Key Points
Through a comprehensive transcriptomic analysis, we discovered 2 major subgroups of myelodysplasia defined by gene expression profiles.
The gene expression–based subgroups had independent prognostic value, which was validated in an external cohort.
Abstract
Myelodysplastic syndromes (MDSs) are a heterogeneous group of clonal hematopoietic disorders with a highly variable prognosis. To identify a gene expression–based classification of myelodysplasia with biological and clinical relevance, we performed a comprehensive transcriptomic analysis of myeloid neoplasms with dysplasia using transcriptome sequencing. Unsupervised clustering of gene expression data of bone marrow CD34+ cells from 100 patients identified 2 subgroups. The first subtype was characterized by increased expression of genes related to erythroid/megakaryocytic (EMK) lineages, whereas the second subtype showed upregulation of genes related to immature progenitor (IMP) cells. Compared with the first so-called EMK subtype, the IMP subtype showed upregulation of many signaling pathways and downregulation of several pathways related to metabolism and DNA repair. The IMP subgroup was associated with a significantly shorter survival in both univariate (hazard ratio [HR], 5.0; 95% confidence interval [CI], 1.8-14; P < .001) and multivariate analysis (HR, 4.9; 95% CI, 1.3-19; P = .02). Leukemic transformation was limited to the IMP subgroup. The prognostic significance of our classification was validated in an independent cohort of 183 patients. We also constructed a model to predict the subgroups using gene expression profiles of unfractionated bone marrow mononuclear cells (BMMNCs). The model successfully predicted clinical outcomes in a test set of 114 patients with BMMNC samples. The addition of our classification to the clinical model improved prediction of patient outcomes. These results indicated biological and clinical relevance of our gene expression–based classification, which will improve risk prediction and treatment stratification of MDS.
Introduction
Myelodysplastic syndrome (MDS) and related myeloid disorders (myelodysplasia) are a heterogeneous group of clonal hematopoietic disorders that are characterized by peripheral blood cytopenias with dysplastic marrow morphology and an increased risk of transformation to acute myeloid leukemia (AML; secondary AML).1,2 Their prognostic profile is highly variable, with survival ranging from a few months to >10 years,3 underscoring the importance of predicting clinical outcomes for treatment stratification. Several scoring systems have been developed on the basis of known prognostic factors, including percentage of marrow blasts, degree of cytopenias, cytogenetic abnormalities, and transfusion requirement.4-8 The effects of gene mutations on clinical outcomes have recently been investigated in large cohorts of myelodysplasia patients and incorporated into a prognostic model.9-11
Gene expression profiling provides a systematic approach for identifying tumor subtypes with prognostic significance and have successfully been applied to the identification of the BCR-ABL1-like subtype of acute lymphoblastic leukemia, activated B-cell–like diffuse large B-cell lymphoma, and basal-like breast cancer.12-15 Recently, several groups performed gene expression profiling of myelodysplasia using microarray platforms, based on which new prognostic models have been proposed.16-19 However, unlike the case with other cancers, these models were not constructed in an unbiased manner and do not represent biologically distinct subsets of patients. Gene expression profiles similar to de novo AML were shown to be associated with leukemic transformation of MDS.19 This raises the possibility that, according to gene expression profiles, myelodysplasia may be subdivided into a subtype with an indolent clinical course and that are at high risk of clonal evolution.
In this study, we performed comprehensive genomic and transcriptomic analyses of 214 patients with myelodysplasia using transcriptome sequencing and targeted-capture sequencing of myelodysplasia-related genes. Through unsupervised class discovery, we have revealed 2 discrete subtypes of myelodysplasia that were characterized by unique gene expression signatures and mutation patterns and, moreover, distinct prognostic profiles.
Methods
Patients and samples
This study was approved by the ethics committees of the Fondazione IRCCS Policlinico San Matteo (Pavia), Karolinska Institutet (Stockholm), and Kyoto University (Kyoto). We enrolled 214 patients with MDS (n = 152), myelodysplastic/myeloproliferative neoplasm (n = 44), and AML with myelodysplasia-related changes (AML-MDS; n = 18) who had been followed at the Department of Hematology, University of Pavia and Fondazione IRCCS Policlinico San Matteo, Pavia (Table 1). Bone marrow mononuclear cells (BMMNCs; n = 165) and/or bone marrow CD34+ cells (n = 100) were obtained from 214 patients, 51 of whom were analyzed for both cell fractions (Figure 1A). CD34+ cells were isolated using magnetic-activated cell sorting separation columns (Miltenyi Biotec, Bergisch Gladbach, Germany).20 Samples were collected 0 to 281 months after diagnosis, between April 2004 and June 2013. No treatment other than supportive care was given prior to sample collection. All patients were reclassified according to the 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia.21 BMMNCs (StemCell Technologies, Vancouver, Canada) and CD34+ cells (Lonza, Basel, Switzerland) of 3 healthy adults each were used as controls.
Patient characteristics
| . | Patients with CD34+ cell samples (n = 100) . | Patients with BMMNC samples only (n = 114) . | ||||
|---|---|---|---|---|---|---|
| First (EMK) subgroup . | Second (IMP) subgroup . | P . | First (EMK) subgroup . | Second (IMP) subgroup . | P . | |
| Number of cases | 61 | 39 | — | 71 | 43 | — |
| Age (y), median (range) | 69 (32–87) | 62 (30–83) | .01 | 67 (30–83) | 67 (39–91) | .46 |
| Sex (male/female) | 38/23 | 27/12 | .52 | 42/27 | 29/18 | 1 |
| Diagnosis, n (%) | ||||||
| MDS | 37 (61) | 22 (56) | — | 57 (80) | 36 (84) | — |
| MDS-SLD | 4 (6.6) | 1 (2.6) | .65 | 5 (7.0) | 1 (2.3) | .41 |
| MDS-RS-SLD | 13 (21) | 1 (2.6) | .008 | 19 (27) | 1 (2.3) | <.001 |
| MDS-MLD | 5 (8.2) | 4 (10) | .73 | 13 (18) | 17 (40) | .02 |
| MDS-RS-MLD | 5 (8.2) | 2 (5.1) | .70 | 9 (13) | 1 (2.3) | .09 |
| MDS with isolated del(5q) | 2 (3.3) | 0 (0) | .52 | 1 (1.4) | 1 (2.3) | 1 |
| MDS-EB-1 | 4 (6.6) | 5 (13) | .31 | 7 (9.9) | 6 (14) | .55 |
| MDS-EB-2 | 4 (6.6) | 9 (23) | .03 | 3 (4.2) | 9 (22) | .009 |
| MDS/MPN | 24 (39) | 6 (15) | — | 13 (18) | 1 (2.3) | — |
| CMML-1 | 19 (31) | 5 (13) | .05 | 8 (11) | 0 (0) | .02 |
| CMML-2 | 1 (1.6) | 0 (0) | 1 | 0 (0) | 0 (0) | 1 |
| MDS/MPN-RS-T | 4 (6.6) | 0 (0) | .15 | 3 (4.2) | 1 (2.3) | 1 |
| MDS/MPN-U | 0 (0) | 1 (2.6) | .39 | 2 (2.8) | 0 (0) | .53 |
| AML-MDS | 0 (0) | 11 (28) | <.001 | 1 (1.4) | 6 (14) | .01 |
| Hemoglobin (g/dL), median (range) | 10.4 (7.0-14.7) | 10.2 (7.0-14.4) | .92 | 9.8 (6.3-15.5) | 9.7 (6.8-13.9) | .43 |
| ANC (×109/L), median (range) | 3.0 (0.35-13) | 1.7 (0.40-32) | .06 | 2.1 (0.20-22) | 1.3 (0.20-11) | .002 |
| Platelet count (×109/L), median (range) | 173 (17.5-895) | 76.0 (15.0-697) | <.001 | 175 (16.0-849) | 102 (12.5-939) | .03 |
| Myeloid/erythroid ratio, median (range) | 2.0 (0.50–10) | 2.0 (0.13–10) | .86 | 2.0 (0.25-10) | 1.5 (0.20-10) | .71 |
| Bone marrow blasts (%), median (range) | 2 (0–15) | 11 (1–90) | <.001 | 2 (0-86) | 4 (0-63) | .003 |
| Bone marrow ring sideroblasts (%), median (range) | 9 (0–94) | 5 (0–78) | .20 | 13 (0–94) | 0.3 (0–81) | .08 |
| IPSS-R in the patients with MDS, n (%) | ||||||
| Very low | 0 (0) | 0 (0) | 1 | 0 (0) | 0 (0) | 1 |
| Low | 15 (25) | 3 (7.7) | .04 | 26 (37) | 10 (23) | .15 |
| Intermediate | 12 (20) | 7 (18) | 1 | 17 (24) | 8 (19) | .64 |
| High | 7 (11) | 5 (13) | 1 | 9 (13) | 10 (23) | .19 |
| Very high | 3 (4.9) | 7 (18) | .04 | 3 (4.2) | 6 (14) | .08 |
| Missing | 0 (0) | 0 (0) | 1 | 2 (2.8) | 2 (4.7) | .63 |
| Treatment during the follow-up period,*n (%) | ||||||
| Allogeneic stem cell transplantation | 3 (4.9) | 7 (18) | .04 | 4 (5.6) | 4 (9.3) | .47 |
| Cytotoxic chemotherapy | 0 (0) | 7 (18) | <.001 | 2 (2.8) | 6 (14) | .05 |
| Azacitidine | 3 (4.9) | 2 (11) | 1 | 3 (4.2) | 4 (9.3) | .42 |
| Response | 1 (33) | 0 (0) | 1 | 0 (0) | 1 (25) | 1 |
| Only supportive care | 56 (92) | 26 (67) | .003 | 63 (89) | 31 (72) | .04 |
| . | Patients with CD34+ cell samples (n = 100) . | Patients with BMMNC samples only (n = 114) . | ||||
|---|---|---|---|---|---|---|
| First (EMK) subgroup . | Second (IMP) subgroup . | P . | First (EMK) subgroup . | Second (IMP) subgroup . | P . | |
| Number of cases | 61 | 39 | — | 71 | 43 | — |
| Age (y), median (range) | 69 (32–87) | 62 (30–83) | .01 | 67 (30–83) | 67 (39–91) | .46 |
| Sex (male/female) | 38/23 | 27/12 | .52 | 42/27 | 29/18 | 1 |
| Diagnosis, n (%) | ||||||
| MDS | 37 (61) | 22 (56) | — | 57 (80) | 36 (84) | — |
| MDS-SLD | 4 (6.6) | 1 (2.6) | .65 | 5 (7.0) | 1 (2.3) | .41 |
| MDS-RS-SLD | 13 (21) | 1 (2.6) | .008 | 19 (27) | 1 (2.3) | <.001 |
| MDS-MLD | 5 (8.2) | 4 (10) | .73 | 13 (18) | 17 (40) | .02 |
| MDS-RS-MLD | 5 (8.2) | 2 (5.1) | .70 | 9 (13) | 1 (2.3) | .09 |
| MDS with isolated del(5q) | 2 (3.3) | 0 (0) | .52 | 1 (1.4) | 1 (2.3) | 1 |
| MDS-EB-1 | 4 (6.6) | 5 (13) | .31 | 7 (9.9) | 6 (14) | .55 |
| MDS-EB-2 | 4 (6.6) | 9 (23) | .03 | 3 (4.2) | 9 (22) | .009 |
| MDS/MPN | 24 (39) | 6 (15) | — | 13 (18) | 1 (2.3) | — |
| CMML-1 | 19 (31) | 5 (13) | .05 | 8 (11) | 0 (0) | .02 |
| CMML-2 | 1 (1.6) | 0 (0) | 1 | 0 (0) | 0 (0) | 1 |
| MDS/MPN-RS-T | 4 (6.6) | 0 (0) | .15 | 3 (4.2) | 1 (2.3) | 1 |
| MDS/MPN-U | 0 (0) | 1 (2.6) | .39 | 2 (2.8) | 0 (0) | .53 |
| AML-MDS | 0 (0) | 11 (28) | <.001 | 1 (1.4) | 6 (14) | .01 |
| Hemoglobin (g/dL), median (range) | 10.4 (7.0-14.7) | 10.2 (7.0-14.4) | .92 | 9.8 (6.3-15.5) | 9.7 (6.8-13.9) | .43 |
| ANC (×109/L), median (range) | 3.0 (0.35-13) | 1.7 (0.40-32) | .06 | 2.1 (0.20-22) | 1.3 (0.20-11) | .002 |
| Platelet count (×109/L), median (range) | 173 (17.5-895) | 76.0 (15.0-697) | <.001 | 175 (16.0-849) | 102 (12.5-939) | .03 |
| Myeloid/erythroid ratio, median (range) | 2.0 (0.50–10) | 2.0 (0.13–10) | .86 | 2.0 (0.25-10) | 1.5 (0.20-10) | .71 |
| Bone marrow blasts (%), median (range) | 2 (0–15) | 11 (1–90) | <.001 | 2 (0-86) | 4 (0-63) | .003 |
| Bone marrow ring sideroblasts (%), median (range) | 9 (0–94) | 5 (0–78) | .20 | 13 (0–94) | 0.3 (0–81) | .08 |
| IPSS-R in the patients with MDS, n (%) | ||||||
| Very low | 0 (0) | 0 (0) | 1 | 0 (0) | 0 (0) | 1 |
| Low | 15 (25) | 3 (7.7) | .04 | 26 (37) | 10 (23) | .15 |
| Intermediate | 12 (20) | 7 (18) | 1 | 17 (24) | 8 (19) | .64 |
| High | 7 (11) | 5 (13) | 1 | 9 (13) | 10 (23) | .19 |
| Very high | 3 (4.9) | 7 (18) | .04 | 3 (4.2) | 6 (14) | .08 |
| Missing | 0 (0) | 0 (0) | 1 | 2 (2.8) | 2 (4.7) | .63 |
| Treatment during the follow-up period,*n (%) | ||||||
| Allogeneic stem cell transplantation | 3 (4.9) | 7 (18) | .04 | 4 (5.6) | 4 (9.3) | .47 |
| Cytotoxic chemotherapy | 0 (0) | 7 (18) | <.001 | 2 (2.8) | 6 (14) | .05 |
| Azacitidine | 3 (4.9) | 2 (11) | 1 | 3 (4.2) | 4 (9.3) | .42 |
| Response | 1 (33) | 0 (0) | 1 | 0 (0) | 1 (25) | 1 |
| Only supportive care | 56 (92) | 26 (67) | .003 | 63 (89) | 31 (72) | .04 |
AML-MDS, AML with myelodysplasia-related changes; ANC, absolute neutrophil count; CMML, chronic myelomonocytic leukemia; MDS-EB, MDS with excess of blasts; MDS-MLD, MDS with multilineage dysplasia; MDS/MPN, myelodysplastic/myeloproliferative neoplasm; MDS/MPN-RS-T, MDS/MPN with ring sideroblasts and thrombocytosis; MDS/MPN-U, MDS/MPN, unclassifiable; MDS-RS-SLD, MDS with ring sideroblasts with single-lineage dysplasia; MDS-SLD, MDS with single-lineage dysplasia; MDS-RS-MLD, MDS with ring sideroblasts with multilineage dysplasia.
Some patients received various treatments (eg, cytotoxic chemotherapy followed by allogeneic stem cell transplantation). This led to the sums of patients exceeding 100%.
Study design and characterization of the gene expression–based subgroups of myelodysplasia. (A) Schematic depicting the study design. Venn diagrams show sources of RNA. The left and right halves indicate outlines of the analyses for bone marrow CD34+ cells and BMMNCs, respectively. Unsupervised clustering was performed on gene expression data of bone marrow CD34+ cells from 100 patients (training set of CD34+ cell samples), revealing 2 distinct subgroups. A regression model was constructed from the training set, followed by validation in an independent cohort (lower left). A regression model to predict the subgroups using BMMNC samples was also constructed from gene expression data of 51 patients with both CD34+ cell and BMMNC samples (training set of BMMNC samples). Prognostic significance of the model was tested in 114 patients with only BMMNC samples. (B) A heatmap shows expression levels of 3141 genes with high variability in 100 CD34+ cell samples. Each row represents 1 gene, and each column represents 1 sample. Gene expression–based subgroups, WHO subtypes, genetic lesions, and patients’ prognosis are shown below the heatmap. AML-MDS, AML with myelodysplasia-related changes; CMML, chronic myelomonocytic leukemia; MDS-EB, MDS with excess blasts; MDS-MLD, MDS with multilineage dysplasia; MDS/MPN-RS-T, MDS/MPN with ring sideroblasts and thrombocytosis; MDS/MPN-U, MDS/MPN, unclassifiable; MDS-RS-SLD, MDS with ring sideroblasts with single lineage dysplasia; MDS-SLD, MDS with single-lineage dysplasia; MDS-RS-MLD, MDS with ring sideroblasts with multilineage dysplasia. (C) A heatmap of expression levels of 7 genes of known prognostic significance in 100 CD34+ cell samples. (D) Expression levels of genes related to specific hematopoietic lineages. The left panel is a heatmap of gene expression levels in 100 CD34+ cell samples. Rows represent genes sorted according to hematopoietic lineages in which they are specifically expressed. Columns represent samples along with their gene expression–based subgroups and WHO subtypes. The middle panel represents mean z scores for each hematopoietic lineage. CLP, common lymphoid progenitor; CMP, common myeloid progenitor; EB, erythroblast; GMP, granulocyte monocyte progenitor; HSC, hematopoietic stem cell; MPP, multipotent progenitor; MEP, megakaryocyte/erythrocyte progenitor; MK, megakaryocyte.
Study design and characterization of the gene expression–based subgroups of myelodysplasia. (A) Schematic depicting the study design. Venn diagrams show sources of RNA. The left and right halves indicate outlines of the analyses for bone marrow CD34+ cells and BMMNCs, respectively. Unsupervised clustering was performed on gene expression data of bone marrow CD34+ cells from 100 patients (training set of CD34+ cell samples), revealing 2 distinct subgroups. A regression model was constructed from the training set, followed by validation in an independent cohort (lower left). A regression model to predict the subgroups using BMMNC samples was also constructed from gene expression data of 51 patients with both CD34+ cell and BMMNC samples (training set of BMMNC samples). Prognostic significance of the model was tested in 114 patients with only BMMNC samples. (B) A heatmap shows expression levels of 3141 genes with high variability in 100 CD34+ cell samples. Each row represents 1 gene, and each column represents 1 sample. Gene expression–based subgroups, WHO subtypes, genetic lesions, and patients’ prognosis are shown below the heatmap. AML-MDS, AML with myelodysplasia-related changes; CMML, chronic myelomonocytic leukemia; MDS-EB, MDS with excess blasts; MDS-MLD, MDS with multilineage dysplasia; MDS/MPN-RS-T, MDS/MPN with ring sideroblasts and thrombocytosis; MDS/MPN-U, MDS/MPN, unclassifiable; MDS-RS-SLD, MDS with ring sideroblasts with single lineage dysplasia; MDS-SLD, MDS with single-lineage dysplasia; MDS-RS-MLD, MDS with ring sideroblasts with multilineage dysplasia. (C) A heatmap of expression levels of 7 genes of known prognostic significance in 100 CD34+ cell samples. (D) Expression levels of genes related to specific hematopoietic lineages. The left panel is a heatmap of gene expression levels in 100 CD34+ cell samples. Rows represent genes sorted according to hematopoietic lineages in which they are specifically expressed. Columns represent samples along with their gene expression–based subgroups and WHO subtypes. The middle panel represents mean z scores for each hematopoietic lineage. CLP, common lymphoid progenitor; CMP, common myeloid progenitor; EB, erythroblast; GMP, granulocyte monocyte progenitor; HSC, hematopoietic stem cell; MPP, multipotent progenitor; MEP, megakaryocyte/erythrocyte progenitor; MK, megakaryocyte.
RNA sequencing
RNA was extracted with TRIzol reagent (Life Technologies, Carlsbad, CA) and treated with DNase I (Qiagen, Hilden, Germany). RNA integrity numbers were confirmed to be >7 using the TapeStation (Agilent Technologies, Palo Alto, CA). The RNA-sequencing libraries were prepared from poly(A)-selected RNA using the NEBNext Ultra RNA Library Prep kit for Illumina (New England BioLabs, Ipswich, MA). Libraries were sequenced using the HiSeq 2000 or 2500 platform according to a standard 100-bp paired-end read protocol (Illumina, San Diego, CA).
Sequencing reads were aligned to the human reference genome (hg19) using RUM version 2.0.4.22 Fusion transcripts were detected by Genomon-fusion (http://genomon.hgc.jp/rna/),23 followed by reverse transcription polymerase chain reaction and direct sequencing of the polymerase chain reaction products. Differential expression analysis was conducted using edgeR version 3.6.8.24 The analysis was confined to those genes expressed at >1 counts per million (CPM) in >5 samples. Generalized linear models were used to compare gene expression data. Correction for multiple testing was done by the Benjamini-Hochberg method, in which q-value < 0.01 was considered significant. High EVI1 expression was defined as CPM >100 and >20 in CD34+ cells and BMMNCs, respectively. ROAST implemented in the R package limma version 3.22.4 was used to find the significantly upregulated pathways, where the pathways defined by the Kyoto Encyclopedia of Genes and Genomes were tested.25 Genes specifically expressed in each hematopoietic stem/progenitor population were adopted from a previous report.26 Genome data have been deposited in the European Genome-phenome Archive (http://www.ebi.ac.uk/ega/) under accession number EGAS00001002346.
Clustering of gene expression data
Consensus clustering was performed based on log2(CPM) values of genes that showed larger dispersions than genes with similar mean expression levels,27 where K-means clustering based on the Ward and the Euclidean distance was repeated 1000 times by randomly sampling 80% of the entire sample set using the R package ConsensusClusterPlus. The number of clusters was determined from the relative change in the area under the cumulative distribution function curve. Clustering was also performed based on a nonnegative matrix factorization algorithm using the R package NMF.
A classifier of the gene expression–based subgroups was constructed using the 100 CD34+ cell samples as a training set (Figure 1A). For applicability to gene expression microarray data, the model only included moderately expressed genes showing 50 to 75 percentile of mean signals among all the genes in 183 MDS patients analyzed on GeneChip Human Genome U133 Plus 2.0 array (Affymetrix).28 We used elastic net logistic regression in which the parameters were determined as a = 0.45 and λ = 0.111 using 10-fold cross-validation on the training set. The mean classification error was 0.05. Another classifier was constructed using BMMNC samples from 51 patients, who had been assigned to the subgroups by the gene expression data of their CD34+ cells (Figure 1A; supplemental Methods, available on the Blood Web site). Since unfractionated BMMNCs contain mature hematopoietic cells and contaminating peripheral blood, variables in the model were selected from highly upregulated genes in the second subgroup of this training cohort. We first performed rigorous differential expression analysis using the generalized linear model likelihood ratio test.24 Elastic net logistic regression was then applied to the 30 most significantly upregulated genes in the second subgroup as compared with the first subgroup and the healthy adults. The parameters of the elastic net were determined as a = 0.7 and λ = 0.0176 using 10-fold cross-validation, with a mean classification error of 0.10.
Targeted DNA sequencing
Genomic DNA was available for 211 of the 214 patients (99%) and was extracted from peripheral blood granulocytes (n = 111), BMMNCs (n = 56), bone marrow polymorphonuclear cells (n = 43), or bone marrow CD34+ cells (n = 1). Nine DNA samples were subjected to whole-genome amplification. Sequencing libraries were prepared from 200 to 1000 ng DNA. Target capture was performed using a SureSelect custom kit (Agilent Technologies). RNA baits were designed using SureDesign (Agilent Technologies) to capture coding exons from 89 known or putative driver genes in myelodysplasia (supplemental Table 1, available on the Blood Web site) and 1674 single-nucleotide polymorphisms.
Libraries were sequenced using the HiSeq 2000 or 2500 platform with a standard 100-bp paired-end read protocol (Illumina). Sequencing reads were aligned to the human reference genome (hg19) using BWA version 0.7.10. Oncogenic variants were identified as previously described.10,29 Genomic copy-number analysis was performed based on the sequencing depths of the target regions compared with those of pooled controls.
Gene expression microarray
Gene expression profiling using the GeneChip Human Genome U133 Plus 2.0 array (Affymetrix) was performed for an independent cohort of 183 MDS patients (Figure 1A; supplemental Table 2).28 Gene expression levels of each gene were obtained through preprocessing by GeneChip Robust Multiarray Analysis (GC-RMA)30 and selection of a representative probe set by the JetSet annotations,31 as described previously.32
Statistical analysis
Numerical and categorical variables were compared using the Mann-Whitney test and Fisher’s exact test, respectively. Overall survival analyses were performed with the Kaplan-Meier method and log-rank test. Patients who had already developed leukemia at the time of sampling were removed from the analysis of leukemia-free survival, with nonleukemic death treated as a competing risk.33 Multivariate analyses were performed using Cox proportional hazards regression for overall survival and competing risk regression for leukemia-free survival, both with stepwise selection based on the Akaike information criterion (AIC) score. Tested variables were the 2 gene expression–based subgroups, age, sex, and the prognostic variables in the revised international prognostic scoring system for MDS (IPSS-R): percentage of marrow blasts, cytogenetic abnormalities, hemoglobin, absolute neutrophil count, and platelet levels. Cytogenetic abnormalities were classified according to the MDS Cytogenetic Scoring System.5 Comparison between models was performed by means of the AIC score and c-index. Analyses were performed using R versions 2.15.3 (model construction using elastic net logistic regression) and 3.0.1 (the other analyses). The R scripts are shown in supplemental Methods.
Results
Identification of unique expression clusters in myelodysplasia
To identify discrete subtypes of myelodysplasia, RNA-sequencing data were analyzed using consensus clustering. Clustering analysis of the data from bone marrow CD34+ cell samples (n = 100) revealed 2 stable clusters (Figure 1B); the clustering stability was decreased for >2 clusters (supplemental Figure 1A-B), which indicated that further subdivision of the 2 clusters was not feasible. Nonnegative matrix factorization, another algorithm for class discovery, also supported a 2-class split of the gene expression data (supplemental Figure 1C-D). By contrast, no stable clusters were detected in the analysis of unfractionated BMMNC samples (n = 165; supplemental Figure 1E-F).
The 2 subgroups had a distinct hematological picture (Table 1). WHO subtypes with increased ring sideroblasts were more enriched in the first subgroup (odds ratio, 6.8; 95% confidence interval [CI], 5.5-8.1; P = .002). In contrast, patients with advanced disease (ie, MDS-EB-1/2 and AML-MDS) were more frequently clustered into the second subgroup (odds ratio, 12; 95% CI, 4.4-32; P < .001). They also had lower platelet counts (median 76.0 × 109/L vs 173 × 109/L; P < .001) and higher percentages of bone marrow blasts (median 11% vs 2%, P < .001) as compared with the first subgroup.
A marked difference between the 2 subgroups was found at the transcriptome level. The patients in the second subgroup showed higher expression of the genes known to be associated with poor prognosis in myeloid malignancies (MSI2, BMI1, SETBP1, BAALC, FLT3, MN1, and ERG; q-value < 0.01) (Figure 1C).34-39 Pathway analysis of differentially expressed genes between both subgroups further characterized their expression profiles. Compared with the first subgroup, 19 and 75 pathways were up- and downregulated in the second subgroup, respectively (supplemental Table 3). Cell signaling accounted for 12 of 19 upregulated pathways (63%) in the second subgroup, including MAPK, phosphatidylinositol 3-kinase, Notch, and JAK/STAT pathways (q-value < 0.01). Of 75 downregulated pathways, 46 (61%) were related to metabolism and 5 (6.7%) were to DNA repair. The 2 subgroups also exhibited contrasting gene expression profiles in terms of the commitment to specific hematopoietic lineages.26 The first subgroup showed increased expression of the sets of genes specifically expressed in erythroblasts and megakaryocyte/erythrocyte progenitors (Figure 1D). Mean expression levels of the erythroid genes were not significantly correlated with bone marrow myeloid/erythroid ratios (r = −0.16; 95% CI, −0.36 to 0.05; P = .13; supplemental Figure 2), suggesting that a strong erythroid signature in the CD34+ fraction did not simply reflect erythroid hyperplasia. In the second subgroup, by contrast, expression of these genes was significantly decreased, even compared with that in the normal individuals (supplemental Figures 3 and 4), whereas genes characteristic of more immature hematopoietic lineages were upregulated (Figure 1D). We hereafter refer to the 2 subgroups as the subgroup enriched with erythroid/megakaryocytic (EMK) signatures and that with immature progenitor (IMP) signatures.
Prognostic significance of the gene expression–based classification
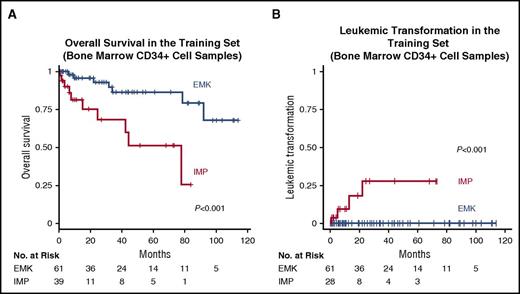
As expected from the association with many adverse prognostic features, the IMP subtype was characterized by poor clinical outcomes; in univariate analysis, the IMP subgroup was significantly associated with an inferior overall survival compared with the EMK subgroup (HR, 5.0; 95% CI, 1.8-14; P < .001) (Figure 2A). In addition to the patients who had already developed leukemia at the time of sampling, the patients who later progressed to leukemia were also limited to the IMP subgroup (Figures 1B and 2B). The negative impact on the survival of the IMP subgroup was still observed after adjustment for the effects of known risk factors using multivariate analysis (HR, 4.9; 95% CI, 1.3-19; P = .02) (Table 2).5
Prognostic significance of the subgroups based on gene expression profiles of bone marrow CD34+ cells. (A) Rates of overall survival in the 2 subgroups for the training set of bone marrow CD34+ cell samples. (B) Cumulative incidence of leukemic transformation in the 2 subgroups for the training set of bone marrow CD34+ cell samples. The patients who had already developed leukemia at the time of sampling were removed from the analysis.
Prognostic significance of the subgroups based on gene expression profiles of bone marrow CD34+ cells. (A) Rates of overall survival in the 2 subgroups for the training set of bone marrow CD34+ cell samples. (B) Cumulative incidence of leukemic transformation in the 2 subgroups for the training set of bone marrow CD34+ cell samples. The patients who had already developed leukemia at the time of sampling were removed from the analysis.
HR for death or leukemic transformation in a multivariate model
| . | HR (95% CI) . | P . |
|---|---|---|
| Patients with CD34+cell samples (n = 100) | ||
| Death | ||
| Hemoglobin (g/dL) | 0.54 (0.37-0.77) | <.001 |
| Bone marrow blast (%) | 1.05 (1.02-1.09) | .003 |
| Age (y) | 1.09 (1.02-1.16) | .009 |
| IMP vs EMK subgroup | 4.87 (1.25-19.0) | .02 |
| Leukemic transformation* | ||
| Patients with BMMNC samples only (n = 114) | ||
| Death | ||
| Bone marrow blast (%) | 1.11 (1.06-1.16) | <.001 |
| Hemoglobin (g/dL) | 0.67 (0.52-0.86) | .001 |
| IMP vs EMK subgroup | 2.90 (1.10-7.62) | .03 |
| Absolute neutrophil count (×109/L) | 1.12 (1.00-1.26) | 0.05 |
| Cytogenetic abnormalities | 2.16 (0.92-5.07) | 0.08 |
| Platelet (×109/L) | 0.996 (0.992-1.001) | .12 |
| Leukemic transformation | ||
| Bone marrow blast (%) | 1.14 (1.08-1.21) | <.001 |
| IMP vs EMK subgroup | 5.86 (1.59-21.6) | .008 |
| Hemoglobin (g/dL) | 0.80 (0.66-0.98) | .03 |
| . | HR (95% CI) . | P . |
|---|---|---|
| Patients with CD34+cell samples (n = 100) | ||
| Death | ||
| Hemoglobin (g/dL) | 0.54 (0.37-0.77) | <.001 |
| Bone marrow blast (%) | 1.05 (1.02-1.09) | .003 |
| Age (y) | 1.09 (1.02-1.16) | .009 |
| IMP vs EMK subgroup | 4.87 (1.25-19.0) | .02 |
| Leukemic transformation* | ||
| Patients with BMMNC samples only (n = 114) | ||
| Death | ||
| Bone marrow blast (%) | 1.11 (1.06-1.16) | <.001 |
| Hemoglobin (g/dL) | 0.67 (0.52-0.86) | .001 |
| IMP vs EMK subgroup | 2.90 (1.10-7.62) | .03 |
| Absolute neutrophil count (×109/L) | 1.12 (1.00-1.26) | 0.05 |
| Cytogenetic abnormalities | 2.16 (0.92-5.07) | 0.08 |
| Platelet (×109/L) | 0.996 (0.992-1.001) | .12 |
| Leukemic transformation | ||
| Bone marrow blast (%) | 1.14 (1.08-1.21) | <.001 |
| IMP vs EMK subgroup | 5.86 (1.59-21.6) | .008 |
| Hemoglobin (g/dL) | 0.80 (0.66-0.98) | .03 |
Failure in convergence due to no leukemic transformation in the EMK subgroup.
The strong effects of the gene expression subtype on survival were validated in an independent cohort.17 Because gene expression levels were analyzed using a different platform (the Affymetrix GeneChip Human Genome U133 Plus 2.0 array), in this external cohort, we developed a classifier of the 2 expression subgroups relying on the expression of a small number of genes so that it can be used across different assay platforms. Elastic net logistic regression was first performed on gene expression data of CD34+ cells in our cohort as a training set (Figure 1A). Elastic net parameters were determined by 10-fold cross-validation, yielding a logistic regression model that predicts the subgroups based on expression levels of 68 genes (Figure 3A; supplemental Table 4). The regression model was then applied to the microarray-based expression data of bone marrow CD34+ cells from the external cohort of 183 patients with MDS.28 According to this model, 116 (63%) and 67 (37%) patients were classified into the EMK and IMP subgroups, respectively (Figure 3B). The gene expression signatures of hematopoietic lineages in the 2 subgroups were largely recapitulated in this validation data set: upregulation of the genes related to erythroid lineages in the EMK subgroup and that of the progenitor signatures in IMP (supplemental Figure 5). Patients assigned to the IMP subtype were shown to have significantly shorter survival than those in the EMK subgroup (HR, 3.0; 95% CI, 1.9-5.1; P < .001) (Figure 3C). The negative impact of the IMP expression profile became more prominent, when the association was tested for leukemic transformation (HR, 6.8; 95% CI, 3.2-14; P < .001) (Figure 3D). The difference in survival and leukemic transformation remained significant even after the effect of bone marrow blasts was adjusted (supplemental Figure 6). These results confirmed the reproducibility of the prognostic value of the expression profiles and the robustness of this set of classifiers, regardless of the assay used for gene expression profiling.
Validation of the prognostic significance of the subgroups in the test set for bone marrow CD34+ cell samples. (A) Heatmap showing expression levels of 68 genes in the regression model for 100 patients with bone marrow CD34+ cell samples. Each row represents 1 gene, and each column represents 1 sample. Gene expression–based subgroups, WHO subtypes, and patients’ prognosis are shown below the heatmap. (B) A heatmap shows expression levels of 68 genes in the regression model for an independent cohort of 183 patients with MDS. (C) Rates of overall survival in the 2 subgroups for the test set of bone marrow CD34+ cell samples. (D) Cumulative incidence of leukemic transformation in the 2 subgroups for the test cohort of bone marrow CD34+ cell samples.
Validation of the prognostic significance of the subgroups in the test set for bone marrow CD34+ cell samples. (A) Heatmap showing expression levels of 68 genes in the regression model for 100 patients with bone marrow CD34+ cell samples. Each row represents 1 gene, and each column represents 1 sample. Gene expression–based subgroups, WHO subtypes, and patients’ prognosis are shown below the heatmap. (B) A heatmap shows expression levels of 68 genes in the regression model for an independent cohort of 183 patients with MDS. (C) Rates of overall survival in the 2 subgroups for the test set of bone marrow CD34+ cell samples. (D) Cumulative incidence of leukemic transformation in the 2 subgroups for the test cohort of bone marrow CD34+ cell samples.
Prediction of the gene expression–based subgroups using BMMNC samples
To facilitate the clinical use of this molecular classification without relying on CD34+ cell selection, we constructed a classifier for the 2 expression subtypes using the data from unfractionated BMMNCs. Among the 100 patients who had been assigned to the EMK or IMP subgroups on the basis of the gene expression data of CD34+ cells, 51 were also analyzed by RNA sequencing for BMMNCs and thus used as a training cohort (Figures 1A and 4A). Through 10-fold crossvalidation on this training set, we developed a logistic regression model to predict the subgroups based on the expression levels of 9 genes (Figure 4B). The model was applied to the gene expression data of BMMNCs in the remaining 114 cases, of whom 71 (62%) and 43 (38%) were predicted to be the EMK and IMP subgroup, respectively (supplemental Figure 7). Compared with the predicted EMK subgroup, the IMP subgroup was associated with a significantly shorter survival in univariate analysis (HR, 4.5; 95% CI, 2.0-10; P < .001) (Figure 4C). Again, association was more pronounced for leukemic transformation (HR, 7.3; 95% CI, 2.0-26; P = .002) (Figure 4D) than for overall survival. Multivariate analysis also demonstrated that the predicted IMP subgroup was independently associated with overall survival (HR, 2.9; 95% CI, 1.1-7.6; P = .03) (Table 2) and leukemic transformation (HR, 5.9; 95% CI, 1.6-22; P = .008) (Table 2). These results indicated the prognostic value of the classification based on the gene expression profiles of BMMNCs.
Prognostic significance of the subgroups based on gene expression profiles of BMMNCs. (A) A schematic depicts how to construct a logistic regression model to predict the subgroups using 10-fold cross-validation on gene expression data. (B) A table shows gene names and their coefficients in a logistic regression model to predict the subgroups based on gene expression profile of BMMNCs. (C) Rates of overall survival in the 2 subgroups for the test set of BMMNC samples. (D) Cumulative incidence of leukemic transformation in the 2 subgroups for the test set of BMMNC samples. The patients who had already developed leukemia at the time of sampling were removed from the analysis.
Prognostic significance of the subgroups based on gene expression profiles of BMMNCs. (A) A schematic depicts how to construct a logistic regression model to predict the subgroups using 10-fold cross-validation on gene expression data. (B) A table shows gene names and their coefficients in a logistic regression model to predict the subgroups based on gene expression profile of BMMNCs. (C) Rates of overall survival in the 2 subgroups for the test set of BMMNC samples. (D) Cumulative incidence of leukemic transformation in the 2 subgroups for the test set of BMMNC samples. The patients who had already developed leukemia at the time of sampling were removed from the analysis.
Genetic alterations in the gene expression subgroups
The 2 expression subgroups also had unique profile of genetic alterations. We interrogated myelodysplasia-related mutations as well as chromosomal lesions identified by targeted-capture sequencing with a mean coverage of 1009× (range, 207-2306×). In total, we identified 313 nonsynonymous single-nucleotide variants, 147 small insertion–deletions in common targets of myeloid neoplasms, and 170 copy-number abnormalities and/or allelic imbalances (supplemental Figure 8). In addition, RNA sequencing detected aberrant gene fusions in 4 patients, of whom 3 had EVI1-containing fusions, including NRIP1-EVI1 (n = 2) and RUNX1-EVI1 (n = 1). Elevated EVI1 expression was also found in an additional 3 patients with no accompanying gene fusions, of whom 1 harbored a 3q26 abnormality (supplemental Figure 9). Of interest, most of these EVI1-overexpressed cases (5 out of 6) were accompanied by mutated SF3B1. All combined, 1 or more genetic lesions were identified in 194 patients with a median of 3 (0-12) lesions per case.
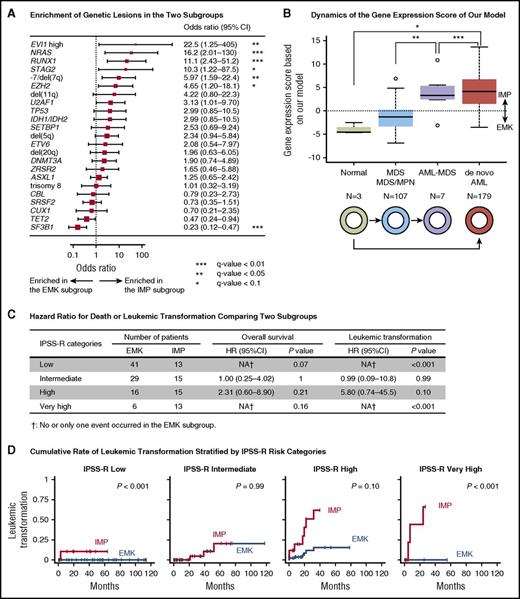
The number of genetic lesions was significantly higher in the IMP subgroup (median, 3 [range, 0-12] vs 2 [0-7], P = .003). The prevalence of individual genetic lesions also showed substantial variation between the 2 expression subtypes. SF3B1 mutation was more frequent in the EMK subgroup (q-value < 0.05; Figure 5A). Enrichment of SF3B1 mutations in the EMK subgroup was observed when the analysis was limited to the patients with bone marrow ring sideroblasts of ≥15% (n = 89) (odds ratio, 5.2; 95% CI, 1.9-14; P = .003). Notable exceptions for this were the SF3B1-mutated patients with EVI1 overexpression, who were all grouped into the IMP subgroup, which is in line with a strong adverse effect of EVI1 overexpression on survival (Figure 5A; supplemental Figure 8).40,41 Effects of genetic alterations on transcriptome profiles were also suggested by higher frequencies of genetic lesions known to be associated with worse prognosis in the IMP subgroup (−7/del(7q) and NRAS and RUNX1 mutations; q-value < 0.05) (Figure 5A).9 In accordance with their higher rate of leukemic transformation, patients in the IMP subgroup were enriched for the type I mutations, including those in FLT3, PTPN11, WT1, IDH1/IDH2, NPM1, and NRAS, which were recently reported to be associated with transformation to secondary AML (supplemental Figure 10).42
Comparison between our gene expression-based classification and other prognostic models. (A) A forest plot of odds ratios for detecting genetic lesions in the IMP subgroup. P values are based on Fisher’s exact test. The square sizes were inversely proportional to the confidence intervals of the estimated odds ratio. (B) A boxplot shows distribution of log odds ratio for the predicted IMP subgroup that was calculated based on our regression model using expression levels of 9 genes. Boxes correspond to healthy adults (n = 3), patients with MDS or myelodysplastic/myeloproliferative neoplasm (MDS/MPN; n = 107), those with AML with myelodysplasia-related changes (AML-MDS; n = 7), and those in a published external AML cohort from The Cancer Genome Atlas (n = 179). *P < .05 (Bonferroni adjusted); **P < .01 (Bonferroni adjusted); ***P < .001 (Bonferroni adjusted). (C) HR of death or leukemic transformation comparing the EMK and IMP subgroups. Number of patients in the EMK and IMP subgroup, HR, its 95% CI, and P values are shown for each IPSS-R risk category. (D) Cumulative incidence of leukemic transformation in MDS patients stratified by categories of IPSS-R.
Comparison between our gene expression-based classification and other prognostic models. (A) A forest plot of odds ratios for detecting genetic lesions in the IMP subgroup. P values are based on Fisher’s exact test. The square sizes were inversely proportional to the confidence intervals of the estimated odds ratio. (B) A boxplot shows distribution of log odds ratio for the predicted IMP subgroup that was calculated based on our regression model using expression levels of 9 genes. Boxes correspond to healthy adults (n = 3), patients with MDS or myelodysplastic/myeloproliferative neoplasm (MDS/MPN; n = 107), those with AML with myelodysplasia-related changes (AML-MDS; n = 7), and those in a published external AML cohort from The Cancer Genome Atlas (n = 179). *P < .05 (Bonferroni adjusted); **P < .01 (Bonferroni adjusted); ***P < .001 (Bonferroni adjusted). (C) HR of death or leukemic transformation comparing the EMK and IMP subgroups. Number of patients in the EMK and IMP subgroup, HR, its 95% CI, and P values are shown for each IPSS-R risk category. (D) Cumulative incidence of leukemic transformation in MDS patients stratified by categories of IPSS-R.
Comparison between our gene expression–based classification and other prognostic models
In view of the enhanced progenitor signatures in the IMP subgroup, it would be of interest to correlate our classifier with the LSC17 score, which has recently been proposed to predict a subset of poor-risk AML based on the expression of 17 genes related to a leukemic stem cell signature.43 None of the genes in the LSC17 score overlapped with our regression model. We first investigated the prognostic impact of the LSC17 score in our test cohort of patients, in which expression profiling was performed using BMMNCs (n = 114). As shown in supplemental Figure 11, patients with higher LSC17 scores (above the median) had a significantly shorter overall survival than those with lower scores (HR, 3.2; 95% CI, 1.4-7.1; P = .003). However, the LSC17 score was outperformed by our classification; the IMP subgroup was associated with a larger HR and a higher accuracy to predict clinical outcomes (Figure 4B). Conversely, when applied to the cohort of 179 AML patients from The Cancer Genome Atlas,44 the LSC17 score was a better predictor of survival than our model (supplemental Figure 12). These results suggest that the IMP subgroup and LSC17-associated expression signatures seem to reflect different aspects of leukemia pathogenesis, including alterations in cell populations (myelodysplasia with low blast counts and full-blown leukemia with increased marrow blasts). Logistic regression odds ratio of being classified as the IMP subgroup (IMP score) dramatically increased during clonal evolution of myelodysplasia, from healthy controls to patients with myelodysplasia and then to those who had experienced leukemic transformation (Figure 5B). Most of the patients with de novo AML had increased IMP scores. Thus, the dramatic increase in the IMP score during leukemic transformation might be the basis of a strong prognostic impact of the IMP subgroup in myelodysplasia. By contrast, the LSC17 score showed a modest, if not statistically significant, increase during clonal evolution of myelodysplasia (supplemental Figure 13), suggesting that unlike the IMP score, the LSC17 score is thought to more reflect an aggressive phenotype of AML than the pathogenesis of AML itself.
Finally, we assessed whether prediction of prognosis is improved by incorporating our gene expression–based classification into the IPSS-R. The analysis was confined to 148 patients with MDS and complete clinical data (Table 1). Although not all samples were obtained at diagnosis, no patients received treatment other than supportive care prior to sample collection. We thus tentatively defined IPSS-R categories based on clinical information available at the time of sampling. Clinical outcomes were first compared between the EMK and IMP subgroups stratified by IPSS-R categories. As compared with the patients in the EMK subgroup, those in the IMP subgroup had a significantly higher risk of leukemic transformation in the IPSS-R low- and very-high-risk groups (Figure 5C-D; supplemental Figure 14). We next made a prognostic model that incorporated our gene expression–based classification into the IPSS-R. This combined model outperformed the IPSS-R only model in predicting overall survival and leukemic transformation (supplemental Figure 15). This indicated an additive effect of the gene expression–based classification in patient prognostication.
Discussion
Myelodysplasia is a highly heterogeneous group of myeloid neoplasms in terms of clinical, pathological, and mutational features, as separately classified into a number of distinct subtypes in the WHO classification system.10,11,21,45 It is rather unexpected that patients were separated into 2 major subgroups with enhanced EMK and IMP signatures based on gene expression profiles. It should be underscored that unlike previous studies,16-18 these 2 myelodysplasia subgroups were identified in a totally unbiased manner based solely on gene expression and without relying on the clinical outcome. Nonetheless, they had strong prognostic value, which was independent of clinical and demographic variables, including percentages of bone marrow blasts. Altered transcriptional programs may precede and predict overt morphological changes and clonal expansion. This may also be consistent with the simple distinction of myelodysplasia between 2 categories: clonal cytopenia before clonal evolution and oligoblastic myelogenous leukemias.46 Importantly, these expression subgroups were largely reproduced in an independent external cohort of patients, even though a different platform was used for gene expression profiling, indicating the robustness of the above classification.
Each subgroup had distinct clinical, genomic, and transcriptomic profiles. The EMK subgroup was associated with the ring sideroblast phenotype, SF3B1 mutation, and the strong erythroid signature. The IMP subgroup was characterized by lower platelet counts, increased marrow blasts, high-risk genomic lesions, and enhanced immature progenitor signatures. Features of the IMP subgroup as compared with the EMK subgroup provide insights into the molecular pathogenesis of disease progression in myelodysplasia. The deregulation of genes related to hematopoietic lineages suggests a severe differentiation block and/or stem cell proliferation in the IMP subgroup. The IMP subgroup also showed upregulation of various cell signaling pathways. Deregulated pathways included Notch, MAPK, phosphatidylinositol 3-kinase, and JAK-STAT signaling, of which activation is known to have a role in hematopoietic differentiation and self-renewal of stem cells.47-50 The importance of cell signaling activation is also supported by the enrichment of NRAS mutation in the IMP subgroup and by the frequent acquisition of mutations in genes encoding signaling molecules, such as NRAS, KRAS, PTPN11, and FLT3, during disease progression of myelodysplasia.42 In contrast, many pathways related to DNA repair and metabolism were downregulated in the IMP subgroup. Impaired DNA repair and damage response in high-risk MDS is consistent with a previous report28 and can accelerate leukemia development. Levels of various metabolites and genes related to metabolism have been shown to change dramatically during differentiation from quiescent hematopoietic stem cells.51,52 Global repression of genes related to metabolism in the IMP subgroup might reflect enrichment of metabolically quiescent stem/progenitor cells. Alternatively, rapid cell proliferation associated with ineffective hematopoiesis in low-risk MDS can be a basis for active metabolism in the EMK subgroup.53,54 Deregulation of several metabolic pathways at the transcriptome level was previously reported in refractory anemia.28 The expression of immunodeficiency, apoptosis, and chemokine signaling pathways, which were previously reported to be significantly deregulated in refractory anemia,28 did not differ significantly between the EMK and IMP subgroups.
These gene expression subtypes were identified from the analysis of purified CD34+ cells, but not whole BMMNCs, suggesting that their characteristic expression profiles represent intrinsic features of leukemic progenitors. Difficulty in identifying stable clusters from global gene expression profiles of unfractionated BMMNCs might be due to mature hematopoietic cells or contaminating peripheral blood cells in BMMNC samples that obscured characteristic gene expression of immature progenitors. The importance of relying on the gene expression of stem/progenitor fractions for better characterization is also underscored by the successful use of the leukemia stem cell signature to predict a prognostic model for AML.43,55 Nevertheless, the 2 subgroups could be successfully predicted on the basis of the expression levels of a small number of genes (n = 9) selected from highly upregulated genes in BMMNC samples of the IMP subgroup. The transcriptomic differences that were consistently detected in more differentiated cell populations suggested the presence of intrinsic biological processes underlying phenotypic differences. The regression model obviates the need for purification of CD34+ progenitor cells, further enhancing the clinical utility of our classification.
In conclusion, we demonstrated that myelodysplasia patients can be classified into 2 distinct clusters, the EMK and IMP subgroups, which have unique genomic, transcriptomic, and clinical features. Our results support the integration of gene expression data into prognostic models of myelodysplasia. Further studies are warranted to assess the relevance of our gene expression–based classification for newly diagnosed myelodysplasia in a prospective setting.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all patients for their participation in this study and Hiroshi Kobayashi for fruitful discussions. They deeply appreciate Elsa Bernard, Elli Papaemmanuil, and Yasuhito Nannya for their help with statistics and computation.
This work was supported by the Project for Development of Innovative Research on Cancer Therapeutics (P-DIRECT, 16cm0106501h0001), Practical Research for Innovative Cancer Control (15Ack0106014h0002, 16ck0106073h0003), and Project for Cancer Research and Therapeutic Evolution (P-CREATE, 16cm0106501h0001) from the Japan Agency for Medical Research and Development (S.O.). This work was also supported by the Japan Society for the Promotion of Science KAKENHI (15J02911 [Y. Shiozawa]; 15H05668 [K.Y.]; 22134006, 26221308, and 15H05909 [S.O.]) and by research grants from Takeda Science Foundation (S.O.), Associazione Italiana per la Ricerca sul Cancro (Special Program Molecular Clinical Oncology 5 per Mille, project 1005 [M.C.] and investigator grant 15356 [L.M.]), Fondazione Regionale Ricerca Biomedica (project 2015-0042 [M.C.]), Bloodwise, UK (A.P. and J.B.), the Swedish Cancer Society, the Research Council of Sweden (E.H.-L.), and the Uehara Memorial Foundation (K.Y.). This research used computational resources of the Human Genome Center, the Institute of Medical Science, the University of Tokyo, Japan, and the K computer provided by the RIKEN Advanced Institute for Computational Science through the High Performance Computing Infrastructure System Research project (hp160219).
Authorship
Contribution: L.M., A.G., A.P., M.K., J.B., E.H.-L., and M.C. collected and provided the specimens; Y. Shiozawa, Y. Sato, and K.Y. performed sequencing experiments; Y. Shiozawa, H.S., T.Y., Y. Shiraishi, K.C., and S.M. developed bioinformatics pipelines; Y. Shiozawa and A.S.-O. performed sequencing data analyses; A.P. and J.B. performed microarray data analyses; Y. Shiozawa, L.M., and H.M. performed clinical analysis; Y. Shiozawa, L.M., S.O., and M.C. generated the figures and tables and wrote the manuscript; M.C. and S.O. co-led the entire project; and all authors participated in discussions and interpretation of the data and results.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mario Cazzola, Department of Molecular Medicine, University of Pavia, Pavia 27100, Italy; e-mail: mario.cazzola@unipv.it; and Seishi Ogawa, Department of Pathology and Tumor Biology, Kyoto University, Kyoto 606-8501, Japan; e-mail: sogawa-tky@umin.ac.jp.
References
Author notes
Y. Shiozawa and L.M. contributed equally to this study.
M.C. and S.O. contributed equally to this study.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal