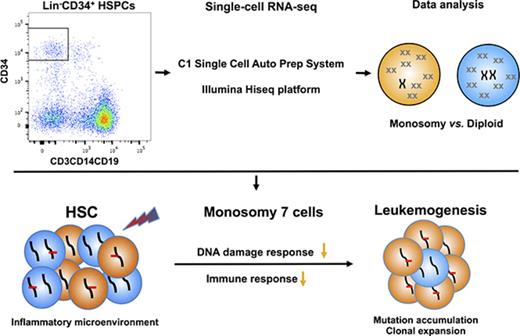
Key Points
We distinguished aneuploid cells from diploid cells within the hematopoietic stem and progenitor cells using scRNA-seq.
Monosomy 7 cells showed downregulated pathways involved in immune response and maintenance of DNA stability.
Abstract
Cancer cells frequently exhibit chromosomal abnormalities. Specific cytogenetic aberrations often are predictors of outcome, especially in hematologic neoplasms, such as monosomy 7 in myeloid malignancies. The functional consequences of aneuploidy at the cellular level are difficult to assess because of a lack of convenient markers to distinguish abnormal from diploid cells. We performed single-cell RNA sequencing (scRNA-seq) to study hematopoietic stem and progenitor cells from the bone marrow of 4 healthy donors and 5 patients with bone marrow failure and chromosome gain or loss. In total, transcriptome sequences were obtained from 391 control cells and 588 cells from patients. We characterized normal hematopoiesis as binary differentiation from stem cells to erythroid and myeloid-lymphoid pathways. Aneuploid cells were distinguished from diploid cells in patient samples by computational analyses of read fractions and gene expression of individual chromosomes. We confirmed assignment of aneuploidy to individual cells quantitatively, by copy-number variation, and qualitatively, by loss of heterozygosity. When we projected patients’ single cells onto the map of normal hematopoiesis, diverse patterns were observed, broadly reflecting clinical phenotypes. Patients’ monosomy 7 cells showed downregulation of genes involved in immune response and DNA damage checkpoint and apoptosis pathways, which may contribute to the clonal expansion of monosomy 7 cells with accumulated gene mutations. scRNA-seq is a powerful technique through which to infer the functional consequences of chromosome gain and loss and explore gene targets for directed therapy.
Introduction
Chromosomal abnormalities are typical in cancer cells.1-5 Various types of chromosomal aberrations are frequently observed in hematologic malignancies, including myelodysplastic syndromes (MDSs) and acute myeloid leukemia (AML),6-8 and they occur as late-evolution events in so-called benign diseases like aplastic anemia (AA) and constitutional bone marrow (BM) failure (BMF) syndromes.9,10 Specific cytogenetic abnormalities are prognostic. In marrow failure, monosomy 7 is associated with refractory cytopenias and progression to acute leukemia and thus poor clinical outcome.2,6-11 Identification of the genetic machinery of chromosome 7 loss would give insight into the pathophysiologic mechanisms of early leukemogenesis.
Efforts to clarify molecular alterations in monosomy 7 syndromes have been of limited value in understanding its development and progression.12-22 In an early study of ours, we identified disease-specific dysregulated genes and pathways using GeneChip analysis of bulk CD34+ hematopoietic stem (HSCs) and progenitor cells (HSPCs) from patients with MDS with monosomy 7.20 However, the absence of specific cell-membrane markers to separate cytogenetically normal and abnormal cells hindered further analysis of dysregulated molecular mechanisms.
Single-cell RNA sequencing (scRNA-seq) allows high-resolution whole-transcriptome profiling of individual cells and direct analysis of subpopulations of cells from a larger heterogeneous population.23-33 Gain or loss of chromosomes should lead to over- or underexpression of genes located on the affected chromosomes.34 Concomitant increases and decreases in chromosome-wide gene expression levels could be used to infer chromosome copy numbers by scRNA-seq.24,35 In identifying aneuploid cells, function may be imputed from transcriptional signatures, and transcriptional programs of individual cells may be compared for aneuploid cells from different patients and between diploid and aneuploid cells of the same patient.
In this study, we performed scRNA-seq analysis of BM-derived CD34+ cells from patients with BMF and cytogenetic abnormalities. We distinguished aneuploid cells from diploid cells using several analytical strategies; we identified molecular signatures in patient cells that had undergone clonal evolution from BMF to premalignancy and in patients presenting with marrow dysplasia. We demonstrate that scRNA-seq is a powerful tool by which to define functional and molecular alterations in human cells with chromosome abnormalities.
Methods
Full descriptions of experimental procedures and analytical methods can be found in the data supplement. Data are available in the Gene Expression Omnibus (accession #GSE99095).
Participants and samples
BM samples were obtained from patients with BMF with aneuploidy by conventional cytogenetics (patients 1-5) after written informed consent in accordance with the Declaration of Helsinki and under Clinicaltrials.gov protocols #NCT00001620 and #NCT00001397, approved by the institutional review boards of the National Heart, Lung, and Blood Institute. We chose 3 patients in whom AA was followed by clonal evolution, which is typically monosomy 7 or trisomy 8; this clinical circumstance allows examination of early leukemogenesis. We also studied 2 patients with de novo MDS: 1 with cytopenias and marrow hypocellularity, and the other with excess myeloblasts. Four healthy donors (31/M, 34/M, 57/F, and 58/M) were controls.
Whole-trancriptome amplification, complementary DNA library preparation, and sequencing
The C1 Single-Cell Auto Prep System (Fluidigm) was employed to perform SMARTer (Clontech) whole-transcriptome amplification (WTA) on up to 96 individual cells, according to the manufacturer’s protocols. WTA products were converted to Illumina sequencing libraries using Nextera XT (Illumina), followed by sequencing on an Illumina HiSequation 2500 or 3000 System using a 75-bp paired-end sequencing strategy.
Data analysis
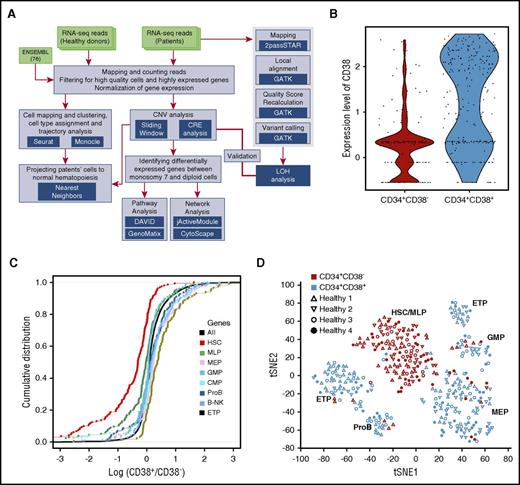
Total reads were mapped to the reference genome with RSubreader, and gene-level read counts were calculated using featureCounts.36 Only data from high-quality cells with >2 million mapped reads, and >2000 captured genes were used in further analysis. The schematic pipeline is depicted in Figure 1A. Cells were mapped and clustered using tSNE and density-based spatial clustering of applications with noise (DBSCAN) on highly variant genes, and an HSPC type was assigned to each cluster based on significance in overlapping between HSPC- and cluster-specific genes (Fisher’s exact test).
Hematopoietic heterogeneity in healthy donors quantified by scRNA-seq. CD34+CD38− and CD34+CD38+ cells from 4 healthy donors (healthy 1-4) were sorted by surface-membrane markers and subjected to analyses. (A) The schematic pipeline consisting of 3 major analytic components: differentiation analysis with cells from healthy donors, identification and characterization of monosomy 7 cells with gene expression, and validation of monosomy identification with loss of reterozygosity (LOH). CNV, copy-number variation; CRE, chromosome relative expression; GATK, Genome Analysis Toolkit. (B) CD38 expression levels in CD34+CD38− and CD34+CD38+ cells. Each dot represents a single cell. y-axis, batch-corrected gene expression levels. (C) Cumulative distribution of fold changes of expression of hematopoietic cell type signature genes between CD34+CD38− and CD34+CD38+ cells. Each dot represents a gene. B-NK, B cell–natural killer cell precursor; CMP, common myeloid progenitor; ETP, earliest thymic progenitor; GMP, granulocyte-monocyte progenitor; MEP, megakaryocytic-erythroid progenitor; MLP, multilymphoid progenitor; ProB, pro–B cell. y-axis, cumulative distribution; x-axis, log (marker gene expression levels in CD34+CD38+ cells/marker gene expression levels in CD34+CD38− cells). (D) t-distributed stochastic neighbor embedding (tSNE) plot of single-cell gene expression data. Single cells from 4 healthy donors (healthy 1-4) are represented by different symbols. Highly variable genes (1024) across all healthy donors were used in tSNE analysis.
Hematopoietic heterogeneity in healthy donors quantified by scRNA-seq. CD34+CD38− and CD34+CD38+ cells from 4 healthy donors (healthy 1-4) were sorted by surface-membrane markers and subjected to analyses. (A) The schematic pipeline consisting of 3 major analytic components: differentiation analysis with cells from healthy donors, identification and characterization of monosomy 7 cells with gene expression, and validation of monosomy identification with loss of reterozygosity (LOH). CNV, copy-number variation; CRE, chromosome relative expression; GATK, Genome Analysis Toolkit. (B) CD38 expression levels in CD34+CD38− and CD34+CD38+ cells. Each dot represents a single cell. y-axis, batch-corrected gene expression levels. (C) Cumulative distribution of fold changes of expression of hematopoietic cell type signature genes between CD34+CD38− and CD34+CD38+ cells. Each dot represents a gene. B-NK, B cell–natural killer cell precursor; CMP, common myeloid progenitor; ETP, earliest thymic progenitor; GMP, granulocyte-monocyte progenitor; MEP, megakaryocytic-erythroid progenitor; MLP, multilymphoid progenitor; ProB, pro–B cell. y-axis, cumulative distribution; x-axis, log (marker gene expression levels in CD34+CD38+ cells/marker gene expression levels in CD34+CD38− cells). (D) t-distributed stochastic neighbor embedding (tSNE) plot of single-cell gene expression data. Single cells from 4 healthy donors (healthy 1-4) are represented by different symbols. Highly variable genes (1024) across all healthy donors were used in tSNE analysis.
Monocle37 was used to reconstruct the differentiation continuum of healthy donor cells. For each patient, individual cells were projected onto the map of normal hematopoietic differentiation based on cell-by-cell comparison of the pattern of global gene expression and localization to the 5 most similar healthy donor cells.
Chromosomal aneuploidy was evaluated by 3 independent methods. First, chromosome CNVs at a location of each gene were estimated using average expression of genes in a sliding window24 that contained the proximate 50 up- and 50 downstream genes. Second, an index, termed CRE, which was originally introduced to call aneuploidy using microarray technology,38 was adapted to scRNA-seq as the fraction of transcripts per million on a given chromosome. CRE value distribution in control cells was used to estimate the false discovery rate. Third, we used degree of LOH in single-nucleotide polymorphisms (SNPs) on each chromosome to estimate the probability of chromosomal loss. Genetic variants were called using Genome Analysis Toolkit best practices for RNA-seq variant calling,39 with STAR v2.3.0e and GRCh37.75. Donor cells were used to build a background model for probability determination.
Results
Profiling of individual hematopoietic cells by scRNA-seq
The experimental workflow for scRNA-seq is depicted in supplemental Figure 1A. We separated subpopulations of BM mononuclear cells by fluorescence-activated cell sorting, in order to enrich for primitive hematopoietic progenitors and increase the homogeneity of the samples. CD34+CD38−Lin(CD3CD14CD19)− and CD34+CD38+Lin(CD3CD14CD19)− populations were obtained for patient 4 and 4 healthy donors; for the other patients (patients 1, 2, 3, and 5), limited cell numbers (because of their BMF) allowed separation only of CD34+ cells that were negative for lineage markers. In total, we sequenced 1202 single-cell libraries and corresponding bulk samples for each participant and retained 391 cells from healthy donors and 588 cells from patients after quality control for further analysis.
Aggregate gene expression profiles of single cells accurately recapitulated bulk gene expression from the same cell population (supplemental Figure 1B). As expected, the gene encoding for CD38 was expressed on CD34+CD38+ cells and not on CD34+CD38− cells from healthy donors (Figure 1B), and there was consistency between CD38 gene expression at the RNA and protein levels. Correlation between individual cells from the same individual showed a broad spread (R = 0.2-0.6; supplemental Figure 1C-D); such cell-to-cell variability is similar to other published scRNA-seq data.24
Single-cell transcription shows heterogeneity of HSPC populations from healthy donors
We first examined scRNA-seq data from healthy donors to determine expression characteristics of individual HSPCs and to provide a reference for patient samples. To assess transcriptional relationships, 391 single-cell transcriptomes were analyzed using highly variable genes (supplemental Figure 2A). There was a clear distinction between CD34+CD38− and CD34+CD38+ cells at the transcript level by hierarchical clustering analysis (supplemental Figure 2B). Genes known to be expressed in stem cells40 were enriched in CD34+CD38− cells, and genes typical of oligopotential or unipotential hematopoietic progenitors were enriched in CD34+CD38+ cells (Figure 1C).
For single cells, we first employed tSNE to visualize high-dimensional data in 2 dimensions while preserving pairwise similarity. The more primitive population of CD34+CD38− cells formed a single cluster, whereas the CD34+CD38+ population, containing differentiating cells, was more heterogeneous. The density-based clustering algorithm in DBSCAN was then used to identify the number of groups, and each cell was automatically assigned to a group (Figure 1D). We assigned to each cluster a specific hematopoietic cell identity by comparing cluster-specific genes with reported lineage signature genes.40 Putative identities could be assigned to clusters of HSCs, multilymphoid progenitors, MEPs, granulocyte-monocyte progenitors, pro–B cells, and earliest thymic progenitors. Collectively, the results from healthy donors suggested that scRNA-seq could be used to resolve human single HSPCs into specific subtypes based upon gene expression profiles.
Aneuploid cells detected from patients’ HSPCs
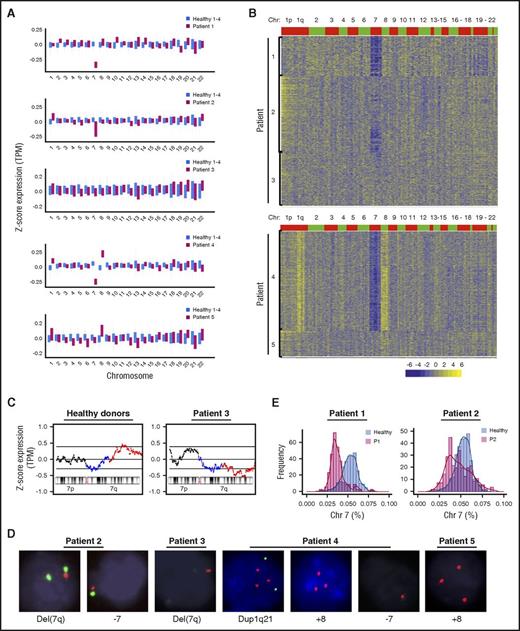
As proof of concept, we applied scRNA-seq to cells from patients with BMF who had cytogenetic abnormalities (Table 1). We first assessed average expression levels based on fractions of reads from each chromosome for healthy donor samples, and as expected, expression levels among them were similar (supplemental Figure 3A). When the same analysis was performed for patient cells, we found lower expression of chromosome 7 genes in patients 1, 2, and 4, higher expression of chromosome 8 genes in patients 4 and 5, and increased expression of chromosome 1 genes in patient 4 (Figure 2A; supplemental Figure 3B). These patterns of lower or higher gene expression were concordant with the known chromosomal aberrations in each patient as determined by clinical cytogenetics. There were no apparent changes in expression of genes by chromosome location for other chromosomes. For higher resolution of subchromosomal CNVs, we used a sliding window method24 to assess mean expression of 101 genes (equivalent to ∼30 MB) sequentially along each chromosome. Healthy donors, whose karyotypes were assumed to be normal diploid, did not show any CNV on any chromosome (supplemental Figure 4A). By comparison with normalized CNV profiles from healthy donors, we verified the presence of CNVs in patients on chromosome 7 (patients 1, 2, and 4), chromosome 8 (patients 4 and 5), and a duplicated region on the long arm of chromosome 1 (patient 4; Figure 2B). We also detected lower copy number of 7q22-36 in patient 3 (Figure 2C), which was present in a small proportion of the patient’s cells by cytogenetics. Results in patient 4 were initially surprising. On previous clinical cytogenetics, the patient had trisomy 8 and duplication of 1q; the sample obtained for scRNA-seq was barely adequate for conventional cytogenetics, which disclosed the chromosome 1 duplication in 3 of only 4 cells examined. By RNA-seq, we observed both the previously reported trisomy 8 and also unsuspected low copy number of chromosome 7 by both our algorithms. Indeed, when fluorescence in situ hybridization was performed on the same marrow specimen, these chromosome abnormalities were validated (Figure 2D).
Clinical and laboratory characteristics of patients with MDS or SAA
| Patient . | Age, y/sex . | Diagnosis . | Hgb, g/dL . | WBC, ×109/L . | ANC, ×109/L . | Platelets, ×109/L . | Somatic mutations . | Prior treatment . | Outcome . | Karyotype . | FISH . | BM cellularity, % . | Blasts, % . | Myeloid precursors, % . | Erythroid precursors, % . | Megakaryocytes . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 54/F | SAA→MDS | 11.5 | 2.07 | 1.08 | 25 | Not detected | ATG + CSA, danazol, HSCT | Death | 45,XX,−7[6]/46,XX[14] | N/A | 10 | 1 | 28 | 59 | Reduced number, dysplastic |
| 2 | 6.5/M | Post-hepatitis SAA→MDS | 10.9 | 3.23 | 0.21 | 33 | Not detected | Danazol | Alive | 45,XY,−7[10]/46,XY[10] | del(7q) [28/254]; monosomy 7 [14/254] | 10-15 | 4 | 34 | 38 | Present, atypical |
| 3 | 56/F | Hypocellular MDS-U (IPSS-R low) | 12.8 | 5.25 | 3.2 | 211 | DNMT3A (31.9%, 2.2%), BCOR (2.1%) | Alemtuzumab, CSA | Alive | 46,XX,del(7)(q22q36)[1]/46,XY[19] | del(7q) [32/208] | 15-20 | 1 | 60 | 29 | Reduced number |
| 4 | 59/M | MDS-RAEB 2 (IPSS-R high) | 7.6 | 1.95 | 1.56 | 62 | ASXL1 (16.7%), KRAS (2.2%), SETBP1 (13.8%), U2AF1 (21%) | Alemtuzumab, danazol | Death | 46,XY,dup(1)(q21q32)[3]/46,XY[1] | Trisomy 8 [118/200]; monosomy 7 [35/200]; dup(1q21) [128/200] | 80 | 15 | 50 | 4 | 50% dysplastic |
| 5 | 67/M | SAA→MDS | 15.4 | 4.71 | 2.5 | 188 | Not detected | ATG, CSA | Alive | 47,XY,+8[13]/46,XY[7] | Trisomy 8 [122/260] | 10 | 0 | 43 | 42 | Reduced number, unremarkable morphology |
| Patient . | Age, y/sex . | Diagnosis . | Hgb, g/dL . | WBC, ×109/L . | ANC, ×109/L . | Platelets, ×109/L . | Somatic mutations . | Prior treatment . | Outcome . | Karyotype . | FISH . | BM cellularity, % . | Blasts, % . | Myeloid precursors, % . | Erythroid precursors, % . | Megakaryocytes . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 54/F | SAA→MDS | 11.5 | 2.07 | 1.08 | 25 | Not detected | ATG + CSA, danazol, HSCT | Death | 45,XX,−7[6]/46,XX[14] | N/A | 10 | 1 | 28 | 59 | Reduced number, dysplastic |
| 2 | 6.5/M | Post-hepatitis SAA→MDS | 10.9 | 3.23 | 0.21 | 33 | Not detected | Danazol | Alive | 45,XY,−7[10]/46,XY[10] | del(7q) [28/254]; monosomy 7 [14/254] | 10-15 | 4 | 34 | 38 | Present, atypical |
| 3 | 56/F | Hypocellular MDS-U (IPSS-R low) | 12.8 | 5.25 | 3.2 | 211 | DNMT3A (31.9%, 2.2%), BCOR (2.1%) | Alemtuzumab, CSA | Alive | 46,XX,del(7)(q22q36)[1]/46,XY[19] | del(7q) [32/208] | 15-20 | 1 | 60 | 29 | Reduced number |
| 4 | 59/M | MDS-RAEB 2 (IPSS-R high) | 7.6 | 1.95 | 1.56 | 62 | ASXL1 (16.7%), KRAS (2.2%), SETBP1 (13.8%), U2AF1 (21%) | Alemtuzumab, danazol | Death | 46,XY,dup(1)(q21q32)[3]/46,XY[1] | Trisomy 8 [118/200]; monosomy 7 [35/200]; dup(1q21) [128/200] | 80 | 15 | 50 | 4 | 50% dysplastic |
| 5 | 67/M | SAA→MDS | 15.4 | 4.71 | 2.5 | 188 | Not detected | ATG, CSA | Alive | 47,XY,+8[13]/46,XY[7] | Trisomy 8 [122/260] | 10 | 0 | 43 | 42 | Reduced number, unremarkable morphology |
Somatic mutations were detected using a commercial panel of 54 genes associated with myeloid neoplasms (supplemental Table 3). Allele fraction of mutations is listed in the brackets after a gene name. Patient 3: DNMT3A Lys826Arg, DNMT3A Arg771Ter, and BCOR Arg976Ter. Patient 4: ASXL1 Met837LeufsTer2, KRAS G12Y, SETBP1 Asp868Asn, and U2AF1 Gln157Arg. CD34+Lin(CD3CD14CD19)− cells or CD34−Lin(CD3CD14CD19)− cells were used for fluorescence in situ hybridization (FISH) test. CD34+CD117+Lin− blasts from patient 4 were also examined by FISH, which showed monosomy 7 [171/200]; del(7q) [27/200].
ANC, absolute neutrophil count; ATG, antithymocyte globulin; CSA, cyclosporine; Hgb, hemoglobin; HSCT, HSC transplantation; IPSS-R, International Prognostic Scoring System–Revised; MDS-U, MDS unclassified; N/A, not available; RAEB 2, refractory anemia with excess blasts 2; SAA, severe AA; WBC, white blood cell.
Detection of CNVs in single cells from patients. (A) Average gene expression for each chromosome in single cells from patients and healthy donors. Average gene expression levels of individual chromosomes from 4 healthy donors were used for comparison. Chromosome mapping read values were median-centered. The top and bottom of the bar represent the 25% and 75% quartiles. (B) Heatmaps of CNV signals obtained by sliding window analysis. scRNA-seq data of patients were normalized against those of healthy donors. Copy-number changes were examined in 22 chromosomes (columns) for patients’ individual cells (rows). (C) CNV signals on chromosome 7 obtained from healthy donors and patient 3 by sliding window analysis. Two black lines show 95% confidence intervals. 7q11-21 and 7q22-36 are indicated in blue and red, respectively. (D) Cytogenetic abnormalities detected by fluorescence in situ hybridization. Probes used to detect specific chromosomes were: CDKN2C and CKS1B for chromosome 1, CEN 8 for chromosome 8, and CEN7 and D7S486 for chromosome 7. (E) Histograms of read ratios on chromosome 7 in patients 1 and 2. Pink, cells from patients; blue, cells from healthy donors; purple, distribution overlap between each patient and healthy donors; y-axis, frequency of cell number; and x-axis, ratios of reads on chromosome 7 in individual cells relative to reads on all chromosomes in the same cell.
Detection of CNVs in single cells from patients. (A) Average gene expression for each chromosome in single cells from patients and healthy donors. Average gene expression levels of individual chromosomes from 4 healthy donors were used for comparison. Chromosome mapping read values were median-centered. The top and bottom of the bar represent the 25% and 75% quartiles. (B) Heatmaps of CNV signals obtained by sliding window analysis. scRNA-seq data of patients were normalized against those of healthy donors. Copy-number changes were examined in 22 chromosomes (columns) for patients’ individual cells (rows). (C) CNV signals on chromosome 7 obtained from healthy donors and patient 3 by sliding window analysis. Two black lines show 95% confidence intervals. 7q11-21 and 7q22-36 are indicated in blue and red, respectively. (D) Cytogenetic abnormalities detected by fluorescence in situ hybridization. Probes used to detect specific chromosomes were: CDKN2C and CKS1B for chromosome 1, CEN 8 for chromosome 8, and CEN7 and D7S486 for chromosome 7. (E) Histograms of read ratios on chromosome 7 in patients 1 and 2. Pink, cells from patients; blue, cells from healthy donors; purple, distribution overlap between each patient and healthy donors; y-axis, frequency of cell number; and x-axis, ratios of reads on chromosome 7 in individual cells relative to reads on all chromosomes in the same cell.
To quantify individual cells with abnormal patterns of gene expression by chromosome, we used CRE analysis. Because monosomy 7 and trisomy 8 were the dominant aneuploidy phenotypes in our patients, we assessed for global expression of these chromosomes. Lower chromosome expression was identified for chromosome 7 in 55% of patient 1’s cells, 26% of patient 2’s cells, and 71% of patient 4’s cells (denoted as monosomy 7 cells in the following text; Figure 2E; supplemental Figure 4B). Higher expression of chromosome 8 genes was present in 67% of patient 4’s cells and 51% of patent 5’s cells (supplemental Figure 4B).
Chromosome loss in aneuploidy results in CNV of RNA transcript levels as well as LOH. When we performed eSNP karyotyping41 for expressed genes to validate CNV results, we found a lower frequency of heterozygous SNPs on chromosome 7 in patients compared with healthy donors (supplemental Figure 5). Furthermore, LOH was evident for chromosome 7 on examination of individual denominated monosomy 7 cells compared with diploid cells from the same patient, as described above (supplemental Figure 6A). There was high correlation (R = 0.72) between CNV and LOH (supplemental Figure 6B), but no correlation between sequencing depth (read count) and LOH score (supplemental Figure 6C).
scRNA-seq analysis reveals distinct differentiation patterns of HSPCs from healthy donors and patients
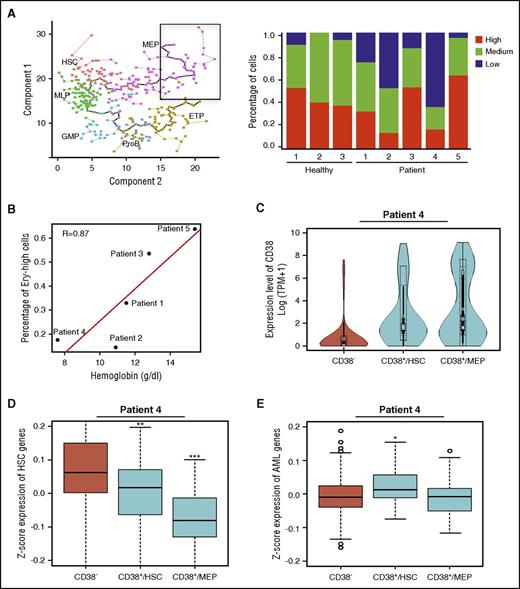
scRNA-seq allows for high-resolution reconstruction of cell differentiation. We performed cell pseudotemporal ordering analysis using scRNA-seq data of healthy donors to deconvolute normal hematopoietic differentiation. Monocle orders cells by maximizing transcriptomic similarity between successive cells. After ordering in the absence of a priori assumptions, each cell was assigned a cell stage based on its expression of known hematopoietic signature genes. We found cells to be ordered along 2 branches of differentiation from HSCs, with 1 branch containing cells expressing genes characteristic of erythroid/megakaryocyte, and the other, lymphoid and myeloid cells (Figure 3A).
Hematopoietic differentiation of marrow cells from healthy donors and patients. (A) Ordering of individual cells from 4 healthy donors (colored circles) into a 2-dimensional independent component space of hematopoietic lineages using Monocle. Lines connecting circles indicate edges of the minimum spanning tree (MST). A solid black line shows the main path of the MST, which provides a backbone for pseudotime ordering of cells by Monocol. ETP, earliest thymic progenitor; GMP, granulocyte-monocyte progenitor; MLP, multilymphoid progenitor; ProB, pro–B cell. (B-D) Mapping of patient cells onto normal MST by the nearest neighbor method (averaging locations of 5 nearest cells on the tree). Hollow circles indicate cells with monosomy 7 or del(7q) from patients 1, 2, 3, and 4 (patient 5 had trisomy 8).
Hematopoietic differentiation of marrow cells from healthy donors and patients. (A) Ordering of individual cells from 4 healthy donors (colored circles) into a 2-dimensional independent component space of hematopoietic lineages using Monocle. Lines connecting circles indicate edges of the minimum spanning tree (MST). A solid black line shows the main path of the MST, which provides a backbone for pseudotime ordering of cells by Monocol. ETP, earliest thymic progenitor; GMP, granulocyte-monocyte progenitor; MLP, multilymphoid progenitor; ProB, pro–B cell. (B-D) Mapping of patient cells onto normal MST by the nearest neighbor method (averaging locations of 5 nearest cells on the tree). Hollow circles indicate cells with monosomy 7 or del(7q) from patients 1, 2, 3, and 4 (patient 5 had trisomy 8).
To deconvolute the differentiation patterns of HSPCs from patients, we mapped patients’ cells onto the normal differentiation continuum. We defined their mathematical coordinates with an average of the 5 most proximate healthy donors’ cells, with proximity based on similarity among their transcriptomes. Patients’ cells showed distinctive hematopoietic development patterns compared with cells from healthy donors (Figure 3B-D), validated using the tSNE algorithm (supplemental Figure 7A-C). There was heterogeneity in differentiation patterns among patients who had only the single cytogenetic abnormality of monosomy 7. Diploid cells from patients also showed abnormal, highly biased patterns of differentiation.
Patterns of patients’ cells were consistent with their individual clinical and laboratory diagnostic characteristics. At sampling, patients 3 and 5 had normal blood counts; by scRNA-seq, their HSPCs differentiated to both erythroid/megakaryocyte and myeloid/lymphoid lineages, similar to the pattern in healthy donors. Patient 2, a child, had experienced posthepatitis severe AA that had rapidly evolved to monosomy 7 MDS with persistent pancytopenia; flow cytometry of his BM cells showed the majority of CD34+ cells to express CD19, consistent with an early lymphoblast phenotype, and one-third of cells within the lymphoid compartment expressed CD10, typical of hematogones seen in children. For this patient, scRNA-seq showed markedly biased lymphoid lineage differentiation. Patients 1 and 4 were pancytopenic, but marrow cellularity in 1 patient was low, and in the other increased. By scRNA-seq, these patients’ cells showed marked skewing to the MEP pathway. Within the MEP compartment, gene signatures of erythropoiesis-specific genes have been assigned to stages of early red blood cell maturation. We identified early-stage erythroid (CD34+CD71+GlyA−) marker genes (eg, CD36)42 within the healthy donors’ most terminally differentiated MEP (Figure 4A). Expression of these early erythroid maturation genes was variable among patients, but low expression correlated with the degree of anemia, even in the presence of marked MEP skewing, as in patients 1 and 4. These results were consistent with impaired erythroid differentiation as detected at the single-cell level (Figure 4A-B).
Impaired hematopoietic differentiation of patients’ cells. (A) Expression of early erythroid maturation genes in MEP cells from healthy donors and patients. Cells encircled in a black box (y location > 15 and x location > 15) in the left panel were analyzed for expression of early erythroid maturation genes; the right panel shows frequency of cells with different expression levels of erythroid genes. Healthy donor 4 was not included because of a limited cell number included in the black square. ETP, earliest thymic progenitor; GMP, granulocyte-monocyte progenitor; MLP, multilymphoid progenitor; ProB, pro–B cell. (B) Correlation of erythroid gene expression levels with hemoglobin concentration. Patient 2 had a hemoglobin concentration of 10.9 g/dL at time of marrow sample collection, which decreased to 9 g/dL 3 months later. (C) CD38 expression levels in cells of patient 4. CD38−, CD34+CD38− cells; CD38+/HSC, CD34+CD38+ cells within the HSC/MLP population (x location < 10 on minimum spanning tree ); and CD38+/MEP, CD34+CD38+ cells within the MEP population (x location ≥ 10). (D-E) Expression levels of HSC genes or AML genes in cells of patient 4. *P < .05, **P < .01, and ***P < .001.
Impaired hematopoietic differentiation of patients’ cells. (A) Expression of early erythroid maturation genes in MEP cells from healthy donors and patients. Cells encircled in a black box (y location > 15 and x location > 15) in the left panel were analyzed for expression of early erythroid maturation genes; the right panel shows frequency of cells with different expression levels of erythroid genes. Healthy donor 4 was not included because of a limited cell number included in the black square. ETP, earliest thymic progenitor; GMP, granulocyte-monocyte progenitor; MLP, multilymphoid progenitor; ProB, pro–B cell. (B) Correlation of erythroid gene expression levels with hemoglobin concentration. Patient 2 had a hemoglobin concentration of 10.9 g/dL at time of marrow sample collection, which decreased to 9 g/dL 3 months later. (C) CD38 expression levels in cells of patient 4. CD38−, CD34+CD38− cells; CD38+/HSC, CD34+CD38+ cells within the HSC/MLP population (x location < 10 on minimum spanning tree ); and CD38+/MEP, CD34+CD38+ cells within the MEP population (x location ≥ 10). (D-E) Expression levels of HSC genes or AML genes in cells of patient 4. *P < .05, **P < .01, and ***P < .001.
Patient 4 showed evidence of early leukemia in the marrow, where 15% of cells were identified as myeloblasts. By scRNA-seq, nearly half of this patient’s CD34+CD38+ cells were aberrantly present within the HSC/multilymphoid progenitor population (Figure 3C; supplemental Figure 7D). CD34+CD38+ cells are expected to lie among differentiated populations. We excluded a possible sorting error for patient 4’s cells as an explanation for abnormal localization by examining for CD38 gene expression within the stem-cell and differentiated cell compartments (Figure 4C); we assessed whether aberrant CD38 stem cells showed known leukemia signatures. We found higher expression of AML signature genes43,44 in these cells, with decreased gene expression levels of the normal stem-cell signature (Figure 4D-E). Thus, we inferred that these cells likely were leukemic myeloid blasts.
Differential gene expression in monosomy 7 cells compared with diploid cells
One of our main aims was to impute functional differences characterizing aneuploid cells. To this end, for each patient we compared monosomy 7 cells with diploid cells for differentially expressed genes, and we analyzed these genes within functional pathways using Genomatix.45 Data from patients 1, 2, and 4 were used [patient 3 was excluded because of a low number of del(7q) cells; patient 5 had trisomy 8; supplemental Table 1].
We first noted that for monosomy 7 cells in general, downregulated genes and gene pathways were prominent, and there were a limited number of upregulated genes. However, there appeared to be marked heterogeneity in gene expression of monosomy 7 cells among patients. Therefore, to examine for common transcriptional features of monosomy 7 cells, we exploited our observed differences in chromosome 7 gene expression in individual cells (Figure 2E), reasoning that cells that were least distinct from diploid patterns might obscure monosomy 7 cell characteristics (the computational method for progressive removal of cells with intermediate chromosome 7 gene expression is described in the data supplement). For patient 4, we assessed the MEP population rather than total cells to exclude cells with blastic properties present in the latter and allow comparisons among predominantly CD38+ cells in all 3 patient cases.
We observed 5 markedly downregulated functional groups in monosomy 7 cells (supplemental Tables 1 and 2): DNA damage checkpoint (genes such as ATR/ATM signaling, BRCA1 and BRCA2 signaling, and Fanconi anemia signaling), cell cycle (regulation of cell-cycle progression by PLK3 and influence of RAS and RHO proteins on G1 to S transition), apoptosis (apoptosis signaling in response to DNA damage and TRAIL signaling), immune response (eg, BCR signaling, TCR signaling, NF-κB cascade, and Toll-like receptor signaling pathway), and hematopoietic differentiation (EPO pathway).
Monosomy 7 gene signature and increased genomic instability in monosomy 7 cells
Because downregulated genes in monosomy 7 cells were similar by function and showed overlap among patients 1, 2, and 4 (Figure 5A), we combined downregulated genes from these 3 patients (data supplement) to obtain a monosomy 7 gene signature (supplemental Table 1). We mapped these genes to a protein-protein interaction network using STRING data,46 followed by in-house software jActiveModulesTopo,47 in order to identify biologically meaningful genes and display their relationships. A network of 94 genes and 188 interactions were identified (supplemental Figure 8), with most genes involved in immune response, DNA damage checkpoint, and apoptosis. When we examined the downregulated genes in the top pathways (supplemental Figure 9; supplemental Table 2) in a protein-protein interaction network, a network of 79 genes and 346 interactions involving the 3 functional groups (annotated as GO0006915 [apoptosis process], GO0006955 [immune response], and GO0042769 and GO2001020 [DNA damage response] by org.Hs.eg.db in Bioconductor) was generated (Figure 5B). Among these genes, 6 were located on chromosome 7, including MCM7, HUS1, RFC2, BRAF, DBNL, and CARD11; another downregulated gene, SAMD9 (on chromosome 7q21.3), was noted (insufficiency of its murine homolog Samd9l produces myeloid malignancy resembling human monosomy 7 disease15 ).
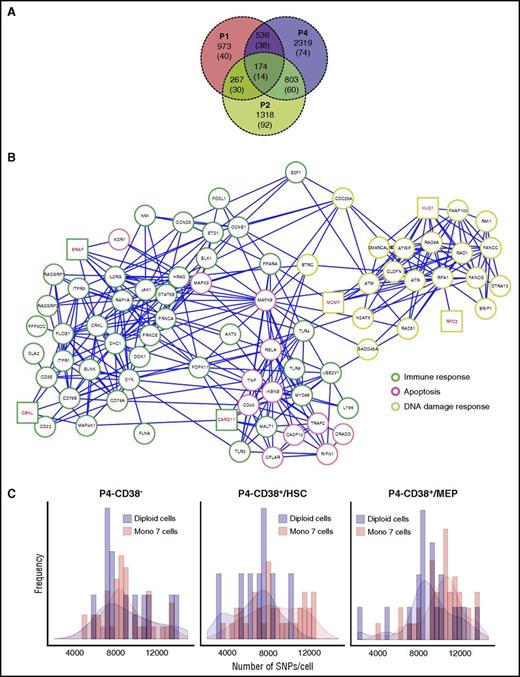
Downregulated genes and increased SNP frequencies in monosomy 7 cells. (A) Venn diagram of downregulated genes in patients 1 and 2 and patient 4 MEP. Patient 4 MEP cells were defined as cells that clustered to the MEP population from supplemental Figure 7D. Numbers in brackets indicate genes located on chromosome 7. (B) Downregulated genes in monosomy 7 CD34+ cells in patients 1 and 2 and patient 4 MEP (P < .05) were combined and subjected to network construction. Genes involved in different signaling pathway groups are highlighted in different border colors. Genes on chromosome 7 are highlighted in red. (C) The number of SNPs per cell in patient 4. CD38−, CD34+CD38− cells; CD38+/HSC, CD34+CD38+ cells within the HSC/multilymphoid progenitor population (x location < 10 on MST); and CD38+/MEP, CD34+CD38+ cells within the MEP population (x location ≥ 10).
Downregulated genes and increased SNP frequencies in monosomy 7 cells. (A) Venn diagram of downregulated genes in patients 1 and 2 and patient 4 MEP. Patient 4 MEP cells were defined as cells that clustered to the MEP population from supplemental Figure 7D. Numbers in brackets indicate genes located on chromosome 7. (B) Downregulated genes in monosomy 7 CD34+ cells in patients 1 and 2 and patient 4 MEP (P < .05) were combined and subjected to network construction. Genes involved in different signaling pathway groups are highlighted in different border colors. Genes on chromosome 7 are highlighted in red. (C) The number of SNPs per cell in patient 4. CD38−, CD34+CD38− cells; CD38+/HSC, CD34+CD38+ cells within the HSC/multilymphoid progenitor population (x location < 10 on MST); and CD38+/MEP, CD34+CD38+ cells within the MEP population (x location ≥ 10).
Because genes involved in maintaining DNA stability were markedly downregulated in monosomy 7 cells, we hypothesized that one consequence might be genomic instability. We generated a catalog of curated SNPs (data supplement) and calculated SNP frequencies in cells from patient 4, who had sufficient cells for reliable analysis, and 3 subpopulations that could serve as references. Similar SNP frequencies were present among diploid cells (Figure 5C), but monosomy 7 cells showed increased SNPs in both CD34+CD38+ cells within the HSC compartment and CD34+CD38+ cells in MEP, compared with corresponding diploid cells and CD34+CD38− monosomy 7 cells, suggesting increased genomic instability in monosomy cells in proliferating cell compartments.
Discussion
We isolated hundreds of individual cells from healthy controls and patients with clinically significant cytogenetic abnormalities and imputed their transcriptomes from RNA sequences by analyzing millions of reads in a variety of computational analytical methods. Using scRNA-seq, we first characterized the pattern of normal human hematopoiesis as binary differentiation from HSCs toward lymphoid/myeloid and erythroid lineages. Second, we distinguished aneuploid cells in mixed cell populations from patients’ BM and inferred abnormal differentiation patterns of patients’ cells by localization onto the map of normal human hematopoiesis. Third, scRNA-seq enabled us to define distinctive gene expression profiles of monosomy 7 HSPCs compared with the corresponding diploid cells from the same patients, providing a provisional molecular signature for this important aneuploid aberration. We believe that ours is the first study to clearly identify molecular characteristics of monosomy 7 HSPCs from patient samples at the level of the single cell.
Methods directed at the measurement of DNA itself, like fluorescence in situ hybridization and CNV techniques, allow accurate identification of individual anueploid cells, and by comparison, quantification of RNA is indirect in its approximation of DNA content. However, scRNA-seq provides robust data relating to DNA transcription, and therefore the functional status of a cell, not available with direct DNA methods. Even with the technical limitation that full-length mRNA sequencing currently only captures 96 cells in a run, it is superior in providing highly informative SNPs and mutations that assure the validity of our CRE approach. Although individual cells obviously cannot be repeatedly assessed, we confirmed the accuracy of nomination of aneuploidy using complementary computational strategies of CRE and LOH, which identified the same set of cells as lacking chromosome 7. We found useful graded removal of cells with intermediate character at the statistical border of aneuploidy and diploidy, which both increased the agreement between LOH and CRE and uncovered common biologically meaningful gene signatures in patients despite their heterogeneous clinical phenotypes. Confidence in our data was also afforded when our detailed view of lineage commitment of HSPCs appeared similar to recently published data sets of ∼1000 cells from 2 normal individuals32 and from parallel scRNA-seq experiments of >10 000 cells performed on a different platform in our own laboratory (supplemental Figure 10).
Three of the most downregulated pathways that we identified in monosomy 7 cells can be meaningfully related to the biology and pathophysiology of BMF. We found downregulation of BCR, TCR, TLR, and NF-κB signaling pathways in monosomy 7 cells in our patients’ samples, consistent with the immune origin of the AA from which aneuploidy had developed. In AA, cytotoxic T cells, their cytokines, and Fas-mediated apoptosis produce immune destruction of hematopoietic cells.48 Patients in this study had undergone immunosuppressive therapy, which led to hematologic improvement but likely did not terminate immune attack on marrow target cells. Functional alteration of critical immune response pathways could result in decreased recognition of monosomy 7 target cells or deficient signal transduction after an immune encounter. Precedents for selection of clonal populations in immune marrow failure include frequent 6p LOH, which decreases HLA expression on target cells49 and the paroxysmal nocturnal hemoglobinuria/aplastic anemia syndrome, in which cells mutated in the PIGA lose expression of many cell-surface proteins and may not be recognized by T cells.50 Supportive of a similar mechanism of immune escape for monosomy 7 is the occasional clinical observation of disappearance of monosomy 7 in conventional cytogenetics after immunosuppressive treatment of patients with MDS.51 Downregulation of the DNA damage response and apoptosis pathways is potentially related to both escape from environmental toxicity and progression of monosomy 7 to MDS and AML. In response to stressful stimuli, such as inflammation and the regenerative demands of cytopenias, HSCs enter cell cycle in enforced proliferation.52 Increased replicative DNA damage would follow, with activation of DNA damage checkpoint pathways, cell-cycle arrest, apoptosis, repair, or cell senescence. Activation of DNA damage checkpoints characterizes BM from patients with MDS, as detected by immunostaining of ATM, Chk2, and gH2AX.53,54 Inactivation of DNA damage checkpoints correlates with RAEB221 and AML,43 as well as disease progression from MDS to acute leukemia.53 Consistent with impaired DNA damage checkpoints in monosomy 7 cells, we observed decreased apoptosis signaling in these cells, and further genomic instability consequent to failure of these regulatory pathways was suggested by markedly increased mutations in monosomy 7 cells in 1 of our patients. On the basis of our experimental results and data from other sources, we outline a hypothesis for monosomy 7 evolution in which a toxic immune microenvironment and resultant regenerative stress allow selection of cells with both reduced sensitivity to inflammatory signals and impaired DNA damage repair; these cells not only have a survival advantage but are genomically unstable and thus the origin of later leukemia (Figure 6). Haploinsufficiency of several genes located on chromosome 7 involved in critical pathways, rather than any specific gene, may contribute to the prevalence of monosomy 7 cells as the predominant clonal event in a wide variety of BMF syndromes.
Proposed mechanism of clonal expansion and DNA damage accumulation in monosomy 7 cells. DNA damage from multiple environmental and intrinsic sources activates DNA damage response, resulting in a number of cellular responses, such as induction of cell-cycle arrest and repair of the lesions. In case of irreparable damage, cells undergo senescence or apoptosis. HSPCs are under an inflammatory microenvironment in BMF, resulting in dysregulated apoptosis signaling (this part of figure is adapted from Gañán-Gómez et al55 ). The frequently overexpressed and/or constitutively activated transmembrane receptors (Fas, tumor necrosis factor [TNF] receptor 1 [TNFR1], TNFR2, Toll-like receptors [TLRs], and interferon-γ (IFN-γ) receptor [IFNGR]), their associated signal transducers, and recruited T cells induce apoptosis of HSCs through different signaling pathways. Dysregulated pathways or genes in monosomy 7 cells are highlighted in orange. Failure to repair DNA damage as well as impaired response to immune signaling in monosomy 7 cells might result in DNA damage accumulation and clonal expansion, and finally lead to leukemogenesis.
Proposed mechanism of clonal expansion and DNA damage accumulation in monosomy 7 cells. DNA damage from multiple environmental and intrinsic sources activates DNA damage response, resulting in a number of cellular responses, such as induction of cell-cycle arrest and repair of the lesions. In case of irreparable damage, cells undergo senescence or apoptosis. HSPCs are under an inflammatory microenvironment in BMF, resulting in dysregulated apoptosis signaling (this part of figure is adapted from Gañán-Gómez et al55 ). The frequently overexpressed and/or constitutively activated transmembrane receptors (Fas, tumor necrosis factor [TNF] receptor 1 [TNFR1], TNFR2, Toll-like receptors [TLRs], and interferon-γ (IFN-γ) receptor [IFNGR]), their associated signal transducers, and recruited T cells induce apoptosis of HSCs through different signaling pathways. Dysregulated pathways or genes in monosomy 7 cells are highlighted in orange. Failure to repair DNA damage as well as impaired response to immune signaling in monosomy 7 cells might result in DNA damage accumulation and clonal expansion, and finally lead to leukemogenesis.
In this pilot study, we selected patients with aneuploidy in the setting of marrow failure purposely to observe cells early after development of a significant chromosome abnormality but before the cascade of molecular and chromosomal changes that typify AML. Whether the gene mutation profiles in individual patients affected the gene expression patterns of these monosomy 7 cells is not known. Nevertheless, despite small numbers of patients, relatively low numbers of cells, and inherent differences in the genetic background of each patient and clinical courses, we obtained novel and provocative biological data that should inform future experimental approaches. We could reliably detect aneuploid cells, identify dysregulated gene pathways and putative molecular signatures, and find evidence of global genomic instability at the single-cell level. Our approach should be useful for the study of other aneuploid syndromes and more complex cytogenetic abnormalities and in following patients serially during disease progression or treatment.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge Delong Liu, Stefan Cordes, Jun Zhu, Gangqing Hu, and Yixing Han for critical review of the manuscript and analysis methods. They thank the patients and healthy volunteers who donated bone marrow for this project. Sequencing and technical support were provided by the DNA Sequencing and Genomics Core of the National Heart, Lung, and Blood Institute.
This research was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health.
Authorship
Contribution: X.Z. designed and performed the experiments, analyzed data, and wrote the manuscript; S.G. did bioinformatics analysis and wrote the manuscript; S.K., X.F., D.M.T., and J. Cooper analyzed data and edited the manuscript; Z.W., Q.L., J. Chen, and M.d.P.F.I. performed experiments; K.K. performed cell sorting; X.W. supervised data analysis and edited the manuscript; and N.S.Y. conceived, designed, and supervised the experiments, analyzed results, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for X.W. is Division of Diabetes, Endocrinology, and Metabolic Diseases, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD.
Correspondence: Xin Zhao, Hematology Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD 20892; e-mail: xin.zhao@nih.gov; zhaoxin@ihcams.ac.cn.
References
Author notes
X.Z. and S.G. contributed equally to this work.


![Figure 6. Proposed mechanism of clonal expansion and DNA damage accumulation in monosomy 7 cells. DNA damage from multiple environmental and intrinsic sources activates DNA damage response, resulting in a number of cellular responses, such as induction of cell-cycle arrest and repair of the lesions. In case of irreparable damage, cells undergo senescence or apoptosis. HSPCs are under an inflammatory microenvironment in BMF, resulting in dysregulated apoptosis signaling (this part of figure is adapted from Gañán-Gómez et al55). The frequently overexpressed and/or constitutively activated transmembrane receptors (Fas, tumor necrosis factor [TNF] receptor 1 [TNFR1], TNFR2, Toll-like receptors [TLRs], and interferon-γ (IFN-γ) receptor [IFNGR]), their associated signal transducers, and recruited T cells induce apoptosis of HSCs through different signaling pathways. Dysregulated pathways or genes in monosomy 7 cells are highlighted in orange. Failure to repair DNA damage as well as impaired response to immune signaling in monosomy 7 cells might result in DNA damage accumulation and clonal expansion, and finally lead to leukemogenesis.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/130/25/10.1182_blood-2017-08-803353/4/m_blood803353f6.jpeg?Expires=1769213993&Signature=sr~956TCDWku8S~YgprBj2E87W9w0o070c0mM2iUp3WfqcHlxc~IyZh~7XXk1mv3yswQuKWK8nP15I42UrbcbzBYk9GcyvYQqzyyY6wSHyqvljZ0IY9fqSkfSp-pK1TvN2ox0KA9zqGffrpcaN24fK54vnqO3d~McQd5L2-u83JcoLrTz1w4yVBxHUxTnS~3I2wfHO41QjEeyYvPVD5QtmTW1yUs2y9TZ4Haj~FLHT6qlJZbQO~TUlzDrnT8z7NDB424Odx512k37qnVc9DXlKqzyLURVsVFLauwTe044JG3ZL0YgGvDAW1GH73z5JsMAb16PIa8VK46d~Tz2IkmUQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal