In the article beginning on page 2642 in the 14 December 2017 issue, there are errors that resulted from a single coding mistake in the construction of the logistic regression model for subgroup prediction based on the gene expression profiles of bone marrow mononuclear cells (BMMNCs). This coding error was identified after publication of the article while the authors were conducting follow-up studies. Even though the coding error affected just a single letter, it led to an erroneous gene list for the model; therefore, the results based on this gene list that needed to be corrected were extensive. However, the resulting errors are restricted to the data concerning BMMNCs and, after the analyses based on the corrected code with some modifications in the model parameter, they do not change the direction or significance of the results, interpretations, and conclusions of the article. The information about CD34+ cells in the article is unchanged.
The corrections are listed here. The errors have been corrected in the online version of the article.
Page 2642, abstract, line 13: “P = .002” should read “P < .001.”
Page 2643, second paragraph under “Clustering of gene expression data”:
In line 8 of the paragraph, “The mean area under the curve for the cross-validation was 0.95” should read “The mean classification error was 0.05.”
In the next-to-last sentence, “(Figure 1A)” should read “(Figure 1A; supplemental Methods, available on the Blood Web site).”
Before the last sentence, the following text should be inserted: “Since unfractionated BMMNCs contain mature hematopoietic cells and contaminating peripheral blood, variables in the model were selected from highly upregulated genes in the second subgroup of this training cohort. We first performed rigorous differential expression analysis using the generalized linear model likelihood ratio test.24 Elastic net logistic regression was then applied to the 30 most significantly upregulated genes in the second subgroup as compared with the first subgroup and the healthy adults.”
The last sentence should read “The parameters of the elastic net were determined as a = 0.7 and λ = 0.0176 using 10-fold cross-validation, with a mean classification error of 0.10.”
Page 2643, under “Statistical analysis”: The last sentence should read “Analyses were performed using R, versions 2.15.3 (model construction using elastic net logistic regression) and 3.0.1 (the other analyses). The R scripts are shown in supplemental Methods.”
Page 2644, Table 1: The data under the heading “Patients with BMMNC samples only (n = 114)” should be updated on the basis of the corrected classification of patients. The corrected Table 1 is shown below.
Patient characteristics
| . | Patients with CD34+ cell samples (n = 100) . | Patients with BMMNC samples only (n = 114) . | ||||
|---|---|---|---|---|---|---|
| . | First (EMK) subgroup . | Second (IMP) subgroup . | P . | First (EMK) subgroup . | Second (IMP) subgroup . | P . |
| Number of cases | 61 | 39 | 71 | 43 | — | |
| Age (y), median (range) | 69 (32-87) | 62 (30-83) | .01 | 67 (30-83) | 67 (39-91) | .46 |
| Sex (male/female) | 38/23 | 27/12 | .52 | 42/27 | 29/18 | 1 |
| Diagnosis, n (%) | ||||||
| MDS | 37 (61) | 22 (56) | — | 57 (80) | 36 (84) | — |
| MDS-SLD | 4 (6.6) | 1 (2.6) | .65 | 5 (7.0) | 1 (2.3) | .41 |
| MDS-RS-SLD | 13 (21) | 1 (2.6) | .008 | 19 (27) | 1 (2.3) | <.001 |
| MDS-MLD | 5 (8.2) | 4 (10) | .73 | 13 (18) | 17 (40) | .02 |
| MDS-RS-MLD | 5 (8.2) | 2 (5.1) | .70 | 9 (13) | 1 (2.3) | .09 |
| MDS with isolated del(5q) | 2 (3.3) | 0 (0) | .52 | 1 (1.4) | 1 (2.3) | 1 |
| MDS-EB-1 | 4 (6.6) | 5 (13) | .31 | 7 (9.9) | 6 (14) | .55 |
| MDS-EB-2 | 4 (6.6) | 9 (23) | .03 | 3 (4.2) | 9 (22) | .009 |
| MDS/MPN | 24 (39) | 6 (15) | — | 13 (18) | 1 (2.3) | — |
| CMML-1 | 19 (31) | 5 (13) | .05 | 8 (11) | 0 (0) | .02 |
| CMML-2 | 1 (1.6) | 0 (0) | 1 | 0 (0) | 0 (0) | 1 |
| MDS/MPN-RS-T | 4 (6.6) | 0 (0) | .15 | 3 (4.2) | 1 (2.3) | 1 |
| MDS/MPN-U | 0 (0) | 1 (2.6) | .39 | 2 (2.8) | 0 (0) | .53 |
| AML-MDS | 0 (0) | 11 (28) | <.001 | 1 (1.4) | 6 (14) | .01 |
| Hemoglobin (g/dL), median (range) | 10.4 (7.0-14.7) | 10.2 (7.0-14.4) | .92 | 9.8 (6.3-15.5) | 9.7 (6.8-13.9) | .43 |
| ANC (×109/L), median (range) | 3.0 (0.35-13) | 1.7 (0.40-32) | .06 | 2.1 (0.20-22) | 1.3 (0.20-11) | .002 |
| Platelet count (×109/L), median (range) | 173 (17.5-895) | 76.0 (15.0-697) | <.001 | 175 (16.0-849) | 102 (12.5-939) | .03 |
| Myeloid:erythroid ratio, median (range) | 2.0 (0.50-10) | 2.0 (0.13-10) | .86 | 2.0 (0.25-10) | 1.5 (0.20-10) | .71 |
| Bone marrow blasts (%), median (range) | 2 (0-15) | 11 (1-90) | <.001 | 2 (0-86) | 4 (0-63) | .003 |
| Bone marrow ring sideroblasts (%), median (range) | 9 (0-94) | 5 (0-78) | .20 | 13 (0-94) | 0.3 (0-81) | .08 |
| IPSS-R in the patients with MDS, n (%) | ||||||
| Very low | 0 (0) | 0 (0) | 1 | 0 (0) | 0 (0) | 1 |
| Low | 15 (25) | 3 (7.7) | .04 | 26 (37) | 10 (23) | .15 |
| Intermediate | 12 (20) | 7 (18) | 1 | 17 (24) | 8 (19) | .64 |
| High | 7 (11) | 5 (13) | 1 | 9 (13) | 10 (23) | .19 |
| Very high | 3 (4.9) | 7 (18) | .04 | 3 (4.2) | 6 (14) | .08 |
| Missing | 0 (0) | 0 (0) | 1 | 2 (2.8) | 2 (4.7) | .63 |
| Treatment during the follow-up period,* n (%) | ||||||
| Allogeneic stem cell transplantation | 3 (4.9) | 7 (18) | .04 | 4 (5.6) | 4 (9.3) | .47 |
| Cytotoxic chemotherapy | 0 (0) | 7 (18) | <.001 | 2 (2.8) | 6 (14) | .05 |
| Azacitidine | 3 (4.9) | 2 (11) | 1 | 3 (4.2) | 4 (9.3) | .42 |
| Response | 1 (33) | 0 (0) | 1 | 0 (0) | 1 (25) | 1 |
| Only supportive care | 56 (92) | 26 (67) | .003 | 63 (89) | 31 (72) | .04 |
| . | Patients with CD34+ cell samples (n = 100) . | Patients with BMMNC samples only (n = 114) . | ||||
|---|---|---|---|---|---|---|
| . | First (EMK) subgroup . | Second (IMP) subgroup . | P . | First (EMK) subgroup . | Second (IMP) subgroup . | P . |
| Number of cases | 61 | 39 | 71 | 43 | — | |
| Age (y), median (range) | 69 (32-87) | 62 (30-83) | .01 | 67 (30-83) | 67 (39-91) | .46 |
| Sex (male/female) | 38/23 | 27/12 | .52 | 42/27 | 29/18 | 1 |
| Diagnosis, n (%) | ||||||
| MDS | 37 (61) | 22 (56) | — | 57 (80) | 36 (84) | — |
| MDS-SLD | 4 (6.6) | 1 (2.6) | .65 | 5 (7.0) | 1 (2.3) | .41 |
| MDS-RS-SLD | 13 (21) | 1 (2.6) | .008 | 19 (27) | 1 (2.3) | <.001 |
| MDS-MLD | 5 (8.2) | 4 (10) | .73 | 13 (18) | 17 (40) | .02 |
| MDS-RS-MLD | 5 (8.2) | 2 (5.1) | .70 | 9 (13) | 1 (2.3) | .09 |
| MDS with isolated del(5q) | 2 (3.3) | 0 (0) | .52 | 1 (1.4) | 1 (2.3) | 1 |
| MDS-EB-1 | 4 (6.6) | 5 (13) | .31 | 7 (9.9) | 6 (14) | .55 |
| MDS-EB-2 | 4 (6.6) | 9 (23) | .03 | 3 (4.2) | 9 (22) | .009 |
| MDS/MPN | 24 (39) | 6 (15) | — | 13 (18) | 1 (2.3) | — |
| CMML-1 | 19 (31) | 5 (13) | .05 | 8 (11) | 0 (0) | .02 |
| CMML-2 | 1 (1.6) | 0 (0) | 1 | 0 (0) | 0 (0) | 1 |
| MDS/MPN-RS-T | 4 (6.6) | 0 (0) | .15 | 3 (4.2) | 1 (2.3) | 1 |
| MDS/MPN-U | 0 (0) | 1 (2.6) | .39 | 2 (2.8) | 0 (0) | .53 |
| AML-MDS | 0 (0) | 11 (28) | <.001 | 1 (1.4) | 6 (14) | .01 |
| Hemoglobin (g/dL), median (range) | 10.4 (7.0-14.7) | 10.2 (7.0-14.4) | .92 | 9.8 (6.3-15.5) | 9.7 (6.8-13.9) | .43 |
| ANC (×109/L), median (range) | 3.0 (0.35-13) | 1.7 (0.40-32) | .06 | 2.1 (0.20-22) | 1.3 (0.20-11) | .002 |
| Platelet count (×109/L), median (range) | 173 (17.5-895) | 76.0 (15.0-697) | <.001 | 175 (16.0-849) | 102 (12.5-939) | .03 |
| Myeloid:erythroid ratio, median (range) | 2.0 (0.50-10) | 2.0 (0.13-10) | .86 | 2.0 (0.25-10) | 1.5 (0.20-10) | .71 |
| Bone marrow blasts (%), median (range) | 2 (0-15) | 11 (1-90) | <.001 | 2 (0-86) | 4 (0-63) | .003 |
| Bone marrow ring sideroblasts (%), median (range) | 9 (0-94) | 5 (0-78) | .20 | 13 (0-94) | 0.3 (0-81) | .08 |
| IPSS-R in the patients with MDS, n (%) | ||||||
| Very low | 0 (0) | 0 (0) | 1 | 0 (0) | 0 (0) | 1 |
| Low | 15 (25) | 3 (7.7) | .04 | 26 (37) | 10 (23) | .15 |
| Intermediate | 12 (20) | 7 (18) | 1 | 17 (24) | 8 (19) | .64 |
| High | 7 (11) | 5 (13) | 1 | 9 (13) | 10 (23) | .19 |
| Very high | 3 (4.9) | 7 (18) | .04 | 3 (4.2) | 6 (14) | .08 |
| Missing | 0 (0) | 0 (0) | 1 | 2 (2.8) | 2 (4.7) | .63 |
| Treatment during the follow-up period,* n (%) | ||||||
| Allogeneic stem cell transplantation | 3 (4.9) | 7 (18) | .04 | 4 (5.6) | 4 (9.3) | .47 |
| Cytotoxic chemotherapy | 0 (0) | 7 (18) | <.001 | 2 (2.8) | 6 (14) | .05 |
| Azacitidine | 3 (4.9) | 2 (11) | 1 | 3 (4.2) | 4 (9.3) | .42 |
| Response | 1 (33) | 0 (0) | 1 | 0 (0) | 1 (25) | 1 |
| Only supportive care | 56 (92) | 26 (67) | .003 | 63 (89) | 31 (72) | .04 |
AML-MDS, AML with myelodysplasia-related changes; ANC, absolute neutrophil count; CMML, chronic myelomonocytic leukemia; MDS-EB, MDS with excess of blasts; MDS-MLD, MDS with multilineage dysplasia; MDS/MPN, myelodysplastic/myeloproliferative neoplasm; MDS/MPN-RS-T, MDS/MPN with ring sideroblasts and thrombocytosis; MDS/MPN-U, MDS/MPN, unclassifiable; MDS-RS-SLD, MDS with ring sideroblasts with single-lineage dysplasia; MDS-SLD, MDS with single-lineage dysplasia; MDS-RS-MLD, MDS with ring sideroblasts with multilineage dysplasia.
Some patients received various treatments (eg, cytotoxic chemotherapy followed by allogeneic stem cell transplantation). This led to the sums of patients exceeding 100%.
Page 2645: In line 6 of the legend to Figure 1, “3142 genes” should read “3141 genes.”
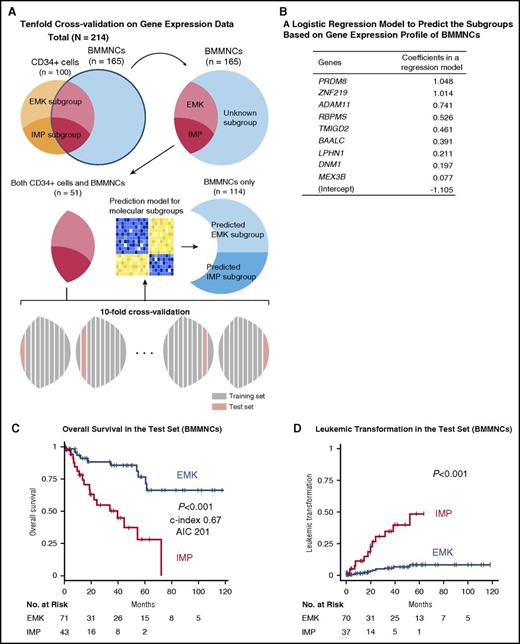
Page 2646, under “Prediction of the gene expression–based subgroups using BMMNC samples”:
In line 9 of the paragraph, “25 genes” should read “9 genes.”
In line 11, “67 (59%) and 47 (41%)” should read “71 (62%) and 43 (38%).”
In line 15, “HR, 7.2; 95% CI, 3.0-17” should read “HR, 4.5; 95% CI, 2.0-10.”
In line 17, “(HR, 12; 95% CI, 2.6-57; P = .001)” should read “(HR, 7.3; 95% CI, 2.0-26; P = .002).”
HR for death or leukemic transformation in a multivariate model
| . | HR (95% CI) . | P . |
|---|---|---|
| Patients with CD34+cell samples (n = 100) | ||
| Death | ||
| Hemoglobin (g/dL) | 0.54 (0.37-0.77) | <.001 |
| Bone marrow blast (%) | 1.05 (1.02-1.09) | .003 |
| Age (y) | 1.09 (1.02-1.16) | .009 |
| IMP vs EMK subgroup | 4.87 (1.25-19.0) | .02 |
| Leukemic transformation* | ||
| Patients with only BMMNC samples (n = 114) | ||
| Death | ||
| Bone marrow blast (%) | 1.11 (1.06-1.16) | <.001 |
| Hemoglobin (g/dL) | 0.67 (0.52-0.86) | .001 |
| IMP vs EMK subgroup | 2.90 (1.10-7.62) | .03 |
| Absolute neutrophil count (×109/L) | 1.12 (1.00-1.26) | .05 |
| Cytogenetic abnormalities | 2.16 (0.92-5.07) | .08 |
| Platelet (×109/L) | 0.996 (0.992-1.001) | .12 |
| Leukemic transformation | ||
| Bone marrow blast (%) | 1.14 (1.08-1.21) | <.001 |
| IMP vs EMK subgroup | 5.86 (1.59-21.6) | .008 |
| Hemoglobin (g/dL) | 0.80 (0.66-0.98) | .03 |
| . | HR (95% CI) . | P . |
|---|---|---|
| Patients with CD34+cell samples (n = 100) | ||
| Death | ||
| Hemoglobin (g/dL) | 0.54 (0.37-0.77) | <.001 |
| Bone marrow blast (%) | 1.05 (1.02-1.09) | .003 |
| Age (y) | 1.09 (1.02-1.16) | .009 |
| IMP vs EMK subgroup | 4.87 (1.25-19.0) | .02 |
| Leukemic transformation* | ||
| Patients with only BMMNC samples (n = 114) | ||
| Death | ||
| Bone marrow blast (%) | 1.11 (1.06-1.16) | <.001 |
| Hemoglobin (g/dL) | 0.67 (0.52-0.86) | .001 |
| IMP vs EMK subgroup | 2.90 (1.10-7.62) | .03 |
| Absolute neutrophil count (×109/L) | 1.12 (1.00-1.26) | .05 |
| Cytogenetic abnormalities | 2.16 (0.92-5.07) | .08 |
| Platelet (×109/L) | 0.996 (0.992-1.001) | .12 |
| Leukemic transformation | ||
| Bone marrow blast (%) | 1.14 (1.08-1.21) | <.001 |
| IMP vs EMK subgroup | 5.86 (1.59-21.6) | .008 |
| Hemoglobin (g/dL) | 0.80 (0.66-0.98) | .03 |
Failure in convergence due to no leukemic transformation in the EMK subgroup.
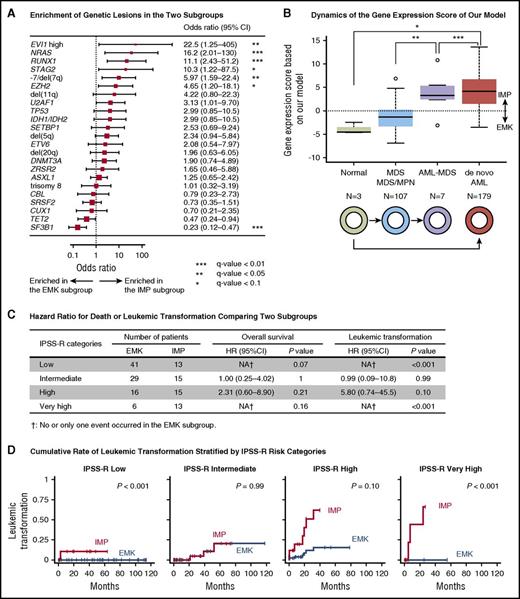
Page 2646, second paragraph under “Genetic alterations in the gene expression subgroups”:
In line 2 of the paragraph, “P = .02” should read “P = .003.”
In lines 4 and 5, “SF3B1 and TET2 mutations were more frequent in the EMK subgroup (q-value < 0.05; Figure 3C)” should read “SF3B1 mutation was more frequent in the EMK subgroup (q-value < 0.05; Figure 5A).”
In lines 8 and 9, “(odds ratio, 4.7; 95% CI, 2.5-6.9; P = .004)” should read “(odds ratio, 5.2; 95% CI, 1.9-14; P = .003).”
Validation of the prognostic significance of the subgroups in the test set for bone marrow CD34+cell samples. (A) Heatmap showing expression levels of 68 genes in the regression model for 100 patients with bone marrow CD34+ cell samples. Each row represents 1 gene, and each column represents 1 sample. Gene expression–based subgroups, WHO subtypes, and patients’ prognosis are shown below the heatmap. (B) A heatmap shows expression levels of 68 genes in the regression model for an independent cohort of 183 patients with MDS. (C) Rates of overall survival in the 2 subgroups for the test set of bone marrow CD34+ cell samples. (D) Cumulative incidence of leukemic transformation in the 2 subgroups for the test cohort of bone marrow CD341 cell samples.
Validation of the prognostic significance of the subgroups in the test set for bone marrow CD34+cell samples. (A) Heatmap showing expression levels of 68 genes in the regression model for 100 patients with bone marrow CD34+ cell samples. Each row represents 1 gene, and each column represents 1 sample. Gene expression–based subgroups, WHO subtypes, and patients’ prognosis are shown below the heatmap. (B) A heatmap shows expression levels of 68 genes in the regression model for an independent cohort of 183 patients with MDS. (C) Rates of overall survival in the 2 subgroups for the test set of bone marrow CD34+ cell samples. (D) Cumulative incidence of leukemic transformation in the 2 subgroups for the test cohort of bone marrow CD341 cell samples.
Page 2647, left column:
In line 3, “TP53” should read “RUNX1.”
In lines 5 and 6 of the first paragraph under “Comparison between our gene expression–based classification and other prognostic models, “Only one gene in the LSC17 score, NYNRIN, overlapped with our regression model” should read “None of the genes included in the LSC17 score overlapped with our regression model.”
Page 2647, right column:
Page 2648, Figure 3: The heatmaps in panels A-B should be modified to show expression levels of the corrected genes. The corrected Figure 3 is shown below.
Page 2648, right column: In the last line, “low-risk group” should read “low- and very–high-risk groups.”
Page 2649, Figure 4: (B) The gene list and the corresponding coefficients should be corrected. (C) The survival curve should be changed on the basis of the corrected classification of patients. (D) The curve should be modified on the basis of the modified classification. The corrected Figure 4 is shown below.
Prognostic significance of the subgroups based on gene expression profiles of BMMNCs. (A) Schematic depicts how to construct a logistic regression model to predict the subgroups using 10-fold cross-validation on gene expression data. (B) A table shows gene names and their coefficients in a logistic regression model to predict the subgroups based on gene expression profile of BMMNCs. (C) Rates of overall survival in the 2 subgroups for the test set of BMMNC samples. (D) Cumulative incidence of leukemic transformation in the 2 subgroups for the test set of BMMNC samples. The patients who had already developed leukemia at the time of sampling were removed from the analysis.
Prognostic significance of the subgroups based on gene expression profiles of BMMNCs. (A) Schematic depicts how to construct a logistic regression model to predict the subgroups using 10-fold cross-validation on gene expression data. (B) A table shows gene names and their coefficients in a logistic regression model to predict the subgroups based on gene expression profile of BMMNCs. (C) Rates of overall survival in the 2 subgroups for the test set of BMMNC samples. (D) Cumulative incidence of leukemic transformation in the 2 subgroups for the test set of BMMNC samples. The patients who had already developed leukemia at the time of sampling were removed from the analysis.
Page 2650, Figure 5: (A) The forest plot should be corrected on the basis of the mutation status of the reclassified patients. (B) The bar plot should be revised to show the gene expression scores based on the corrected regression model. (C) The data should be corrected to show the statistics calculated on the basis of the corrected classification of patients. (D) The curves should be modified according to the corrected classification. In the figure legend, in line 3, “25 genes” should read “9 genes.” In lines 5 and 6, “*P < .05 (Bonferroni adjusted); ***P < .001 (Bonferroni adjusted).” should read “*P < .05 (Bonferroni adjusted); **P < .01 (Bonferroni adjusted); ***P < .001 (Bonferroni adjusted).” The corrected Figure 5 is shown below.
Comparison between our gene expression–based classification and other prognostic models. (A) A forest plot of odds ratios for detecting genetic lesions in the IMP subgroup. P values are based on Fisher’s exact test. The square sizes were inversely proportional to the confidence intervals of the estimated odds ratio. (B) A boxplot shows distribution of log odds ratio for the predicted IMP subgroup that was calculated based on our regression model using expression levels of 9 genes. Boxes correspond to healthy adults (n = 3), patients with MDS or myelodysplastic/myeloproliferative neoplasm (MDS/MPN; n = 107), those with AML with myelodysplasia-related changes (AML-MDS; n = 7), and those in a published external AML cohort from The Cancer Genome Atlas (n = 179). *P < .05 (Bonferroni adjusted); **P < .01 (Bonferroni adjusted); ***P < .001 (Bonferroni adjusted). (C) HR of death or leukemic transformation comparing the EMK and IMP subgroups. Number of patients in the EMK and IMP subgroup, HR, its 95% CI, and P values are shown for each IPSS-R risk category. (D) Cumulative incidence of leukemic transformation in MDS patients stratified by categories of IPSS-R.
Comparison between our gene expression–based classification and other prognostic models. (A) A forest plot of odds ratios for detecting genetic lesions in the IMP subgroup. P values are based on Fisher’s exact test. The square sizes were inversely proportional to the confidence intervals of the estimated odds ratio. (B) A boxplot shows distribution of log odds ratio for the predicted IMP subgroup that was calculated based on our regression model using expression levels of 9 genes. Boxes correspond to healthy adults (n = 3), patients with MDS or myelodysplastic/myeloproliferative neoplasm (MDS/MPN; n = 107), those with AML with myelodysplasia-related changes (AML-MDS; n = 7), and those in a published external AML cohort from The Cancer Genome Atlas (n = 179). *P < .05 (Bonferroni adjusted); **P < .01 (Bonferroni adjusted); ***P < .001 (Bonferroni adjusted). (C) HR of death or leukemic transformation comparing the EMK and IMP subgroups. Number of patients in the EMK and IMP subgroup, HR, its 95% CI, and P values are shown for each IPSS-R risk category. (D) Cumulative incidence of leukemic transformation in MDS patients stratified by categories of IPSS-R.
Page 2651, right column:
In lines 7 to 10, “Nevertheless, the 2 subgroups could be successfully predicted on the basis of the expression levels of a small number of genes (n = 25) in BMMNCs. Deregulation of these genes was thought to persist in more differentiated cells” should read “Nevertheless, the 2 subgroups could be successfully predicted on the basis of the expression levels of a small number of genes (n = 9) selected from highly upregulated genes in BMMNC samples of the IMP subgroup.”
The following sentence should be added at the end of the first paragraph of “Acknowledgments”: “They deeply appreciate Elsa Bernard, Elli Papaemmanuil, and Yasuhito Nannya for their help with statistics and computation.”
In the online data supplement, there were errors in the following items: supplemental Figures 7, 8, 10, 12, 14, and 15 and supplemental Table 4. The errors have been corrected, and a supplemental methods section has been added.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal