Key Points
High sIgM level is a potential key factor associated with poorer clinical outcome in CLL.
Genetic and epigenetic features influence sIgM levels and function in CLL.
Abstract
Chronic lymphocytic leukemia (CLL) with unmutated (U-CLL) or mutated (M-CLL) immunoglobulin gene heavy-chain variable region (IGHV) displays different states of anergy, indicated by reduced surface immunoglobulin M (sIgM) levels and signaling, consequent to chronic (super)antigen exposure. The subsets also differ in the incidence of high-risk genetic aberrations and in DNA methylation profile, preserved from the maturational status of the original cell. We focused on sIgM expression and function, measured as intracellular Ca2+ mobilization following stimulation, and probed correlations with clinical outcome. The relationship with genetic features and maturation status defined by DNA methylation of an 18-gene panel signature was then investigated. sIgM levels/signaling were higher and less variable in U-CLL than in M-CLL and correlated with disease progression between and within U-CLL and M-CLL. In U-CLL, increased levels/signaling associated with +12, del(17p) or NOTCH1 mutations. In M-CLL, there were fewer genetic lesions, although the methylation maturation status, generally higher than in U-CLL, varied and was increased in cases with lower sIgM levels/signaling. These features revealed heterogeneity in M-CLL and U-CLL with clear clinical correlations. Multivariate analyses with phenotype, genetic lesions, or DNA methylation maturation status identified high sIgM levels as a new potential independent factor for disease progression. Multiple influences on sIgM include the cell of origin, the clonal history of antigen encounter in vivo, and genetic damage. This simple marker compiles these different factors into an indicator worthy of further investigations for prediction of clinical behavior, particularly within the heterogeneous M-CLL subset.
Introduction
The B-cell receptor (BCR) is the essential functional unit for most normal and neoplastic B cells.1,2 In chronic lymphocytic leukemia (CLL), it is key to survival and proliferation, and is now a therapeutic target of very effective inhibitors of BCR-associated kinases, including SYK, BTK, or phosphoinositide 3′-kinase isoform p110δ (PI3Kδ).1-6
The molecular characteristics of the tumor surface immunoglobulin indicate that CLL consists of 2 major subsets. The one with unmutated (U-CLL) immunoglobulin gene heavy-chain variable regions (IGHV) has arisen from pre-germinal center CD5+ B cells and the subset with mutated (M-CLL) IGHV has arisen from postfollicular CD5+ B cells which have undergone somatic hypermutation.7-9 The nature of the B cell of origin clearly influences tumor behavior, with U-CLL having a worse prognosis than M-CLL.10,11
The functional characteristics of the tumor surface immunoglobulin M (sIgM) indicate that circulating CLL cells from both subsets are characterized by a degree of anergy. This is defined by variably reduced sIgM, but not sIgD, levels and signaling capacity consequent to chronic (super)antigenic exposure.1,2,12 Analysis of a small cohort of patients indicated that the 2 subsets differ in mean sIgM levels/signaling capacity, being higher in U-CLL cells than in the more anergized M-CLL.13-18 The outcome may be that U-CLL cases have a larger proliferative component than M-CLL, explaining more rapid tumor progression in U-CLL.1,2,15 Consistently, U-CLL appear to respond more profoundly to BCR-associated kinase inhibitors (BIs) than M-CLL,19 whereas duration of the lymphocytosis in the circulation appears more prolonged in M-CLL than U-CLL.20 However, sIgM expression and function is heterogeneous between patients,13 and the clinical meaning of this heterogeneity has not yet been examined.
DNA methylation is emerging as another key to variable CLL behavior.21-26 Genome-wide DNA methylation studies have uncovered 2 major methylation subtypes along with a third intermediate group.23,25,27 These groups largely represent the degree of epigenetic programming experienced by the B cell of origin in CLL, and are termed low-, intermediate-, and high-programmed CLLs (LP-CLLs, IP-CLLs, and HP-CLLs, respectively).23,27 Maturation of DNA methylation patterns is generally concordant with the degree of IGHV mutation, with LP-CLLs and HP-CLLs mostly composed of U-CLLs and M-CLLs, respectively.25,27 Variability exists particularly within M-CLL,25 potentially reflecting a range of maturation of the cell of origin.27 A “methylation maturation score” using a panel of selected gene regions has been shown to efficiently represent overall maturation,27 further dividing M-CLL into at least 2 subcategories (IP-CLL and HP-CLL) with different clinical behavior.23,25,27
Genetic alterations also influence outcome, response to (immuno)chemotherapy, and clonal evolution.28-31 In particular, those associated with poorer outcome are markedly enriched in U-CLL.32-35 Trisomy 12 (+12), 11q(ATM) deletion (del(11q)), and 17p(TP53) deletion (del(17p)) stratify CLL patients into separate prognostic categories with different survivals,33 and integration with mutations including NOTCH1 ΔCT_7544-7545 (NOTCH1ΔCT) or those of SF3B1 help further refine prognosis.33 Also, although BIs seem dramatically effective in all genetic categories including del(17p),36 supporting a dominant role of BCR signaling over genetics for clinical efficacy,37 BI-induced tumor lymphocytosis appears shorter in +12,20,38 opening the question of whether there is any link between specific genetic lesions and BCR characteristics.
In this study, we investigated the links between anergy, deduced global DNA methylation, and genetics in CLL. We confirmed that increased sIgM associated with more rapid progression and inferior survival, and revealed heterogeneity in M-CLL. We also showed a correlation with a more aggressive genetic profile particularly in U-CLL. Within M-CLL, a strong inverse correlation was found between sIgM and DNA methylation maturation, indicating that profound anergy is associated with a more mature profile.
Materials and methods
CLL patients and samples
The study included samples from a series of 270 consecutive patients with previously untreated sIgM+/D+ CLL recruited in the lymphoproliferative disorder study at time of initial evaluation at the Department of Hematology of the Southampton University Hospital Trust from January 2001 to May 2015. Diagnosis of CLL was according to the 2008 International Workshop on Chronic Lymphocytic Leukemia (IWCLL2008)/National Cancer Institute (NCI) criteria and confirmed by a flow cytometry “Matutes score” > 3 in all cases.39,40 For clinical association studies, 235 CLL patients with full sIgM/D levels/signaling analysis and clinical history with a minimum follow-up of 12 months were studied (supplemental Table 1, available on the Blood Web site). Median follow-up of patients that were alive was 99 months. No patients were lost at follow-up. The lymphoproliferative disorder study was approved by the institutional review boards at the University of Southampton (228/02/t). All patients provided informed consent prior to inclusion in the study.
Phenotypic, signaling capacity, and immunogenetic studies
Peripheral blood mononuclear cells (PBMCs) were prepared and stored in liquid nitrogen. Prior to each assay, cells were thawed, washed, and allowed to recover in complete RPMI 1640 medium (supplemented with 10% fetal calf serum, 2 mM glutamine, and 1 mM sodium pyruvate) for 1 hour at 37°C.41 Phenotypic, signaling capacity, and immunogenetic studies were performed using established internal standard operating procedures.13,42
For phenotypic analyses, sIgM and sIgD levels were determined on the CD19+/CD5+ CLL cells using soluble rabbit F(ab′)2 phycoerythrin-conjugated anti-human IgM or fluorescein isothiocyanate–conjugated anti-human IgD or control polyclonal antibodies (DAKO, Ely, United Kingdom) and peridinin-chlorophyll-protein–cyanine 5.5–conjugated anti-CD5 and allophycocyanin-conjugated anti-CD19 monoclonal antibodies (Biolegend, London, United Kingdom). Surface staining was carried out on 1 × 106 PBMCs in 100 μL of fluorescence-activated cell sorting (FACS) buffer on ice for 30 minutes in all cases. Cells were washed in 2 mL of FACS buffer and resuspended in 300 μL of FACS buffer and a total of 10 000 events were acquired before analysis. Expression of CD38, ζ-chain-associated protein of 70 kDa (ZAP-70), and CD49d was analyzed as reported.43 Cutoff points for CD38, ZAP-70, and CD49d positivity were 30%, 20% and 30%, respectively. Mean fluorescence intensity (MFI) was calculated as [test antibody geometric mean − control antibody geometric mean] for all markers included in the study.
Signaling capacity was measured as percentage intracellular Ca2+ [iCa2+] mobilization following stimulation.13 Briefly, 107 PBMC/mL were incubated with 4 μM Fluo3-AM (Invitrogen, Paisley, United Kingdom) and 0.02% (vol/vol) Pluronic F-127 (Sigma, Poole, United Kingdom) for 30 minutes at 37°C. Cells were then washed and resuspended at 5 × 106 cells per mL at room temperature, warmed to 37°C for 5 minutes prior to acquisition for 35 seconds to obtain the background fluorescence (unstimulated cells). Following addition of 20 μg/mL goat F(ab′)2 anti-human IgM or IgD (Southern Biotechnology, Cambridge, United Kingdom) or control antibodies. Data were acquired for 10 minutes. Maximum calcium release was observed within the first 2 minutes in all circumstances. Treatment with 1 μM ionomycin (Sigma) was used to confirm viability of samples and exclude negative artifacts. Percent iCa2+ mobilization was calculated as [peak (all events) − mean Y (unstimulated cells)/%CD19+ cells] × 100, where %CD19+ cells was the percentage of CD19+ cells in the live lymphocyte gate of the test sample.
A FACSCalibur flow cytometer (Becton Dickinson, Oxford, United Kingdom) was used for acquisition in all circumstances. Analysis of all phenotypic and signaling profiles was performed and uniformly reviewed by 2 independent researchers (I.T. and I.H.) using FlowJo software v9.5.2 (Tree Star, Ashland, Oregon).
The full IGHV-IGHD-IGHJ-IGHCM constant region rearrangements were amplified from complementary DNA in all circumstances and directly sequenced bidirectionally using our primers from leader to constant IGHCM region.13,43,44 Sequences were aligned to ImMunoGeneTics directories, and considered mutated if homology to the corresponding germ line gene was <98%. Tumors, using IGHCG or IGHCA rearrangements (which required amplification with a primer specific to the IGHCG or IGHCA constant region) and/or expressing IgG or IgA on the CLL cells by flow cytometry, were excluded from the study.
Genetic studies
Interphase fluorescence in situ hybridization (FISH) was performed at the Wessex Regional Genetics Laboratory in Salisbury, using the probes (Vysis) LSI13 and LSID13S319 for del13q14 (del13q), CEP12 for chromosome 12 aneuploidy (+12), LSIp53 for del17p13 (del(17p)), and LSIATM for del11q22-q23 (del(11q)). NOTCH1ΔCT mutation was sought by amplification refractory mutation system (ARMS) polymerase chain reaction (PCR) and Sanger sequencing.45 In the CLL harboring the NOTCH1ΔCT, mutated allele frequency was determined by digital droplet PCR (ddPCR).46 Briefly, 50 ng of DNA and 70 μL of oil were loaded into a cartridge (Bio-Rad, Milan, Italy) to form 20 000 monodispersed 1 nL of surfactant-stabilized droplets per sample. Droplets were transferred into a 96-well PCR plate (40 μL per well). Droplet PCR amplification was performed using the primePCR ddPCR_mutation_assay_NOTCH1_p.P2514fs*4 human kits for wild-type or mutant c.7541_7542delCT (Bio-Rad) and a Veriti DX thermal cycler (Applied Biosystems). Fluorescence of amplified products was read using a QX200 droplet reader (Bio-Rad) and analyzed with QuantaSoft software (Bio-Rad). Frequency of the NOTCH1ΔCT allele in the tumor population was calculated as [(ΔCT droplets/total droplets) × (% CLL cells per sample)], where CT is the cycle threshold. SF3B1 mutations were sought by high-resolution melting (HRM) PCR and confirmed by Sanger sequencing, as previously described.47
DNA methylation studies
Genomic DNA isolated CLL samples were bisulfite-converted using the EZ-DNA Methylation kit (Zymo Research). Targeted DNA methylation analysis was performed using MassARRAY (Agena Biosciences) by PCR amplification of 18 genomic regions. Briefly, regions were selected from previous genome-wide analyses using Infinium 450K array data that were differentially methylated between CLL samples and, when combined, retained high discriminatory power to separate the 3 CLL DNA methylation subtypes.27 To determine LP-CLL, IP-CLL, and HP-CLL DNA methylation subtypes, CpG methylation values were averaged per genomic region and then used to subgroup patients by consensus clustering.27 Reduction of amplicon methylation data to a singular value representing the methylation maturation score (MMS) per sample was calculated by subtracting from 1 the methylation value (range, 0-1.0) for amplicons associated with hypomethylation programming and then calculating the mean methylation for all 18 amplicons.
Statistical analyses and clinical association studies
Supplemental Table 1 summarizes the clinical variables recorded at presentation. The clinical variables recorded at follow-up were date of progression requiring treatment of the first time and date of lymphocyte doubling from diagnosis according to the IWCLL2008/NCI guidelines40 and date of death. Time to progression requiring first treatment (TTFT), time to lymphocyte doubling (LDT), and overall survival (OS) were measured from date of CLL diagnosis to date of progressive and/or symptomatic disease requiring treatment according to IWCLL/NCI guidelines (TTFT),40 to date of lymphocyte doubling (LDT), or to date of death or last follow-up (OS). TTFT was used as primary end point, whereas OS was used as a secondary end point to avoid the chemotherapy and kinase inhibitors as confounders. We used our previous cutoffs of 50% and 5% to distinguish patients with high/low sIgM/D (MFI) or signaling capacity (iCa2+ mobilization %), respectively.13 These cutoffs corresponded to the best cutoffs by receiver operating characteristic and Youden t tests when treatment was used as a state variable.
Categorical variables were compared by the χ2 test or the Fisher exact test when appropriate. Continuous variables were compared by the Mann-Whitney nonparametric test for 2 or k independent samples. All statistical tests were 2-sided. Statistical significance was defined as P value < .05. Survival analysis was done by the Kaplan-Meier method using log-rank statistics. Multivariate analysis was done by Cox proportional hazard regression. A variable was entered into the model if the probability was ≤0.05. Statistical analyses were performed with the Statistical Package for the Social Sciences (SPSS) software v.22.0 (Chicago, IL) and Graphpad Prism 6 software (La Jolla, CA).
Results
U-CLL and M-CLL have different levels of signal-responsive sIgM
sIgM levels and signaling capacity were investigated in 270 CLL patients. Levels of sIgM were broadly variable (range, 3-918; median, 50; coefficient of variation [CV], 144%), as was sIgM signaling capacity (range, 0-100; median, 25; CV, 87%). A significant correlation was present between sIgM levels and signaling capacity (r = 0.55; P < .0001; 95% confidence interval [CI], 0.44-0.64).
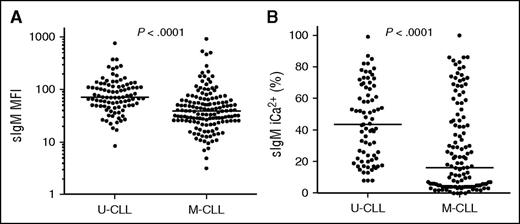
Levels were less variable and significantly higher (P < .0001, Figure 1A) in U-CLL (range, 8-781; median, 72) than in M-CLL (range, 3-918; median, 39). Signaling capacity was also variable and significantly different between the 2 subsets (range, 8-99; median, 43 in U-CLL; range, 0-100; median, 17 in M-CLL, P < .0001, Figure 1B). These data validated previous findings in a separate smaller cohort of patients.13 Of interest, the subset of CLL using IGHV3-21 (n = 9), which typically has an aggressive course,48-50 also had significantly higher sIgM levels and signaling capacity than non-IGHV3-21 M-CLL, irrespective of IGHV3-21 mutational status (supplemental Figure 1). In M-CLL, there was a significant group of cases with no detectable ability to signal (<5% iCa2+ flux) and these correlated with very low expression, indicating a deeply anergic group not found in U-CLL. In each subset, a significant correlation between levels and signaling was maintained, although this was more evident in the M-CLL subset (r = 0.38, P = .0012 in U-CLL; r = 0.52, P = .0001 in M-CLL).
sIgM expression and signaling capacity in CLL subgroups. sIgM expression (MFI) and sIgM signaling capacity (iCa2+ %) were analyzed by flow cytometry in 270 patients with CLL. The patient cohort was divided by IGHV mutational status to assess (A) sIgM expression and (B) anti-IgM signaling capacity. Horizontal lines indicate mean values. The statistical significance of difference was analyzed using the Mann-Whitney test.
sIgM expression and signaling capacity in CLL subgroups. sIgM expression (MFI) and sIgM signaling capacity (iCa2+ %) were analyzed by flow cytometry in 270 patients with CLL. The patient cohort was divided by IGHV mutational status to assess (A) sIgM expression and (B) anti-IgM signaling capacity. Horizontal lines indicate mean values. The statistical significance of difference was analyzed using the Mann-Whitney test.
sIgD levels (range, 2-471; median, 33; CV, 114%) and signaling capacity (range, 1-100; median, 52; CV, 58%) were less variable and no significant differences were observed between U-CLL and M-CLL (supplemental Figure 2A), nor did the variations show robust correlations with sIgM levels.
Levels of functional sIgM predict progression of CLL
TTFT was used as a primary indicator of natural progression to investigate the role of sIgM in 235 CLL patients (supplemental Table 1). Both high sIgM expression and high sIgM signaling associated with significantly more rapid progression (Figure 2A-B). IGHV status is an independent prognostic factor of progression and this previously unpublished cohort at the Cancer Sciences Unit (CSU) confirmed its relevance (supplemental Figure 3). To understand potential individual relevance of BCR-associated characteristics and explore significance of sIgM variability in each U-CLL and M-CLL subset, we initially performed a multivariate Cox regression adjusted for IGHV status, sIgM levels, and sIgM signaling. This revealed that sIgM levels and signaling predicted progression of CLL in a fashion-independent from IGHV status (Table 1). Hence, we analyzed sIgM within U-CLL or M-CLL separately (Figure 2C-F). High levels associated with more aggressive behavior within U-CLL or M-CLL (Figure 2C,E). High and low signaling also separated U-CLL and M-CLL in 2 categories with different outcome (Figure 2D,F). The differences were most evident within M-CLL, of which the subset with high sIgM levels/signaling appeared to have a progression apparently as rapid as the U-CLL with low sIgM levels/signaling (supplemental Figure 4). This overlap also highlights the fact that, although the cell of origin has a major influence on tumor behavior, other influences on the BCR can, in a minority of cases, lead to a convergent clinical outcome.
The significance of sIgM levels and signaling for TTFT in CLL. The previous cutoffs of 50 (MFI) and 5% (iCa2+) were used to distinguish patients with high or low sIgM levels and signaling capacity, respectively.13 These cutoffs corresponded to the best cutoffs by receiver operating characteristic and Youden t tests when treatment was used as a state variable. (A) Patients with sIgM expression above (sIgM MFI high, dotted line) or below (sIgM MFI low, continuous line) the MFI cutoff of 50 were investigated for time to progression from diagnosis to requirement of treatment for the first time (TTFT). (B) Patients with sIgM signaling capacity above (sIgM high-signaler, dotted line) or below (sIgM low-signaler, continuous line) the cutoff of 5% were investigated for progression requiring treatment. Association between sIgM levels (C,E) and signaling capacity (D,F) with TTFT was also investigated in U-CLL and M-CLL, respectively. Survival analysis was performed by Kaplan-Meier algorithm.
The significance of sIgM levels and signaling for TTFT in CLL. The previous cutoffs of 50 (MFI) and 5% (iCa2+) were used to distinguish patients with high or low sIgM levels and signaling capacity, respectively.13 These cutoffs corresponded to the best cutoffs by receiver operating characteristic and Youden t tests when treatment was used as a state variable. (A) Patients with sIgM expression above (sIgM MFI high, dotted line) or below (sIgM MFI low, continuous line) the MFI cutoff of 50 were investigated for time to progression from diagnosis to requirement of treatment for the first time (TTFT). (B) Patients with sIgM signaling capacity above (sIgM high-signaler, dotted line) or below (sIgM low-signaler, continuous line) the cutoff of 5% were investigated for progression requiring treatment. Association between sIgM levels (C,E) and signaling capacity (D,F) with TTFT was also investigated in U-CLL and M-CLL, respectively. Survival analysis was performed by Kaplan-Meier algorithm.
Cox multivariate analysis of BCR-associated parameters for TTFT
| . | SE . | P . | HR (95% CI) . |
|---|---|---|---|
| sIgM MFI > 50 | 0.215 | .000 | 2.586 (1.698-3.938) |
| sIgM iCa2+ ≥ 5% | 0.292 | .045 | 1.796 (1.013-3.181) |
| IGHV ≥ 98% | 0.202 | .000 | 2.476 (1.668-3.675) |
| . | SE . | P . | HR (95% CI) . |
|---|---|---|---|
| sIgM MFI > 50 | 0.215 | .000 | 2.586 (1.698-3.938) |
| sIgM iCa2+ ≥ 5% | 0.292 | .045 | 1.796 (1.013-3.181) |
| IGHV ≥ 98% | 0.202 | .000 | 2.476 (1.668-3.675) |
Number of cases entered in the multivariate analysis = 206; number of events = 112.
HR, hazard ratio; SE, standard error.
When sIgD was analyzed, no significantly different survivals could be documented between CLL with high vs low levels/signaling (supplemental Figure 2B).
sIgM levels and signaling associate with poor-risk genetic lesions, which are enriched in U-CLL
We then investigated links between IgM features and specific FISH lesions (isolated del13q, +12, del(11q), del(17p), n = 189), or mutations of NOTCH1 (n = 220) or SF3B1 (n = 189). U-CLL were enriched for poor-risk genetic lesions compared with M-CLL (supplemental Table 1; supplemental Figure 5) as expected.33
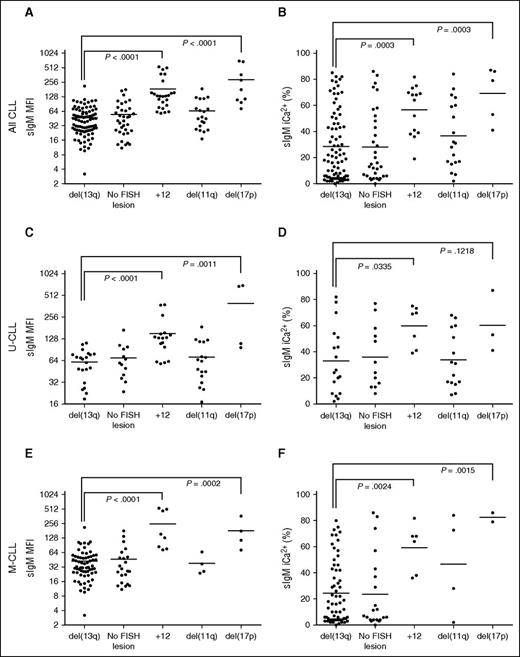
Analysis of FISH lesions revealed that CLL subsets harboring +12 or del(17p) had significantly higher sIgM levels and signaling capacity than del13q (Figure 3A-B). The associations held even when U-CLL or M-CLL were analyzed separately (Figure 3C-F).
sIgM levels or signaling and FISH lesions in CLL. CLL samples were investigated for sIgM expression (MFI) and signaling capacity (iCa2+ %) by flow cytometry. The CLL patient cohort were divided by FISH lesions according to Dӧhner hierarchical model. Association between (A) sIgM levels, or (B) signaling capacity with FISH lesions was investigated. Association between sIgM levels (C,E) and signaling capacity (D,F) with FISH lesions was also investigated in the U-CLL and M-CLL patient cohort, respectively. Horizontal bars indicate mean values. Statistical analysis were done by comparing isolated del13q vs each other individual FISH category using the Mann-Whitney test (2-tailed, 95% CI). P values are represented only for statistically significant differences.
sIgM levels or signaling and FISH lesions in CLL. CLL samples were investigated for sIgM expression (MFI) and signaling capacity (iCa2+ %) by flow cytometry. The CLL patient cohort were divided by FISH lesions according to Dӧhner hierarchical model. Association between (A) sIgM levels, or (B) signaling capacity with FISH lesions was investigated. Association between sIgM levels (C,E) and signaling capacity (D,F) with FISH lesions was also investigated in the U-CLL and M-CLL patient cohort, respectively. Horizontal bars indicate mean values. Statistical analysis were done by comparing isolated del13q vs each other individual FISH category using the Mann-Whitney test (2-tailed, 95% CI). P values are represented only for statistically significant differences.
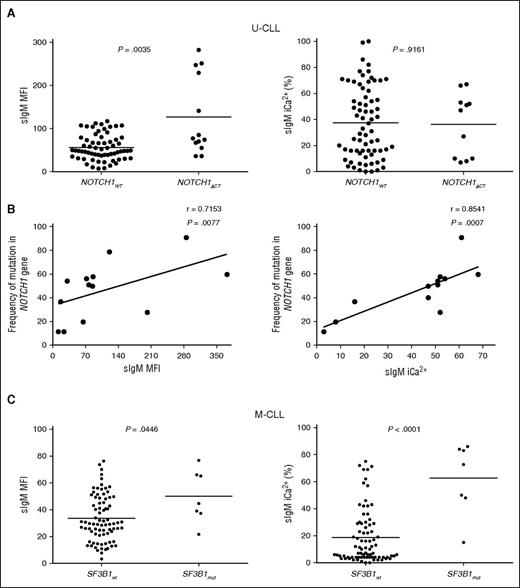
NOTCH1ΔCT was identified in 13 of 220 patients. Eleven of thirteen CLL with NOTCH1ΔCT were U-CLL. We interrogated the U-CLL cohort for associations of NOTCH1ΔCT status with sIgM levels/signaling. Although only mean sIgM level was higher in NOTCH1ΔCT U-CLL, NOTCH1ΔCT U-CLL appeared to cluster in 2 separate groups with high and low sIgM levels or signaling capacity (Figure 4A). Subsequent analysis by ddPCR revealed variable NOTCH1ΔCT allele frequency (range, 11.4-90.8), and the variability strongly correlated with sIgM levels and signaling capacity (Figure 4B).
sIgM levels or signaling and NOTCH1 or SF3B1 mutations in CLL. Associations or correlations of NOTCH1ΔCT with sIgM levels or signaling capacity were sought following identification of NOTCH1ΔCT by ARMS PCR and Sanger sequencing or following determination of NOTCH1ΔCT allele frequency by digital PCR in the tumor population. (A) Associations NOTCH1ΔCT with sIgM levels or signaling capacity. (B) Correlation analyses between NOTCH1ΔCT allele frequency and sIgM expression and signaling was assessed by digital PCR in those cases that scored NOTCH1ΔCT mutants by ARMS PCR/sequencing. (C) SF3B1 mutational status was determined by HRM-PCR and confirmed by Sanger sequencing. Cases with and without SF3B1 mutation were investigated for association with sIgM expression and sIgM-mediated signaling responses. Horizontal bars indicate mean values. Statistical analyses for associations were performed using the Mann-Whitney test (2 tailed, 95% CI). Statistical analyses for correlations were performed using the Spearman rank correlation test.
sIgM levels or signaling and NOTCH1 or SF3B1 mutations in CLL. Associations or correlations of NOTCH1ΔCT with sIgM levels or signaling capacity were sought following identification of NOTCH1ΔCT by ARMS PCR and Sanger sequencing or following determination of NOTCH1ΔCT allele frequency by digital PCR in the tumor population. (A) Associations NOTCH1ΔCT with sIgM levels or signaling capacity. (B) Correlation analyses between NOTCH1ΔCT allele frequency and sIgM expression and signaling was assessed by digital PCR in those cases that scored NOTCH1ΔCT mutants by ARMS PCR/sequencing. (C) SF3B1 mutational status was determined by HRM-PCR and confirmed by Sanger sequencing. Cases with and without SF3B1 mutation were investigated for association with sIgM expression and sIgM-mediated signaling responses. Horizontal bars indicate mean values. Statistical analyses for associations were performed using the Mann-Whitney test (2 tailed, 95% CI). Statistical analyses for correlations were performed using the Spearman rank correlation test.
SF3B1 mutations were found in 12 of 189 CLL (5 of 62 U-CLL and 7 of 127 M-CLL). In U-CLL, no differential sIgM levels/signaling were seen, possibly due to significant enrichment with other genetic lesions in U-CLL (supplemental Table 1; Figure 4). Conversely in M-CLL, the samples with SF3B1 mutations (n = 7) had higher sIgM levels/signaling if compared with those with isolated del13q or no +12/ NOTCH1ΔCT/del(17p) (n = 87) (Figure 4C).
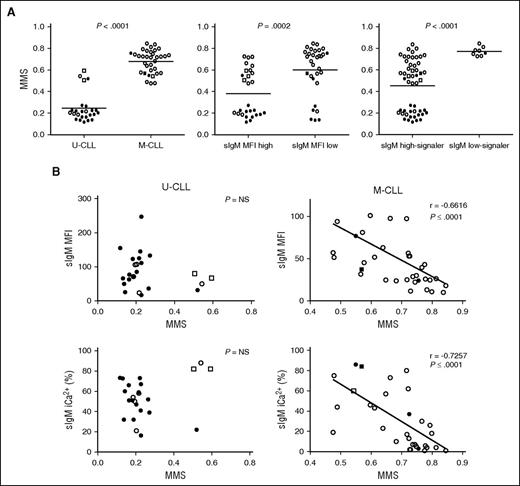
sIgM levels or signaling capacity and DNA methylation maturation status in CLL. DNA methylation profiling of 18 selected regions was determined by MassARRAY in 57 CLL samples and a MMS was calculated. (A) The associations between MMS and IGHV status (U-CLL vs M-CLL), sIgM level status (sIgM MFI high vs sIgM MFI low), and signaling capacity (sIgM high-signaler vs low-signaler) are represented. Horizontal bars indicate mean values. Statistical analysis was performed using the Mann-Whitney test (2 tailed, 95% CI). (B) Correlation analysis between MMS and sIgM levels and signaling capacity in U-CLL and in M-CLL. Statistical analyses were performed using the Spearman rank correlation test. In all panels, empty symbols represent CLL with isolated del13q or negative FISH; black filled symbols represent CLL with either +12 or del(11q) or del(17p) or NOTCH1ΔCT or SF3B1 mutation. Square symbols represent IGHV3-21+ CLL. NS, not significant.
sIgM levels or signaling capacity and DNA methylation maturation status in CLL. DNA methylation profiling of 18 selected regions was determined by MassARRAY in 57 CLL samples and a MMS was calculated. (A) The associations between MMS and IGHV status (U-CLL vs M-CLL), sIgM level status (sIgM MFI high vs sIgM MFI low), and signaling capacity (sIgM high-signaler vs low-signaler) are represented. Horizontal bars indicate mean values. Statistical analysis was performed using the Mann-Whitney test (2 tailed, 95% CI). (B) Correlation analysis between MMS and sIgM levels and signaling capacity in U-CLL and in M-CLL. Statistical analyses were performed using the Spearman rank correlation test. In all panels, empty symbols represent CLL with isolated del13q or negative FISH; black filled symbols represent CLL with either +12 or del(11q) or del(17p) or NOTCH1ΔCT or SF3B1 mutation. Square symbols represent IGHV3-21+ CLL. NS, not significant.
DNA methylation maturation status inversely associates with sIgM levels/signaling capacity in M-CLL
The DNA MMS, which represents the degree of CLL epigenetic maturity, was analyzed in 66 CLL samples, which were selected based on availability of material and tumor cell purity >85%, as previously described.27 The immunogenetic characteristics, sIgM/D level and signaling capacity, and genetic characteristics of the individual samples are described in supplemental Table 2. Correlations between MMS and sIgM were sought in the 57 CLL with full FISH/genetic details available from this selection (supplemental Table 2). U-CLL had a lower MMS (range, 0.12-0.54; median, 0.20) than M-CLL (range, 0.48-0.85; median, 0.68; Figure 5A) as expected from previous findings.27 A lower MMS was found in CLL with higher sIgM levels or signaling capacity than in CLL with low sIgM levels or signaling capacity, respectively (Figure 5A). The cases with an intermediate MMS (0.5 ± 0.1) were enriched in CLL using IGHV3-21, as expected.23,25 IGHV3-21+ CLL were all strong signalers and explained 50% (2 of 4) of the exceptions with higher MMS in U-CLL. No significant correlations between sIgM levels or signaling and methylation maturation were found if U-CLL only were analyzed (Figure 5B). Analysis of M-CLL revealed a strikingly high correlation between maturation of methylation and reduction of sIgM levels (r = 0.67, P < .001) or signaling capacity (r = 0.73, P < .001), and the great majority of these M-CLL carried no genetic lesions (Figure 5B).
Level of sIgM may be an independent prognostic factor of TTFT in CLL
High sIgM levels associated not only with more rapid TTFT, but also with advanced stage, with ZAP-70 ≥20%, CD38 ≥30%, CD49d ≥30% and with shorter LDT and shorter OS (supplemental Figures 6-7). TTFT was used as end point to determine the potential role of sIgM as a clinical prognostic parameter against either known phenotypic, or genetic or methylation-risk categories verified by univariate analysis in this cohort (supplemental Table 3).
When a multivariate analysis adjusted for high sIgM levels and phenotypic markers, CD38 ≥30%, ZAP-70 ≥20%, high sIgM levels scored as the strongest prognostic parameter of short TTFT (Table 2). A multivariate analysis adjusted for high sIgM levels, U-IGHV status, and genetic factors (+12/no FISH lesions, del(11q), and del(17p)/NOTCH1ΔCT) demonstrated that high sIgM levels were also a powerful prognostic factor of TTFT independent from genetics (Table 3).
Cox multivariate analysis of high sIgM and phenotypic prognostic parameters associated with TTFT
| . | SE . | P . | HR (95% CI) . |
|---|---|---|---|
| sIgM MFI > 50 | 0.211 | .000 | 2.501 (1.654-3.782) |
| CD38 ≥ 30% | 0.205 | .011 | 1.682 (1.127-2.513) |
| ZAP-70 ≥ 20% | 0.192 | .003 | 1.781 (1.223-2.594) |
| . | SE . | P . | HR (95% CI) . |
|---|---|---|---|
| sIgM MFI > 50 | 0.211 | .000 | 2.501 (1.654-3.782) |
| CD38 ≥ 30% | 0.205 | .011 | 1.682 (1.127-2.513) |
| ZAP-70 ≥ 20% | 0.192 | .003 | 1.781 (1.223-2.594) |
Number of cases entered in the multivariate analysis = 222; number of events = 124.
Abbreviations are explained in Table 1.
Cox multivariate analysis of high sIgM and molecular prognostic parameters associated with TTFT
| . | SE . | P . | HR (95% CI) . |
|---|---|---|---|
| sIgM MFI > 50 | 0.223 | .015 | 1.718 (1.109-2.662) |
| IGHV ≥ 98% | 0.227 | .004 | 1.917 (1.228-2.993) |
| +12/FISH− | 0.223 | .001 | 2.099 (1.355-3.251) |
| del(11q) | 0.330 | .048 | 1.918 (1.005-3.663) |
| del(17p)/NOTCH1ΔCT* | 0.306 | .493 | 1.233 (0.678-2.245) |
| . | SE . | P . | HR (95% CI) . |
|---|---|---|---|
| sIgM MFI > 50 | 0.223 | .015 | 1.718 (1.109-2.662) |
| IGHV ≥ 98% | 0.227 | .004 | 1.917 (1.228-2.993) |
| +12/FISH− | 0.223 | .001 | 2.099 (1.355-3.251) |
| del(11q) | 0.330 | .048 | 1.918 (1.005-3.663) |
| del(17p)/NOTCH1ΔCT* | 0.306 | .493 | 1.233 (0.678-2.245) |
Number of cases entered in the multivariate analysis = 184; number of events = 108.
Abbreviations are explained in Table 1.
Because no difference in TTFT was observed between patients with NOTCH1ΔCT and del(17p), these lesions were pooled together for multivariate survival analyses (also refer to supplemental Table 3).
For clinical correlations with DNA methylation status, analyses were done against the 3 discrete LP-CLL, IP-CLL, and HP-CLL categories.27 All LP-CLL patients grouped into U-CLL (19 of 23 U-CLL) and were sIgM signalers, whereas M-CLL were either IP-CLL (14 of 43 M-CLL) or HP-CLL (29 of 43 M-CLL) with high or low sIgM levels/signaling, consistent with the previously described broader methylation heterogeneity in M-CLL than U-CLL (supplemental Figure 8).25,27 Hence, we focused on M-CLL with a multivariate analysis adjusted for sIgM levels (high vs low) and methylation status (IP vs HP) to understand relative relevance of these 2 parameters specifically in M-CLL. The analysis revealed a similarly independent value of high sIgM and IP-CLL status in predicting disease progression, despite the low numbers entered (n = 43, supplemental Figure 8).
Discussion
The overall data add new support to the critical role of anergy in CLL. We now observe and report that it is the degree of anergy operating on sIgM, but not on sIgD, which appears to associate most strongly with slower disease progression in patients. This observation reveals intrasubset variability which adds more prognostic information.
Evidence for interaction of CLL cells with putative (super)antigen in vivo is provided by the downregulation of sIgM expression in the tumor cells, which can be reversed in vitro and during circulation following engagement in tissue sites.13,51 In healthy individuals, the natural levels of sIgM in B cells are higher than in CLL,52,53 and in memory B cells are higher than in naive B cells.54 This is not reflected in the leukemic counterparts, M-CLL and U-CLL. The reason for this is that expression levels in CLL are modulated by events in vivo. The major perturbation is antigen-induced anergy which operates in both subsets but appears more profound in M-CLL.1 This is the likely determinant of the differential disease behavior between subsets. However, in both subsets, the degree of anergy is variable, being especially broad in M-CLL. Dissecting this variability within M-CLL now reveals that the factor which most associates with disease progression is the level of sIgM.
An additional influence on disease progression is genetic damage. Here, we learn that sIgM levels/signaling are different in different genetic categories in CLL, suggesting influences also by intrinsic tumor-related factors. This is particularly evident in U-CLL where genetic lesions with inferior prognosis including +12, del(17p) and/or NOTCH1ΔCT associate with higher sIgM. CLL cells with loss of TP53 and NOTCH1ΔCT mutation have a survival and proliferation advantage over those without these lesions.55 The +12 translocation has also been associated with upregulation of levels and signaling of integrins including CD49d and CD38.56 Our data show an association between sIgM with CD49d or CD38 levels in both U-CLL and M-CLL.
Global DNA methylation status of CLL cells is closely related to the B cell of origin and is remarkably preserved posttransformation during disease course.25,57,58 The degree of DNA methylation maturity in CLL (as defined by parallel changes occurring in normal B-cell development) can be used to conveniently divide CLL into 3 subgroups across U-CLL and M-CLL namely LP-CLL, IP-CLL, and HP-CLL.25,27 The highest degree of DNA methylation variability is seen not only among M-CLLs, composed mainly of the HP-CLL methylation subgroup, but also includes many IP-CLLs, as highlighted by the MMS. Analysis of the corresponding sIgM levels now reveals a remarkable inverse relationship between sIgM expression/function and methylation maturation in M-CLL. It appears that cells derived from more mature B cells within the spectrum of M-CLL may be more susceptible to induction of anergy. This could be intrinsic to the normal counterpart of memory-like B cells. Alternatively, it could be due to the nature of the autoantigens.1
An association with DNA methylation maturity and increasingly favorable outcome has been reported and is confirmed here by a significant increase of TTFT in HP-CLLs compared with IP-CLLs (supplemental Figure 8).23,25,27 We now show that both the reduced sIgM and the higher MMS within M-CLL associated with a slower progression of disease. These results add that not only do IP-CLLs differ from HP-CLLs regarding their correlation with prognostic markers and clinical outcome, but also rather simply with sIgM levels and signaling capacity.
Our study highlights the potentially relevant clinical role of sIgM levels in CLL progression. In this cohort, 55% of patients required chemoimmunotherapy, and 16% of them (of which 75% were U-CLL) were treated with inhibitors of BCR pathway-associated kinases that prolonged patients’ survivals.6 Hence, we chose TTFT as the primary end point of disease progression. Our analysis of sIgM levels in separate multivariate analyses with known phenotypic, genetic, or methylation prognostic markers of progression revealed the independent role of sIgM levels. Our data highlight the potential utility of sIgM to identify those CLL with more aggressive behavior, although their routine use in a clinical setting will require standardization and validation in independent cohorts.
In conclusion, we emphasize the importance of sIgM as an indicator of tumor cell origin and behavior. sIgM levels and function appear to reflect the critical factors operating on CLL in vivo, including genetic damage. Not only does it include features of the cell of origin but it is also a meaningful marker of interaction with antigen. This simple marker could now be worthy of further investigations to verify its role to assist prediction of tumor behavior between and within the major subsets, particularly within the heterogeneous M-CLL subset.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Andy S. Davies, Andrew S. Duncombe, Kathy Potter, and Carina Mundy for sample identification, collection, and storage in the South Coast Tissue Bank (Bloodwise grant 12021) and all the patients who consented to participate in this study.
This work was supported by Cancer Research UK (CRUK centre grant C34999/A18087), the Southampton Cancer Research UK Centre and National Institute for Health Research Experimental Cancer Medicine Centre (ECMC), University of Southampton, Bloodwise (grants 16003, 14037, and 12021), the Keanu Eyles Haematology Fellowship for the Cancer Immunology Centre, the Gilead UK & Ireland Oncology Fellowship Programme 2016, the German Federal Ministry of Education and Research CancerEpiSys network (BMBF 031 6049C), and the Virtual Helmholtz Institute (VH-VI-404). I.H. was supported by the ECMC C24563/A15581 grant.
Authorship
Contribution: A.D., S.D., I.T., and I.H. performed research and analyzed data; L.C., M.L., M.R.-Z., and J.S. contributed to genetic analyses; P.W.J. contributed to identification and provision of well-characterized biological samples; C.P. contributed to methylation studies; A.J.S. and G.P. contributed to interpretation of data; C.C.O. performed, analyzed, and interpreted methylation studies and co-wrote the manuscript; F.F. designed the study, and analyzed and interpreted data; and F.K.S. and F.F. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Francesco Forconi, Cancer Sciences Unit, University of Southampton, Cancer Research UK Centre, Somers Cancer Research Building, MP824, Southampton General Hospital, Southampton SO16 6YD, United Kingdom; e-mail: f.forconi@soton.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal