Key Points
CHD7 interacts with CBFβ-SMMHC through RUNX1 and modulates their gene expression regulation.
CHD7 is important for CBFB-MYH11 leukemogenesis in the mouse model.
Abstract
Inversion of chromosome 16 is a consistent finding in patients with acute myeloid leukemia subtype M4 with eosinophilia, which generates a CBFB-MYH11 fusion gene. Previous studies showed that the interaction between CBFβ-smooth muscle myosin heavy chain (SMMHC; encoded by CBFB-MYH11) and RUNX1 plays a critical role in the pathogenesis of this leukemia. Recently, it was shown that chromodomain helicase DNA-binding protein-7 (CHD7) interacts with RUNX1 and suppresses RUNX1-induced expansion of hematopoietic stem and progenitor cells. These results suggest that CHD7 is also critical for CBFB-MYH11–induced leukemogenesis. To test this hypothesis, we generated Chd7f/fMx1-CreCbfb+/56M mice, which expressed the Cbfb-MYH11 fusion gene and deactivated Chd7 in hematopoietic cells upon inducing Cre with polyinosinic-polycytidylic acid. The Lin–Sca1–c-Kit+ (LK) population was significantly lower in Chd7f/fMx1-CreCbfb+/56M mice than in Mx1-CreCbfb+/56M mice. In addition, there were fewer 5-bromo-2′-deoxyuridine–positive cells in the LK population in Chd7f/fMx1-CreCbfb+/56M mice, and genes associated with cell cycle, cell growth, and proliferation were differentially expressed between Chd7f/fMx1-CreCbfb+/56M and Mx1-CreCbfb+/56M leukemic cells. In vitro studies showed that CHD7 interacted with CBFβ-SMMHC through RUNX1 and that CHD7 enhanced transcriptional activity of RUNX1 and CBFβ-SMMHC on Csf1r, a RUNX1 target gene. Moreover, RNA sequencing of c-Kit+ cells showed that CHD7 functions mostly through altering the expression of RUNX1 target genes. Most importantly, Chd7 deficiency delayed Cbfb-MYH11–induced leukemia in both primary and transplanted mice. These data indicate that Chd7 is important for Cbfb-MYH11–induced leukemogenesis by facilitating RUNX1 regulation of transcription and cellular proliferation.
Introduction
Inversion of chromosome 16 (inv(16)) is associated with acute myeloid leukemia (AML) subtype M4 with eosinophilia and generates a CBFB-MYH11 fusion gene. CBFB-MYH11 encodes the CBFβ-smooth muscle myosin heavy chain (SMMHC) chimeric protein, which makes up most of CBFβ (165 amino acids of a total of 187 amino acids) and the C-terminal coiled-coil domain of the SMMHC.1-3 Mouse model studies have shown that CBFβ-SMMHC is necessary but not sufficient for leukemogenesis.4,5
CBFβ binds with RUNX1 (also known as AML1), a key hematopoietic transcription factor, leading to stabilization of the RUNX1-DNA interaction and enhanced gene expression regulation.6,7 CBFβ-SMMHC is thought to initiate leukemogenesis by blocking normal hematopoietic differentiation through inhibition of RUNX1. In vitro studies have shown that CBFβ-SMMHC may serve as a transcriptional repressor and that CBFβ-SMMHC may sequester RUNX1 in the cytoplasm.7-9 In vivo, Cbfb-MYH11 heterozygous knockin mice (Cbfb+/MYH11) have a complete block in definitive hematopoiesis and severe central nervous system hemorrhaging at embryonic day 12.5 (E12.5), which contributes to lethality by E13.5.10 This phenotype is very similar to that of Runx1 null (Runx1−/−) or Cbfb null (Cbfb−/−) mice,11-15 suggesting that CBFβ-SMMHC acts as a dominant repressor of RUNX1 and CBFβ functions during embryogenesis.
To more directly test whether RUNX1 is important for leukemogenesis by CBFβ-SMMHC, we generated mice expressing Cbfb-MYH11 but having reduced RUNX1 activity. The results showed that RUNX1 activity is required for Cbfb-MYH11–induced differentiation defects during both primitive and definitive hematopoiesis. Importantly, we found that insufficient RUNX1 activity delayed Cbfb-MYH11–induced leukemia.16 Chromatin immunoprecipitation sequencing experiments revealed that in the CBFB-MYH11–expressing ME-1 cells, 89% of CBFβ-SMMHC binding regions co-localize with RUNX1 binding sites.17 These results indicate that RUNX1 activity is required for CBFB-MYH11–induced leukemogenesis. Thus, molecules that modulate RUNX1 activity may also be important for CBFB-MYH11–induced leukemogenesis.
Chromodomain helicase DNA-binding protein-7 (CHD7) is a chromatin remodeling factor that regulates transcription in an adenosine triphosphate (ATP)–dependent manner18 and also in a cell type and tissue-specific manner.19 De novo heterozygous mutations of CHD7 cause the inherited CHARGE syndrome and potentially a subgroup of Kallmann syndrome.18,20,21 Mutations and copy number variations of CHD7 have been found in hematologic, lung, renal, gastrointestinal, gynecologic, and central nervous system cancers.22 Recent results showed that CHD7 interacts with RUNX1, and hematopoietic-specific Chd7 deficiency in mice with Vav1-Cre resulted in myeloid lineage expansion and increased granulocytes and/or monocytes, as well as increased numbers of Lin–Sca1+c-Kit+ (LSK) cells,23 which are similar to hematopoietic defects in Runx1-deficient mice,24 suggesting that CHD7 is an important modulator of the activity of RUNX1. Because our previous results have shown that RUNX1 activity is required for CBFB-MYH11–induced leukemogenesis,16 Chd7 may also be important for Cbfb-MYH11–induced functional changes in vivo, including leukemogenesis.
To test this hypothesis, we generated Cbfb-MYH11 knockin mice with homozygous Chd7 deletion. We found that Chd7 deficiency led to altered transcriptional activity of the RUNX1/CBFβ-SMMHC complex, inhibition of the proliferation ability of the Cbfb-MYH11– expressing leukemic cells, and delayed leukemogenesis by Cbfb-MYH11. These results indicate that Chd7 is important for Cbfb-MYH11–induced leukemogenesis and may be a good target for developing novel treatments of inv(16) leukemia.
Methods
Animals and plasmids
All animals used in this study were approved by the National Human Genome Research Institute Animal Care and Use Committee, and all the procedures performed followed relevant National Institutes of Health guidelines and regulations. Cbfb-MYH11 conditional knockin (Cbfb+/56M)25 , Mx1-Cre mice26 , and Chd7 conditional knockout (Chd7f/f) mice23 have been previously described. All mice were genotyped by polymerase chain reaction (PCR) with gene-specific primers (sequences available upon request) using tail-snip DNA prepared with DNeasy Blood and Tissue Kit (Qiagen). Eight- to 12-week-old mice and their littermate controls were injected intraperitoneally with 250 mg of poly(I:C) (pIpC; InvivoGen) to induce the expression of Cbfb-MYH11 and/or knockout of Chd7 every other day for 3 doses. To accelerate leukemia development, these mice were treated with N-ethyl-N-nitrosourea (ENU) (100 mg/kg) 2 weeks before pIpC injection as previously described.4 All mice were observed for leukemia development for 12 months.
Flag-tagged PCMV6-CHD7 and Flag-tagged MigR1-CHD7 plasmids were gifts from Peter Scambler (Institute of Child Health, University College, London, London, United Kingdom). RUNX1 cDNA was cloned into PCDNA3.1(+) and PEGFP-C1 vector respectively. CBFB, and CBFB-MYH11 cDNAs were cloned in to PGEM vector (Promega).
Assays
Peripheral blood cells, spleen and bone marrow cells from mice were isolated and stained as previously described for flow cytometry assay.16 See supplemental Methods, available on the Blood Web site, for details regarding the antibodies used in this study. Cell proliferation was determined by BrdU incorporation assay with a BrdU Flow kit (BD Biosciences), and Cell apoptosis was determined by Annexin V Apoptosis Detection Kit (BD Biosciences) according to the manufacturer's instruction. Annexin V+/7−AAD− cells were defined as apoptotic cells.
Coimmunoprecipitation and Western blot analysis were performed with standard protocols. Nuclear extracts were prepared from 293T cells which were transiently transfected with the indicated plasmids as previously described.27 Immunofluorescence staining of transfected 293T cells and Cbfb-MYH11 leukemic cells from mice were performed with standard protocols. The CSF1R promoter reporter assay was performed as previously described.28
Quantitative PCR was performed by using Power SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer's instructions. Chd7 unexcised primers were used to detect unexcised Chd7 flox allele; Exon5/6 primers were used to detect unexcised Cbfb-MYH11 flox allele. Genomic control primers were used to as internal control for genomic DNA. Primers are listed in Supplemental Table 1. RNA-sequencing (RNA-seq) was performed on messenger RNA (mRNA) isolated from c-Kit+ bone marrow cells of mice treated with poly(I:C) for two weeks. The RNA-seq data set has been deposited at Gene Expression Omnibus (accession number GSE102388). Microarray analysis was performed on mRNA isolated from c-Kit1 leukemic cells. The microarray data set has been deposited to Gene Expression Omnibus (accession number GSE103367). Details on all of the above procedures and data analysis are provided in supplemental Methods.
Statistical analysis
Data were analyzed by using GraphPad Prism. Results are expressed as mean ± standard error of the mean. Differences between 2 groups were tested with a Student t test. The survival times of mice were analyzed with the Kaplan-Meier method and log-rank test. A value of P < .05 was considered statistically significant.
Results
Generation of Chd7f/fMx1-CreCbfb+/56M mice
To determine the functional significance of Chd7 in Cbfb-MYH11–induced leukemia, we crossed Cre-based conditional Cbfb-MYH11 knockin mice (Mx1-CreCbfb+/56M)25 with Cre-based conditional Chd7 knockout mice (Chd7f/f)23 to generate Chd7f/fMx1-CreCbfb+/56M mice. These mice would express Cbfb-MYH11 but not Chd7 after poly(I:C) treatment to induce the expression of Cre recombinase. The excision efficiency in the bone marrow cells 4 weeks after poly(I:C) treatment was nearly complete for both the Cbfb-MYH11 and Chd7 floxed alleles (supplemental Figure 1A). Moreover, the Cbfb-MYH11 or the Chd7 flox allele was completely excised in >95% of the colonies of methylcellulose-cultured bone marrow cells (supplemental Figure 1B). These results suggested high induction efficiency of Cbfb-MYH11 expression and Chd7 deletion treatment with poly(I:C).
Chd7 deficiency delays Cbfb-MYH11– induced leukemia
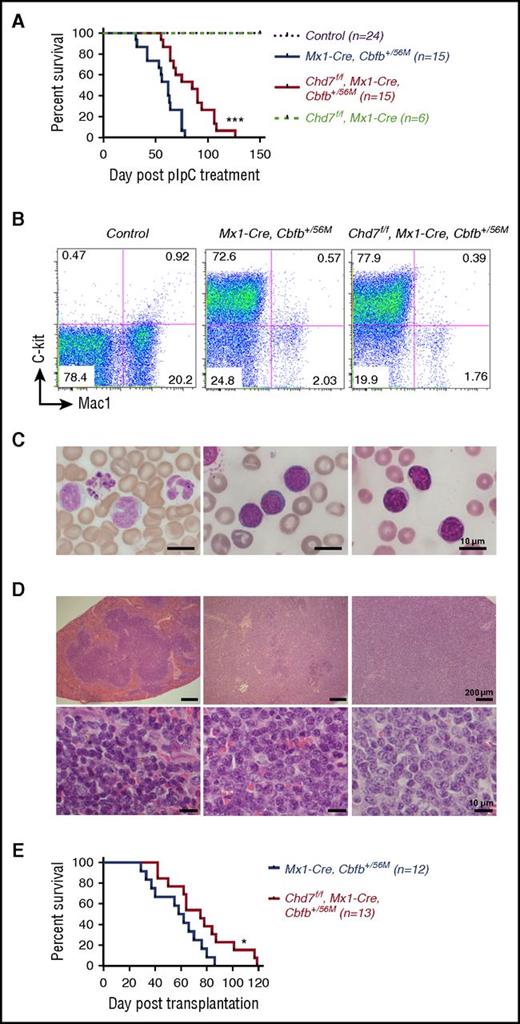
We treated adult Mx1-CreCbfb+/56M (n = 15), Chd7f/fMx1-CreCbfb+/56M (n = 15), Chd7f/fMx1-Cre (n = 6), and their littermate control mice (wild-type; Mx1-Cre or other genotypes without Mx1-Cre, n = 24) with low-dose (100 mg/kg) ENU to induce additional mutations that would accelerate leukemia development as shown previously,4 followed by poly(I:C) treatment 2 weeks later. As expected, Mx1-CreCbfb+/56M mice succumbed to AML with a median survival of 62 days (Figure 1A). Chd7f/fMx1-CreCbfb+/56M mice also developed AML (Figure 1A) but with a significantly longer survival time (85 days; P < .001). No leukemia was observed in the Chd7f/fMx1-Cre mice or the control mice (Figure 1A).
Chd7 deficiency delays Cbfb-MYH11–induced leukemia. (A-D) Mice were treated with ENU to induce mutations and then with poly(I:C) to induce the expression of Cbfb-MYH11 and/or Chd7 deficiency, and leukemia development in these mice was monitored. (A) Kaplan-Meier survival curves of mice with indicated genotypes during a 5-month observation of leukemia development. (B) Representative FACS plots of peripheral blood cells stained with c-Kit and Mac1. (C) Representative Wright-Giemsa–stained peripheral blood smears. (D) Representative hematoxylin and eosin–stained spleen sections (upper panel, ×50; bottom panel, ×400) from control, end-stage Mx1-CreCbfb+/56M, and Chd7f/fMx1-CreCbfb+/56M mice. (E) Kaplan-Meier survival curves of mice transplanted with 1 million spleen cells from end-stage Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M mice. *P < .05; ***P < .001, each comparing the Chd7f/fMx1-CreCbfb+/56M group with the Mx1-CreCbfb+/56M group.
Chd7 deficiency delays Cbfb-MYH11–induced leukemia. (A-D) Mice were treated with ENU to induce mutations and then with poly(I:C) to induce the expression of Cbfb-MYH11 and/or Chd7 deficiency, and leukemia development in these mice was monitored. (A) Kaplan-Meier survival curves of mice with indicated genotypes during a 5-month observation of leukemia development. (B) Representative FACS plots of peripheral blood cells stained with c-Kit and Mac1. (C) Representative Wright-Giemsa–stained peripheral blood smears. (D) Representative hematoxylin and eosin–stained spleen sections (upper panel, ×50; bottom panel, ×400) from control, end-stage Mx1-CreCbfb+/56M, and Chd7f/fMx1-CreCbfb+/56M mice. (E) Kaplan-Meier survival curves of mice transplanted with 1 million spleen cells from end-stage Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M mice. *P < .05; ***P < .001, each comparing the Chd7f/fMx1-CreCbfb+/56M group with the Mx1-CreCbfb+/56M group.
At the end stage, both Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M mice showed marked leukocytosis, severe thrombocytopenia, and progressive anemia (supplemental Figure 2A) and had a large c-Kit+ population in the peripheral blood (Figure 1B; supplemental Figure 2C). Morphologic examination of the peripheral blood showed the presence of many blast cells (Figure 1C). Both groups were characterized by marked splenomegaly, with infiltration of c-Kit+ blasts and a decrease in mature hematopoietic cells (Figure 1D; supplemental Figure 2B-C). Similar results were observed in the bone marrow, lungs, and liver (supplemental Figure 2C-D). All together, these results suggested that Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M mice developed a similar type of leukemia.
To exclude the possibility that the delayed leukemia development in Chd7f/fMx1-CreCbfb+/56M mice was the result of inefficient excision of the Chd7 or the Cbfb-MYH11 flox allele, quantitative PCR was performed to determine the excision efficiency of Chd7 and Cbfb-MYH11 flox alleles in c-Kit+ leukemic cells in end-stage mice. The data confirmed that the excisions were complete (supplemental Figure 3A).
To further determine whether Chd7 deficiency delays the development of leukemia, we performed transplantations with spleen cells isolated from end-stage leukemic mice. Even though all the recipients transplanted with cells from Mx1-CreCbfb+/56M or Chd7f/fMx1-CreCbfb+/56M mice developed leukemia, recipients transplanted with cells from Chd7f/fMx1-CreCbfb+/56M leukemic mice had a longer survival than those transplanted with cells from Mx1-CreCbfb+/56M mice (median survival, 75 vs 60 days; P < .05; Figure 1E). Again, the excisions of Chd7 allele and Cbfb-MYH11 flox alleles were complete in leukemic cells (supplemental Figure 3B). To exclude the possibility that the delayed development of leukemia in recipients receiving cells from Chd7f/fMx1-CreCbfb+/56M mice was the result of lower engraftment capacity of the cells, we performed a homing assay, and the results showed no difference between leukemic cells from Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M mice in terms of percentage of donor-derived cells in the spleen and bone marrow of the recipients (supplemental Figure 4). Taken together, these results suggest that Chd7-deficient Cbfb-MYH11 leukemic cells are transplantable and that Chd7 deficiency delays Cbfb-MYH11–induced leukemia.
Chd7 deficiency delays leukemia initiation induced by Cbfb-MYH11
As we showed previously, Cbfb-MYH11 expression in adult mice causes definitive hematopoiesis defects before leukemic transformation, and these defects include an increase of lineage negative (Lin–) cells and Lin–c-Kit+Sca1– (LK) cells in the bone marrow.25 More recently, we showed that Runx1 insufficiency rescued the increase of Lin– cells by Cbfb-MYH11,16 which may be associated with the ability of Runx1 deficiency to delay leukemia induced by Cbfb-MYH11. We were therefore interested in determining whether Chd7 deficiency had a similar effect.
At 2 weeks after poly(I:C) treatment (before leukemia started), examination of bone marrow LK, LSK, granulocyte-macrophage progenitor (GMP; Lin–Sca1+c-Kit+CD34+FcγII/IIIhigh), common myeloid progenitor (CMP; Lin–Sca1+c-Kit+CD34+FcγII/III+), and megakaryocyte-erythroid progenitor (MEP; Lin–Sca1+c-Kit+CD34–FcγII/III–) populations (supplemental Figure 5A-B), in vitro colony-forming ability (supplemental Figure 5C), and the proliferation ability of the LK and LSK populations (supplemental Figure 5D) demonstrated a slight difference between wild-type and Chd7f/fMx1-Cre mice, suggesting that CHD7 has limited effect on hematopoiesis. In addition, there was no difference between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M mice in terms of bone marrow LK and LSK populations (supplemental Figure 5A-B). Interestingly, within the LK population, unlike control and Chd7f/fMx1-Cre mice, an abnormal myeloid progenitor (AMP) population (Lin–c-Kit+Sca1–CD34–FcγRII/III+), which has a CMP-like differentiation potential and the ability to induce leukemia,16,25 accumulated in both Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M mice. The AMP population had similar percentage (supplemental Figure 5A-B) and morphology (supplemental Figure 5E) in both groups of mice. Altogether, the above results suggest that Chd7 deficiency does not affect the differentiation block induced by Cbfb-MYH11. Interestingly, more than 95% of c-Kit+ leukemic cells in both Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M leukemic mice are CD34– FcγRII/III+, similar to AMP, suggesting that the leukemia cells arose from this AMP population (supplemental Figure 5F). In conclusion, these results suggested that Chd7 deficiency has no effect on Cbfb-MYH11–induced hematopoietic defects before leukemia transformation.
The percentage of c-Kit+Sca1– cells in the peripheral blood was significantly increased in both Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M mice compared with control mice (Figure 2A) by 3 weeks after ENU and poly(I:C) treatments, indicating that leukemia was developing in these mice. However, the percentage of the c-Kit+Sca1– cells in peripheral blood was significantly lower in Chd7f/fMx1-CreCbfb+/56M mice than in Mx1-CreCbfb+/56M mice (Figure 2A). In the bone marrow, Chd7f/fMx1-CreCbfb+/56M mice also had a lower percentage of the Lin– LK population and the AMP population when compared with Mx1-CreCbfb+/56M mice (Figure 2B-C). Conversely, the LSK population was not significantly different between these 2 groups of mice (Figure 2B-C). These results suggest that Chd7 deficiency slows the expansion of the AMP population and the initiation of leukemia.
Chd7 deficiency delays leukemia initiation induced by Cbfb-MYH11. (A-C) The indicated groups of mice were treated with ENU to induce additional mutations and with poly(I:C) to induce the expression of Cbfb-MYH11 and/or Chd7 deficiency. Three weeks after poly(I:C) treatment, mice were euthanized and flow cytometry assays were performed. (A) Leukemia development monitored by measuring the c-Kit+/Sca1– population in peripheral blood (PB). (B) Representative FACS plots of bone marrow cells gated on single cells (upper plots), Lin– cells (middle plots), and LK cells (bottom plots). (C) Bar graph showing the percentages (mean ± standard error of the mean [SEM]) of Lin–, LK, LSK, AMP, CMP, GMP, and MEP compartments in the bone marrow of mice of the indicated genotypes. *P < .05; **P < .01; ***P < .001; ****P < .0001, each compared with the control group except for (#), for which the comparisons were between the Chd7f/fMx1-CreCbfb+/56M group and the Mx1-CreCbfb+/56M group.
Chd7 deficiency delays leukemia initiation induced by Cbfb-MYH11. (A-C) The indicated groups of mice were treated with ENU to induce additional mutations and with poly(I:C) to induce the expression of Cbfb-MYH11 and/or Chd7 deficiency. Three weeks after poly(I:C) treatment, mice were euthanized and flow cytometry assays were performed. (A) Leukemia development monitored by measuring the c-Kit+/Sca1– population in peripheral blood (PB). (B) Representative FACS plots of bone marrow cells gated on single cells (upper plots), Lin– cells (middle plots), and LK cells (bottom plots). (C) Bar graph showing the percentages (mean ± standard error of the mean [SEM]) of Lin–, LK, LSK, AMP, CMP, GMP, and MEP compartments in the bone marrow of mice of the indicated genotypes. *P < .05; **P < .01; ***P < .001; ****P < .0001, each compared with the control group except for (#), for which the comparisons were between the Chd7f/fMx1-CreCbfb+/56M group and the Mx1-CreCbfb+/56M group.
Chd7 deficiency inhibits proliferation of Cbfb-MYH11 Lin– and LK populations
We then investigated the possibility that CHD7 regulates proliferation and apoptosis of the preleukemic population. BrdU incorporation assay was performed and showed that there were significantly fewer BrdU+ of the Lin– and LK populations in Chd7f/fMx1-CreCbfb+/56M mice than in Mx1-CreCbfb+/56M mice (Figure 3A-D). Conversely, no significant difference in apoptosis was observed between these 2 groups of mice (supplemental Figure 6). These results suggested that Chd7 deficiency inhibits proliferation of Lin– cells and LK cells, which contribute to the delayed leukemia initiation induced by the Cbfb-MYH11 oncogene.
Chd7 deficiency inhibits proliferation of Lin–and LK populations in Cbfb-MYH11 mice. (A-D) The indicated groups of mice were treated with ENU to induce additional mutations and with poly(I:C) to induce the expression of Cbfb-MYH11 and/or Chd7 deficiency. Three weeks after poly(I:C) treatment, mice were euthanized after being treated with BrdU for 1 hour, and a BrdU incorporation assay was performed. (A-B) Representative FACS plots of cells stained for 7-aminoactinomycin D (7-AAD) and BrdU that were gated on (A) Lin– population and (B) LK population. (C-D) The percentages of BrdU+ cells in (C) Lin– or (D) LK cell population are shown (mean ± SEM). Three mice each for control and the Chd7f/fMx1-Cre groups and 6 mice each for Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M groups were used in the experiments. *P < .05.
Chd7 deficiency inhibits proliferation of Lin–and LK populations in Cbfb-MYH11 mice. (A-D) The indicated groups of mice were treated with ENU to induce additional mutations and with poly(I:C) to induce the expression of Cbfb-MYH11 and/or Chd7 deficiency. Three weeks after poly(I:C) treatment, mice were euthanized after being treated with BrdU for 1 hour, and a BrdU incorporation assay was performed. (A-B) Representative FACS plots of cells stained for 7-aminoactinomycin D (7-AAD) and BrdU that were gated on (A) Lin– population and (B) LK population. (C-D) The percentages of BrdU+ cells in (C) Lin– or (D) LK cell population are shown (mean ± SEM). Three mice each for control and the Chd7f/fMx1-Cre groups and 6 mice each for Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M groups were used in the experiments. *P < .05.
CHD7 interacts with RUNX1/CBFβ and RUNX1/CBFβ-SMMHC complexes
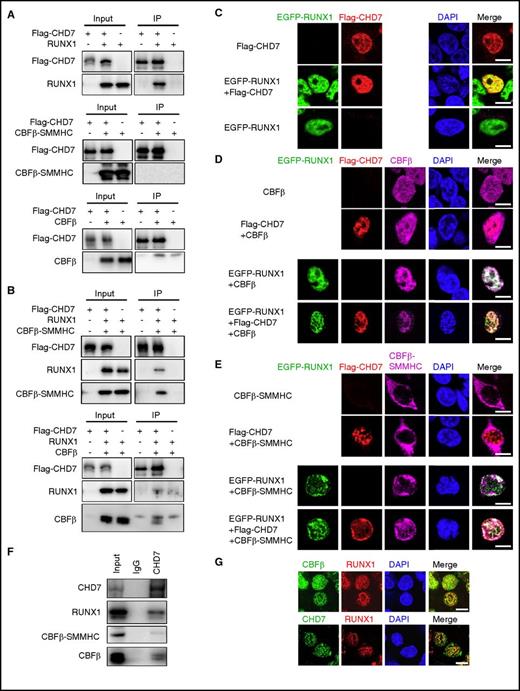
To further explore the mechanism of delayed leukemogenesis by Cbfb-MYH11 in the Chd7-deleted mice, we determined whether CHD7 interacted with CBFβ-SMMHC. Co-immunoprecipitation assay showed that, as expected, CHD7 interacted with RUNX1 (Figure 4A). Although CHD7 was not able to interact with CBFβ or CBFβ-SMMHC directly (Figure 4A), in the presence of RUNX1, CHD7 could immunoprecipitate CBFβ or CBFβ-SMMHC (Figure 4B). In addition, immunofluorescence microscopy assay showed that CHD7 was co-localized with RUNX1 in the nucleus when 293T cells were transfected with these 2 proteins together (Figure 4C). But CHD7 did not co-localize with CBFβ or CBFβ-SMMHC, suggesting that CHD7 did not directly interact with CBFβ or CBFβ-SMMHC in the transfected 293T cells (Figure 4D-E). However, when CHD7, RUNX1, and CBFβ or CBFβ-SMMHC were cotransfected, CBFβ and CBFβ-SMMHC localized to the nuclei, and there were clear co-localization signals among CHD7, RUNX1, and CBFβ and among CHD7, RUNX1, and CBFβ-SMMHC (Figure 4D-E). The interaction among CHD7, RUNX1, and CBFβ/CBFβ-SMMHC was also confirmed with endogenous proteins. As shown in Figure 4F, CHD7 could immunoprecipitate RUNX1, CBFβ, and CBFβ-SMMHC in the CBFβ-SMMHC–expressing cell line ME-1.29 In Figure 4G, clear co-localization signals were observed between RUNX1 and CBFβ/CBFβ-SMMHC (upper panel) and between RUNX1 and CHD7 (lower panel) in primary leukemia cells isolated from the Mx1-CreCbfb+/56M leukemic mice. Taken together, these results suggest that CHD7 assembles in the same complex with RUNX1/CBFβ or RUNX1/CBFβ-SMMHC in a RUNX1-dependent manner.
CHD7 interacts with RUNX1, RUNX1/CBFβ, and RUNX1/CBFβ-SMMHC complex. (A-B) 293T cells were transfected with indicated plasmids, and co-immunoprecipitation assays were performed to detect the interaction among these proteins. Flag-tagged CHD7 was immunoprecipitated with anti-Flag M2 beads, and western blot was performed with the indicated antibodies. (A) CHD7 pulls down RUNX1 but not CBFβ or CBFβ-SMMHC. (B) CHD7 pulls down CBFβ and CBFβ-SMMHC in the presence of RUNX1. (C-E) 293T cells were transfected with the indicated plasmids, and immunofluorescence assay was performed to detect the interaction between the indicated proteins. The labels on the left of the photomicrographs indicate the transfected plasmids; labels on the top indicate the observed proteins at the appropriate microscope filter settings. (F) Co-immunoprecipitation assay was performed in ME-1 cells with anti-CHD7 antibody, and western blot was performed with the indicated antibodies. (G) Immunofluorescent staining of leukemic cells from the spleen of an end-stage Mx1-CreCbfb+/56M mouse to detect the co-localization between RUNX1 and CHD7 (top panel) and between RUNX1 and CBFβ/CBFβ-SMMHC (bottom panel). Scale bars, 10 μm. DAPI, 4′,6-diamidino-2-phenylindole; IgG, immunoglobulin G; IP, immunoprecipitate.
CHD7 interacts with RUNX1, RUNX1/CBFβ, and RUNX1/CBFβ-SMMHC complex. (A-B) 293T cells were transfected with indicated plasmids, and co-immunoprecipitation assays were performed to detect the interaction among these proteins. Flag-tagged CHD7 was immunoprecipitated with anti-Flag M2 beads, and western blot was performed with the indicated antibodies. (A) CHD7 pulls down RUNX1 but not CBFβ or CBFβ-SMMHC. (B) CHD7 pulls down CBFβ and CBFβ-SMMHC in the presence of RUNX1. (C-E) 293T cells were transfected with the indicated plasmids, and immunofluorescence assay was performed to detect the interaction between the indicated proteins. The labels on the left of the photomicrographs indicate the transfected plasmids; labels on the top indicate the observed proteins at the appropriate microscope filter settings. (F) Co-immunoprecipitation assay was performed in ME-1 cells with anti-CHD7 antibody, and western blot was performed with the indicated antibodies. (G) Immunofluorescent staining of leukemic cells from the spleen of an end-stage Mx1-CreCbfb+/56M mouse to detect the co-localization between RUNX1 and CHD7 (top panel) and between RUNX1 and CBFβ/CBFβ-SMMHC (bottom panel). Scale bars, 10 μm. DAPI, 4′,6-diamidino-2-phenylindole; IgG, immunoglobulin G; IP, immunoprecipitate.
CHD7 regulates RUNX1/CBFβ and RUNX1/CBFβ-SMMHC transactivation activity
We then tested the effect of CHD7 on the transcriptional activity of CBFβ-SMMHC with a luciferase reporter assay in which the expression of luciferase was driven by the promoter of CSF1R, the gene encoding the macrophage colony-stimulating factor receptor.30 As can be seen in Figure 5A, RUNX1/CBFβ activated this reporter activity, which was repressed by CBFβ-SMMHC. Interestingly, CHD7 enhanced transactivation of the CSF1R reporter by RUNX1/CBFβ and RUNX1/CBFβ-SMMHC (Figure 5A). Representative protein expression levels of the transfected constructs for this reporter assay are shown in Figure 5B. Taken together, these results suggest that CHD7 interacts with RUNX1 and regulates RUNX1/CBFβ and RUNX1/CBFβ-SMMHC transactivation activity.
CHD7 regulates RUNX1/CBFβ and RUNX1/CBFβ-SMMHC transactivation activity. (A) Luciferase reporter assay in 293T cells transfected with a CSF1R promoter-driven luciferase reporter plasmid and plasmids encoding the indicated proteins. Relative activities (mean ± SEM) were determined on the basis of 3 independent experiments. (B) Representative expression levels of the transfected proteins for this reporter assay. *P < .05.
CHD7 regulates RUNX1/CBFβ and RUNX1/CBFβ-SMMHC transactivation activity. (A) Luciferase reporter assay in 293T cells transfected with a CSF1R promoter-driven luciferase reporter plasmid and plasmids encoding the indicated proteins. Relative activities (mean ± SEM) were determined on the basis of 3 independent experiments. (B) Representative expression levels of the transfected proteins for this reporter assay. *P < .05.
Effect of Chd7 deficiency on Cbfb-MYH11–induced gene expression changes by RNA-seq
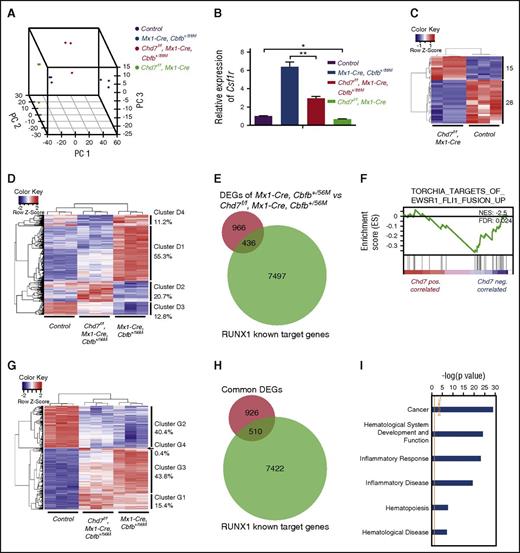
To further understand how CHD7 regulates RUNX1/CBFβ-SMMHC transactivation activity, we examined global gene expression changes in Chd7f/fMx1-CreCbfb-MYH11 mice at the preleukemia stage (2 weeks after treatment with poly(I:C)) with RNA-seq. Three-dimensional principal component analysis showed a clear separation of the 4 tested groups: Mx1-CreCbfb+/56M, Chd7f/fMx1-CreCbfb+/56M, Chd7f/fMx1-Cre, and the control (Figure 6A). Consistent with the enhanced CSF1R-luciferase activity when CHD7 was overexpressed in cultured 293T cells (Figure 5A), the expression of Csf1r was lower in Chd7f/fMx1-CreCbfb+/56M cells when compared with Mx1-CreCbfb+/56M cells and lower in Chd7f/fMx1-Cre cells compared with control cells (Figure 6B).
Effect of Chd7 knockout on Cbfb-MYH11–induced gene expression changes in preleukemic cells. (A-I) RNA-seq was performed on c-Kit+ bone marrow cells isolated from poly(I:C)-treated mice (n = 3 for each genotype). (A) Three-dimensional principal component analysis plots showing clear separations among these 4 genotype groups. (B) Relative expression levels of Csf1r in this data set (mean ± SEM). *P < .05; **P < .01. (C) Heatmap representation of unsupervised hierarchical clustering of DEGs between control and Chd7f/fMx1-Cre cells. Numbers on the right indicate DEGs in each of the 2 expression clusters. (D) Heatmap representation of unsupervised hierarchical clustering of DEGs between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M cells. Overall, 4 main clusters were identified, with the percentage of DEGs within each cluster labeled to the right. (E) Venn diagrams representing the overlap between DEGs in panel D (after conversion to human equivalent genes; supplemental Table 3) and RUNX1 target genes in ME-1 cells.17,32 (F) GSEA identified the TORCHIA_TARGETS_OF_EWSR1_FLI1_FUSION_UP gene set as being negatively correlated with DEGs upregulated in Chd7f/fMx1-CreCbfb+/56M cells. FDR, false discovery rate; NES, normalized enrichment scores. (G) Heatmap representation of unsupervised hierarchical clustering of overlapped DEGs between control vs Mx1-CreCbfb+/56M and control vs Chd7f/fMx1-CreCbfb+/56M (supplemental Figure 7C). In general, 4 main clusters were identified, with the percentage of DEGs within each cluster labeled to the right. (H) Venn diagrams representing the overlapped DEGs shown in panel G (after conversion to human equivalent genes; supplemental Table 6) with RUNX1 target genes in ME-1 cells.17,32 (I) Canonical pathways and disease functions associated with overlapped DEGs shown in panel G, identified by IPA. DEGs: Padj < .05; absolute fold changes ≥2.
Effect of Chd7 knockout on Cbfb-MYH11–induced gene expression changes in preleukemic cells. (A-I) RNA-seq was performed on c-Kit+ bone marrow cells isolated from poly(I:C)-treated mice (n = 3 for each genotype). (A) Three-dimensional principal component analysis plots showing clear separations among these 4 genotype groups. (B) Relative expression levels of Csf1r in this data set (mean ± SEM). *P < .05; **P < .01. (C) Heatmap representation of unsupervised hierarchical clustering of DEGs between control and Chd7f/fMx1-Cre cells. Numbers on the right indicate DEGs in each of the 2 expression clusters. (D) Heatmap representation of unsupervised hierarchical clustering of DEGs between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M cells. Overall, 4 main clusters were identified, with the percentage of DEGs within each cluster labeled to the right. (E) Venn diagrams representing the overlap between DEGs in panel D (after conversion to human equivalent genes; supplemental Table 3) and RUNX1 target genes in ME-1 cells.17,32 (F) GSEA identified the TORCHIA_TARGETS_OF_EWSR1_FLI1_FUSION_UP gene set as being negatively correlated with DEGs upregulated in Chd7f/fMx1-CreCbfb+/56M cells. FDR, false discovery rate; NES, normalized enrichment scores. (G) Heatmap representation of unsupervised hierarchical clustering of overlapped DEGs between control vs Mx1-CreCbfb+/56M and control vs Chd7f/fMx1-CreCbfb+/56M (supplemental Figure 7C). In general, 4 main clusters were identified, with the percentage of DEGs within each cluster labeled to the right. (H) Venn diagrams representing the overlapped DEGs shown in panel G (after conversion to human equivalent genes; supplemental Table 6) with RUNX1 target genes in ME-1 cells.17,32 (I) Canonical pathways and disease functions associated with overlapped DEGs shown in panel G, identified by IPA. DEGs: Padj < .05; absolute fold changes ≥2.
Compared with genes in the control mice, only 43 genes in Chd7f/fMx1-Cre mice were differentially expressed with Padj < .05 and absolute fold changes ≥2 (Figure 6C; supplemental Table 2), suggesting a limited effect of Chd7 on normal hematopoiesis, which is consistent with the fluorescence-activated cell sorting (FACS) data that a slight difference was observed between control and Chd7f/fMx1-Cre bone marrow cells 2 weeks after poly(I:C) injection (supplemental Figure 5A-B). But compared with Mx1-CreCbfb+/56M mice, 1617 genes in Chd7f/fMx1-CreCbfb+/56M mice were differentially expressed (Figure 6D; supplemental Table 3), suggesting a synergistic effect between Chd7 deficiency and Cbfb-MYH1 expression. Interestingly, 68.1% of these differentially expressed genes (DEGs; clusters D1 and D3) had attenuated changes in expression levels in Chd7f/fMx1-CreCbfb+/56M cells compared with Mx1-CreCbfb+/56M cells (Figure 6D; supplemental Figure 7A; supplemental Table 4). These results suggest that Chd7 mostly modulates the magnitude rather than the direction of gene expression changes by Cbfb-MYH11, even though FACS data showed no significant difference between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M bone marrow cells at the preleukemia stage (supplemental Figure 5A-B). Consistent with the observation that most of the CHD7 target sites are associated with active gene enhancer elements,31 most (66.4%) of the DEGs between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M cells were downregulated by Chd7 loss (Figure 6D; supplemental Table 3). In addition, 31.1% of these DEGs overlapped with RUNX1 target genes in ME-1 cells17,32 (Figure 6E; supplemental Table 3).
Gene set enrichment analysis (GSEA) of the DEGs between Chd7f/fMx1-CreCbfb+/56M and Mx1-CreCbfb+/56M cells shows that only 7 gene sets are significantly enriched (supplemental Table 5). Among them, the TORCHIA_TARGETS_OF_EWSR1_FLI1_FUSION_UP gene set, which contains genes upregulated in leukemic progenitor cells expressing the EWSR-FLI1 fusion gene, is negatively correlated with DEGs upregulated in Chd7f/fMx1-CreCbfb+/56M cells (Figure 6F; supplemental Table 3), suggesting that Chd7 deficiency delays Cbfb-MYH11–induced leukemia at least in part through inhibiting EWSR-FLI1 activity on gene expression. Ingenuity pathway analysis (IPA) showed that genes associated with hematologic system development and function, hematopoiesis, and hematologic disease were significantly enriched in DEGs between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M mice (supplemental Figure 7B).
Because Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M mice developed a similar leukemia (Figure 1) and expressed similar cell surface markers at the preleukemia stage (supplemental Figure 5A-B), we identified a common set of DEGs (n = 1710) in Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M cells that should contain genes critical for Cbfb-MYH11–induced leukemogenesis (supplemental Figure 7C ; supplemental Table 6). Among these 1710 DEGs common to both genotypes were Il1rl1 and Csf2rb,which were previously shown to be expressed in Cbfb-MYH11–expressing preleukemic cells33 (supplemental Table 6). Interestingly, 85.2% of these common DEGs (clusters G2 and G3) between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M mice had attenuated changes in expression levels in the Chd7f/fMx1-CreCbfb+/56M cells compared with Mx1-CreCbfb+/56M cells (Figure 6G; supplemental Figure 7D), suggesting Chd7 mainly regulates the magnitude rather than the direction of gene expression changes. In addition, consistent with the result that Runx1 is required for Cbfb-MYH11–induced defects, we found that 35.5% of these common DEGs overlapped with RUNX1 target genes in ME-1 cells17,32 (Figure 6H). Moreover, IPA showed that genes associated with cancer, hematologic system development and function, hematopoiesis, and hematologic disease were significantly enriched in these overlapped DEGs (Figure 6I). Taken together, these results suggest that CHD7 regulates RUNX1/CBFβ-SMMHC transactivation activity for leukemia initiation.
CHD7 alters expression levels of genes associated with cell cycle and proliferation in leukemic cells
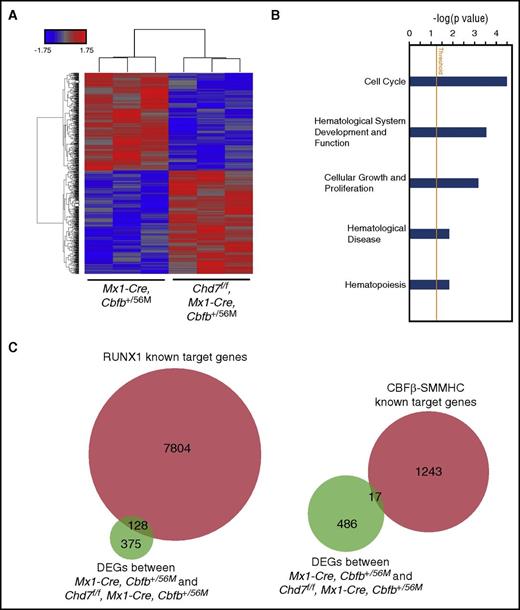
We also performed microarray analysis on c-Kit+ leukemic cells to determine gene expression differences between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M mice. Compared with genes in Mx1-CreCbfb+/56M mice, 634 genes in Chd7f/fMx1-CreCbfb+/56M mice were differentially expressed with Padj < .05 and absolute fold changes ≥1.2; 247 genes were upregulated and 387 genes were downregulated in the Chd7f/fMx1-CreCbfb+/56M mice (Figure 7A; supplemental Table 7). IPA analysis showed that genes associated with cell cycle, hematologic system development and function, cell growth, and proliferation were significantly enriched in these DEGs (Figure 7B). IPA analysis also predicts that JAK-STAT and CDKN1A pathways are inhibited by DEGs in Chd7f/fMx1-CreCbfb+/56M leukemic cells (supplemental Table 8). Again, many of these DEGs (25.4%) are potential targets of RUNX1, whereas only 3.4% of these DEGs are CBFβ-SMMHC target genes (Figure 7C) identified in the inv(16) leukemia cell line ME-1.17,32 However, only 33 DEGs were found between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M leukemic cells when more stringent criteria were used (Padj < .05; absolute fold changes ≥2; supplemental Table 7), with 45.4% and 18.2% of them overlapping with RUNX1 and CBFβ-SMMHC target genes in ME-1 cells, respectively. These results demonstrated that leukemia cells in the Chd7f/fMx1-CreCbfb+/56M mice had a gene expression profile similar to that in Mx1-CreCbfb+/56M mice.
CHD7 regulates expression of genes associated with cell cycle and proliferation in leukemia cells. (A-C) Genome-wide RNA expression profiling by microarray on splenic c-Kit+ leukemic cells from end-stage leukemic mice. (A) Heatmap of DEGs (Padj < .05; absolute fold changes ≥1.2) between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M leukemic cells. (B) Canonical pathways and disease functions associated with DEGs between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M leukemic cells, identified by IPA. (C) Venn diagram representing the overlap between these DEGs and RUNX1 (left panel) or CBFβ-SMMHC (right panel) target genes in ME-1 cells.17,32
CHD7 regulates expression of genes associated with cell cycle and proliferation in leukemia cells. (A-C) Genome-wide RNA expression profiling by microarray on splenic c-Kit+ leukemic cells from end-stage leukemic mice. (A) Heatmap of DEGs (Padj < .05; absolute fold changes ≥1.2) between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M leukemic cells. (B) Canonical pathways and disease functions associated with DEGs between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M leukemic cells, identified by IPA. (C) Venn diagram representing the overlap between these DEGs and RUNX1 (left panel) or CBFβ-SMMHC (right panel) target genes in ME-1 cells.17,32
Discussion
It has been shown that RUNX1 activity is required for CBFB-MYH11–induced leukemogenesis. Therefore, molecules that modulate RUNX1 activity may also be important for CBFB-MYH11–induced leukemogenesis. Recently, it was demonstrated that CHD7 interacted with RUNX1 to regulate hematopoiesis,23 suggesting that CHD7 may also be important for leukemogenesis by CBFβ-SMMHC. In this study, we performed experiments to test this hypothesis.
In vitro studies showed that CHD7 was not only a partner of the RUNX1/CBFβ complex, but it was also associated with the RUNX1/CBFβ-SMMHC complex in a RUNX1-dependent manner. RNA-seq analysis showed that many genes affected by CHD7 deficiency were RUNX1 target genes, suggesting that CHD7 modulates the transactivation activity of RUNX1. However, formal functional demonstration of CHD7 regulation of RUNX1 transactivation activity would require perturbing RUNX1 expression in the Chd7f/fMx1-CreCbfb+/56M and Mx1-CreCbfb+/56M cells.
The gene expression profile at the preleukemic stage (2 weeks after poly(I:C)) was distinctly different between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M mice. It is possible that such large difference in gene expression was the result of the cellular composition of the c-Kit+ cells, which were used for RNA-seq. However, this is not likely because no difference was observed between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M mice in terms of the percentage of LK, LSK, GMP, CMP, MEP, and AMP compartments by FACS analysis, or in terms of the morphology of the AMPs in these 2 groups of mice.
Notably, as shown in Figure 6D,G, many DEGs had reduced magnitude of gene expression changes in the Chd7f/fMx1-CreCbfb+/56M cells compared with Mx1-CreCbfb+/56M cells, suggesting that CHD7 only modulates other transcription factors such as RUNX1 rather than regulating gene expressions directly. Moreover, at the preleukemic stage, 35.5% of the common DEGs between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M cells, which are likely responsible for leukemia development, overlapped with RUNX1 target genes in ME-1 cells (Figure 6H). At leukemic stage, 25.4% DEGs in the Chd7f/fMx1-CreCbfb+/56M mice were potential RUNX1 target genes in ME-1 cells. These results suggest that CHD7 alters RUNX1 target gene expression (Figure 7C). Altogether, our results support the notion that CHD7 most likely functions through interacting with the RUNX1/CBFβ-SMMHC complex and altering the expression of its target genes. However, CHD7 has been reported to function as an ATP-dependent chromatin remodeler, so many of the DEGs may result from indirect effects of altered chromatin state. Additional experiments are needed to formally demonstrate that the effects of CHD7 loss are through its ability to bind RUNX1, such as perturbing Runx1 in the context of Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M mice.
At the preleukemic stage, the proliferation ability of the LK cells from Mx1-CreCbfb+/56M cells is similar to that from Chd7f/fMx1-CreCbfb+/56M cells. Consistently, genes associated with proliferation or cell cycle were not significantly enriched when the RNA-seq data were analyzed with GSEA and IPA. Conversely, IPA analysis showed that genes associated with cell cycle, hematologic system development and function and cell growth and proliferation were significantly enriched in the DEGs between Mx1-CreCbfb+/56M and Chd7f/fMx1-CreCbfb+/56M leukemic cells, supporting the observation that Chd7 deficiency inhibited the proliferation of the leukemic cells. A similar effect of Chd7 on proliferation was also observed in murine neural stem/progenitor cells,34 olfactory neural stem cells,35 and inner ear neuroblasts.36 Therefore, we postulate that the ability of Chd7 deficiency to delay Cbfb-MYH11–induced leukemogenesis in both primary and transplanted mice was at least in part through inhibiting the proliferation of leukemia-initiating cells and leukemic cells.
Currently, the treatment of inv(16) AML is nonselective cytotoxic chemotherapy, which results in a good initial response,37 but approximately 50% of patients will eventually relapse.38 Because of the importance of CBFβ-SMMHC in leukemogenesis, several efforts were made to develop targeted inhibitors for CBFβ-SMMHC activity, such as inhibitors that disrupt the CBFβ-SMMHC and RUNX1 interaction39,40 and inhibitors for HDAC8, which is also required for CBFβ-SMMHC activity.41 Our results show that CHD7 also contributes to CBFβ-SMMHC activity, providing another potential candidate target for treating this type of leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Irene C. Ginty, Abdel G. Elkahloun, Stephen Wincovitch, Stacy Anderson, Martha Kirby, Chung-Tsai Lee, Guadalupe Lopez, and the National Institutes of Health (NIH) Intramural Sequencing Center for their technical help. This work used the computational resources of the NIH High Performance Computing Biowulf cluster (http://hpc.nih.gov).
This work was supported by the NIH Intramural Research Program of the National Human Genome Research Institute, and by grants R01 HL089969 and R01 HL091724 from the NIH National Heart, Lung, and Blood Institute (N.A.S.).
Authorship
Contribution: T.Z. designed and performed experiments, analyzed data, and wrote the paper; L.Z., R.K.H., Y.L., and L.A. performed experiments; E.M.K. performed RNA sequencing and microarray data analysis; J.H. and N.A.S. contributed the Chd7 knockout mice; and P.P.L. designed the experiments, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: P. Paul Liu, National Human Genome Research Institute, National Institutes of Health, 49 Convent Dr, Building 49, Room 4A38, Bethesda, MD 20892; e-mail: pliu@mail.nih.gov.

![Figure 2. Chd7 deficiency delays leukemia initiation induced by Cbfb-MYH11. (A-C) The indicated groups of mice were treated with ENU to induce additional mutations and with poly(I:C) to induce the expression of Cbfb-MYH11 and/or Chd7 deficiency. Three weeks after poly(I:C) treatment, mice were euthanized and flow cytometry assays were performed. (A) Leukemia development monitored by measuring the c-Kit+/Sca1– population in peripheral blood (PB). (B) Representative FACS plots of bone marrow cells gated on single cells (upper plots), Lin– cells (middle plots), and LK cells (bottom plots). (C) Bar graph showing the percentages (mean ± standard error of the mean [SEM]) of Lin–, LK, LSK, AMP, CMP, GMP, and MEP compartments in the bone marrow of mice of the indicated genotypes. *P < .05; **P < .01; ***P < .001; ****P < .0001, each compared with the control group except for (#), for which the comparisons were between the Chd7f/fMx1-CreCbfb+/56M group and the Mx1-CreCbfb+/56M group.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/130/22/10.1182_blood-2017-04-780106/4/m_blood780106f2.jpeg?Expires=1765884327&Signature=1sK0XEh-6B~-NJOCxydOMcqDyqSh8TtQw23InNRCQ6zhMit9TxvGQcgAc7PtRMoLa8VOvk31V~cx~QQOugZNJ0Uc6o98t-Gvi5S2iyUJ9GnUqQ8qazMKoaCxlZf3aQFtUH3hkC9kmKQdhnFbj5xQ2AOfIdz8RPjLjigon2rqfXLceorIEPYnm2vclkD4FwJZsD2~SxaAiMrH9tGCkZOmSSXYg9Ibu8iJhyPL~uyzpXCYT5FTcEY9-kQCJ7t3N9d974u1ejVi6hJTK4KnW03I0kNqrgMD64VBddglWhAkJEtd61X60vW78Woyg~OI~mngR87qDcMPv2eju3eC8XzXKA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal