Key Points
CSF3R was the most frequently mutated gene identified in this CEBPAbi AML cohort analyzed by next-generation sequencing.
CEBPAbi AML that have a characteristic transcriptomic profile are more sensitive to JAK inhibitors than CEBPAwt AML.
Abstract
In this study, we analyzed RNA-sequencing data of 14 samples characterized by biallelic CEBPA (CEBPAbi) mutations included in the Leucegene collection of 415 primary acute myeloid leukemia (AML) specimens, and describe for the first time high frequency recurrent mutations in the granulocyte colony-stimulating factor receptor gene CSF3R, which signals through JAK-STAT proteins. Chemical interrogation of these primary human specimens revealed a uniform and specific sensitivity to all JAK inhibitors tested irrespective of their CSF3R mutation status, indicating a general sensitization of JAK-STAT signaling in this leukemia subset. Altogether, these results identified the co-occurrence of mutations in CSF3R and CEBPA in a well-defined AML subset, which uniformly responds to JAK inhibitors and paves the way to personalized clinical trials for this disease.
CEBPA (CCAAT/enhancer binding protein α) encodes a 42-kDa transcription factor essential for differentiation of myeloid progenitor cells. CEBPA regulates expression of the granulocyte colony-stimulating factor receptor (CSF3R) gene, which plays a prominent role in granulocyte differentiation.1 Homozygous deletion of CEBPA in mouse hematopoietic cells leads to a selective loss of CSF3R expression and results in a complete block of neutrophil differentiation.1,2 Acquired mutations in CSF3R are present in a majority of chronic neutrophilic leukemias and atypical (BCR-ABL1–negative) chronic myeloid leukemias,3 which are neoplasms affecting the granulocytic lineage. CSF3R mutations comprise either the membrane-proximal missense mutations or C-terminal truncating mutations proposed to lead to ligand independence and ligand hypersensitivity, respectively.4 CSF3R mutations have also been described in patients with congenital neutropenia treated with granulocyte colony-stimulating factor therapy upon acute myeloid leukemia (AML) transformation,5,6 but they have only been reported in <1% to 2% of AML.3,7,8
Two major categories of collaborating CEBPA mutations have been described in human AML: (1) frameshift (FS) insertions or deletions affecting the N-terminal region resulting in a loss of the 42 kDa protein and overexpression of a shorter 30 kDa CEBPA protein proposed to exhibit a dominant negative activity,9 and (2) in frame mutations in the C-terminal region that alter the basic leucine zipper domain.10 CEBPA-mutated AML carrying mutations in both alleles (CEBPA biallelic [CEBPAbi] AML) represent a distinct subgroup characterized by a normal karyotype (NK) and favorable prognosis.11,12 In most specimens, a combination of N-terminal FS and C-terminal in frame mutations are observed, hereafter called typical CEBPAbi AML. Other combinations of CEBPAbi mutations, hereafter called atypical CEBPAbi AML, were also described.13,14
Gene expression studies have shown that CEBPAbi samples, but not specimens with CEBPA monoallelic mutations, have a distinctive gene expression profile (GEP),15-18 and analyses of small series indicated a possibility that this profile is shared by some atypical CEBPAbi AML.15 Mutations in genes such as GATA2, WT1, and TET2 have been described thus far in CEBPAbi specimens,19-22 including 6 specimens reported in The Cancer Genome Atlas cohort.7
We previously used comparative transcriptomic approaches to report the mutational and transcriptional landscapes of MLL,23 EVI1,24 NUP98-NSD1,25 and CBF26 AML subgroups included in the Leucegene cohort, and also demonstrated that chemical interrogation of a mutation could identify new therapeutic targets in AML.23 We hereby describe RNA-sequencing analysis of the 14 CEBPAbi AML specimens included in our collection, and report new activating signaling mutations in this disease, which revealed the sensitivity of this subgroup to compounds that specifically affect the JAK-STAT signaling pathway.
Methods
Human leukemia and normal samples
The Leucegene project is an initiative approved by the Research Ethics Boards of the Université de Montréal and Maisonneuve-Rosemont Hospital. As part of this project, RNA sequencing of 415 primary AML specimens from various cytogenetic groups was performed, including 110 samples that were also characterized by exome sequencing, as previously described.23 All leukemia samples and paired normal DNA specimens were collected and characterized by the Quebec Leukemia Cell Bank (BCLQ). Normal bone marrow (BM) samples were obtained from the BCLQ and from Lonza, and cord blood from Héma-Québec.23
Next-generation sequencing (NGS) and mutation validations
Sequencing was performed as previously described.23 Sequence data were mapped to the reference genome hg19 according to RefSeq annotations (University of California Santa Cruz; April 16th, 2014). Variants were all identified using CASAVA 1.8.2 or km (https://bitbucket.org/iric-soft/km) approaches according to the previously reported pipeline.24,25 All variants present in 80 genes mutated in myeloid cancers or in acute leukemias were investigated (see supplemental Table 1, available on the Blood Web site). Acquired or germ line origin of these variants not present in the Catalogue of Somatic Mutations in Cancer database were all confirmed by Sanger sequencing of nontumoral DNA from mouth swabs or saliva. Other genes with recurrent variants (ie, in 3 or more CEBPAbi samples) were also analyzed in nontumoral DNA. Samples with CEBPA variant coverage <10× were confirmed by tumor DNA Sanger sequencing. Samples from the CEBPAbi group with no detectable WT1 mutations were also analyzed by tumor complementary DNA sequencing of WT1 exons 6-10 (based on NM_001198551). NRAS, KRAS, and PTPN11 mutations were detected at a variant allele frequency (VAF) ≥5%, and FLT3-internal tandem duplications with a VAF ≥10% were reported.
For the analysis of variants in JAK-related genes, all variants in coding regions of genes in supplemental Table 2 identified using CASAVA 1.8.2 (≥8 variant reads and ≥20× coverage) are reported, after filtering out variants also present in normal samples (n = 67 sequenced populations) and those present in samples resistant to ruxolitinib (IC50 >100 nM). For enrichment calculation, frequency of variant in samples sensitive to ruxolitinib (IC50 <100 nM) was divided by the frequency in normal individuals based on the Single Nucleotide Polymorphism database version 137. The frequency of variants in normal individuals that are not cataloged in the Single Nucleotide Polymorphism database was arbitrarily set at 1/1000. Enrichment was not calculated (NA) for variants present in only 1 sensitive sample.
Normal myeloid populations
The following populations were sorted from normal BM on a BD Aria II cell sorter using the corresponding panels as denoted here. Promyelocytes: propidium iodide (PI)−/CD34−/CD16−/CD11b−/CD33+/CD15+; myelocytes: PI−/CD34−/CD16−/CD11b+/CD13−/CD15+/CD33+; metamyelocytes: PI−/CD34−/CD16int/CD11b+/CD33int/CD15+; and band and segmented granulocytes: PI−/CD34−/CD16high/CD11b+/CD33int/CD13+/CD15+. Granulocytes were sorted from normal peripheral blood by gating on SSC++CD33int cells. Fresh normal BM was purchased from Lonza. The following fluorescence-activated cell sorter antibodies were used: CD33 PE (555450; BD Biosciences), CD34 allophycocyanin (555824; BD Biosciences), CD11b PE-Cy5 (555389; BD Biosciences), CD16 Pacific Blue (558122; BD Biosciences), CD13 allophycocyanin-Cy7 (301710; BD Biosciences), and CD15 FITC (555401; BD Biosciences).
Cell culture and chemical screen
Preparation of cell culture from frozen AML mono-nucleated cells and chemical screen were performed as previously described23,27 using serum-free media supplemented with cytokines, 500 nM SR1 (Alichem) and 500 nM UM729 (Institute for Research in Immunology and Cancer [IRIC]). Compounds were added to seeded cells in serial dilutions (8 dilutions, 1:3, 10 μM down to 4.5 nM) in duplicate wells. The exception was daunorubicin for which dilutions from 1 μM to 0.45 nM were performed. Control wells received dimethyl sulfoxide (0.1%) only. Cell viability was evaluated after 6-day culture using the CellTiterGlo assay (Promega) according to the manufacturer’s instructions. Percentage of inhibition for dose-response curves was calculated as 100 – (100 × [mean luminescence (compound)/mean luminescence (dimethyl sulfoxide)]). IC50 values were calculated using ActivityBase SARview Suite. Dose-response curves were generated using nonlinear regression in GraphPad Prism 4.03. For cases where compounds failed to inhibit AML cell survival/proliferation, IC50 values were arbitrarily reported at the highest dose tested (10 000 nM).
Statistical analyses
Fisher’s exact test was used in the analysis of contingency tables. Analysis of differential gene expression was performed using the Wilcoxon rank-sum test, and the false discovery rate method was applied for global gene analysis as previously described.23
Gene ontology enrichment analysis was performed by computing overlaps with curated gene sets from MSigDB (via a hypergeometric test) using the provided tool on the Broad Institute website. Principal component analyses (PCA) were performed on log10 transformed reads per kilobase per million (RPKM) values using prcomp function in the stats package and visualized using ggbiplot package in R version 3.1.2. Differences in response to small molecules between genetic groups were evaluated using a Wilcoxon rank-sum test performed on IC50 values in R version 3.1.2.
Results and discussion
Fourteen AML samples with CEBPAbi mutations were identified in the Leucegene cohort, including 7 typical and 7 atypical CEBPAbi AML, comprising 3.4% (14/415) of this collection. Positions of mutations are illustrated in Figure 1A and detailed in supplemental Table 3. Baseline characteristics of cohorts are indicated in Table 1. CEBPAbi samples were significantly associated with intermediate risk cytogenetics, French-American-British (FAB) M1 morphology, and higher white blood cell counts.
Gene expression signature of CEBPAbi AML reclassifies atypical CEBPAbi AML. (A) Primary structure of CEBPA and positions of mutations in CEBPAbi samples. (B) Comparative analyses of differentially expressed genes in a typical CEBPAbi AML subgroup. Diamonds correspond to the 95 most differentially expressed genes listed in supplemental Table 4. Genes with low expression in both groups, defined by a mean (log10 [(RPKM + 0.0001) × 10 000]) <4 (corresponding approximately to 1 RPKM) were not included in this analysis. (C) PCA performed on the entire cohort (n = 415) using the 95-gene signature that characterizes typical CEBPAbi AML (orange dots). Atypical CEBPAbi samples (light blue dots) that cluster with typical CEBPAbi AML (defined by the dashed line) are grouped under GEP+ CEBPAbi AML, whereas others are termed GEP− CEBPAbi AML. (D) Classification of CEBPAbi AML based on CEBPA mutations and gene expression. (E) Representation of CEBPA mutations in atypical GEP+ or GEP− CEBPAbi AML. (F) HOXA9 expression in GEP+ and GEP− CEBPAbi AML samples. (G) Gene expression of CEBPA (left) and CSF3R (right) in the entire 415 AML sample cohort. C-ter, C-terminal; CEBPAmono, monoallelic CEBPA mutation; IF, inframe; Mid, middle; MS, missense; NS, nonsense; N-ter, N-terminal; PC, principal component.
Gene expression signature of CEBPAbi AML reclassifies atypical CEBPAbi AML. (A) Primary structure of CEBPA and positions of mutations in CEBPAbi samples. (B) Comparative analyses of differentially expressed genes in a typical CEBPAbi AML subgroup. Diamonds correspond to the 95 most differentially expressed genes listed in supplemental Table 4. Genes with low expression in both groups, defined by a mean (log10 [(RPKM + 0.0001) × 10 000]) <4 (corresponding approximately to 1 RPKM) were not included in this analysis. (C) PCA performed on the entire cohort (n = 415) using the 95-gene signature that characterizes typical CEBPAbi AML (orange dots). Atypical CEBPAbi samples (light blue dots) that cluster with typical CEBPAbi AML (defined by the dashed line) are grouped under GEP+ CEBPAbi AML, whereas others are termed GEP− CEBPAbi AML. (D) Classification of CEBPAbi AML based on CEBPA mutations and gene expression. (E) Representation of CEBPA mutations in atypical GEP+ or GEP− CEBPAbi AML. (F) HOXA9 expression in GEP+ and GEP− CEBPAbi AML samples. (G) Gene expression of CEBPA (left) and CSF3R (right) in the entire 415 AML sample cohort. C-ter, C-terminal; CEBPAmono, monoallelic CEBPA mutation; IF, inframe; Mid, middle; MS, missense; NS, nonsense; N-ter, N-terminal; PC, principal component.
Characteristics of CEBPAbi and non-CEBPAbi AML cohorts
| . | CEBPAbi (n = 14) . | Non-CEBPAbi (n = 401) . | P . |
|---|---|---|---|
| T-AML | 0 | 26 (6.5%) | — |
| Gender | |||
| Male | 9 (64.3%) | 226 (56.4%) | — |
| Female | 5 (35.7%) | 175 (43.6%) | — |
| Age (y) | |||
| Median | 54.5 | 58 | — |
| Range | 27-84 | 17-87 | — |
| WBC count (× 109/L) | |||
| Median | 88 | 31 | .04 |
| Range | 4-177 | 0.8-361.2 | — |
| Karyotype | |||
| Intermediate | 12 (85.7%) | 207 (51.6%) | .006 |
| NK | 7 (50%) | 125 (31.2%) | — |
| Non-intermediate | 2 (14.3%) | 192 (47.9) | — |
| Undetermined | 0 | 2 (0.5%) | — |
| FAB subtype | |||
| M0 | 0 | 27 (6.7%) | — |
| M1 | 8 (57.1%) | 107 (26.7%) | .03 |
| M2 | 3 (21.4%) | 49 (12.2%) | — |
| M3 | 0 | 15 (3.7%) | — |
| M4 | 0 | 57 (14.2%) | — |
| M5 | 0 | 66 (16.5%) | — |
| M6 | 0 | 10 (2.5%) | — |
| M7 | 0 | 3 (0.7%) | — |
| Not classifiable | 3 (21.4%) | 67 (16.7%) | — |
| Type of CEBPA mutation | |||
| Biallelic typical | 7 (50%) | — | — |
| Biallelic atypical | 7 (50%) | — | — |
| Monoallelic | — | 11 (2.7%) | — |
| . | CEBPAbi (n = 14) . | Non-CEBPAbi (n = 401) . | P . |
|---|---|---|---|
| T-AML | 0 | 26 (6.5%) | — |
| Gender | |||
| Male | 9 (64.3%) | 226 (56.4%) | — |
| Female | 5 (35.7%) | 175 (43.6%) | — |
| Age (y) | |||
| Median | 54.5 | 58 | — |
| Range | 27-84 | 17-87 | — |
| WBC count (× 109/L) | |||
| Median | 88 | 31 | .04 |
| Range | 4-177 | 0.8-361.2 | — |
| Karyotype | |||
| Intermediate | 12 (85.7%) | 207 (51.6%) | .006 |
| NK | 7 (50%) | 125 (31.2%) | — |
| Non-intermediate | 2 (14.3%) | 192 (47.9) | — |
| Undetermined | 0 | 2 (0.5%) | — |
| FAB subtype | |||
| M0 | 0 | 27 (6.7%) | — |
| M1 | 8 (57.1%) | 107 (26.7%) | .03 |
| M2 | 3 (21.4%) | 49 (12.2%) | — |
| M3 | 0 | 15 (3.7%) | — |
| M4 | 0 | 57 (14.2%) | — |
| M5 | 0 | 66 (16.5%) | — |
| M6 | 0 | 10 (2.5%) | — |
| M7 | 0 | 3 (0.7%) | — |
| Not classifiable | 3 (21.4%) | 67 (16.7%) | — |
| Type of CEBPA mutation | |||
| Biallelic typical | 7 (50%) | — | — |
| Biallelic atypical | 7 (50%) | — | — |
| Monoallelic | — | 11 (2.7%) | — |
Samples classified under “typical CEBPAbi” mutations have a combination of heterozygous N-terminal FS or nonsense and C-terminal in frame mutations. Other combinations are considered “atypical CEBPAbi.”
P values are based on 2-tailed Fisher’s exact test or Wilcoxon rank-sum test.
T-AML, therapy-related AML; WBC, white blood cell counts.
Typical CEBPAbi AML shows a uniform GEP
The typical CEBPAbi specimens are best characterized by a total of 95 genes (Figure 1B; supplemental Table 4). CEBPAbi AML was marked by low expression of HOXA and HOXB genes, MEIS1 and CPNE8, a profile partially comparable to that determined for t(8;21) specimens (supplemental Figure 1). Likewise, both leukemia subgroups express high levels of TRH, CD96, UGT2B11, MYO18B, and LSGN, whereas other genes, such as SHD, were most specific to CEBPAbi AML. This signature is partially consistent but also additive to other signatures published, such as AML cluster 4 reported by Valk et al.16
Gene expression and mutation profile help categorize atypical CEBPAbi AML
Using the 95-gene signature derived from typical CEBPAbi AML, we next performed a PCA on the entire Leucegene cohort and confirmed the distinctive GEP of typical CEBPAbi samples compared with other leukemias, and to specimens with CEBPA monoallelic mutations (orange and dark blue dots in Figure 1C). This analysis also showed that 4 out of 7 atypical CEBPAbi samples clustered with the CEBPAbi (light blue dots in dashed zone, Figure 1C), suggesting they are transcriptionally similar to typical CEBPAbi leukemia (hereafter grouped under GEP+ and schematized in Figure 1D). No recurrent CEBPA mutation pattern was found in the 4 atypical CEBPAbi GEP+ specimens, thus suggesting that expression profiling may help to categorize such patients (Figure 1D-E). HOXA (eg, HOXA9) gene expression may be sufficient to identify CEBPAbi GEP+ specimens in clinical settings, ie, any HOXAlow atypical CEBPA specimen is likely similar to typical CEBPAbi AML (Figure 1F). The 3 CEBPAbi GEP− samples were associated with additional genetic rearrangements and/or mutations, which are more typical of non–CEBPA-mutated AML (supplemental Table 5). CEBPA expression is higher in all CEBPAbi AML, but more significantly in CEBPAbi GEP+ AML, in line with the published positive auto-regulatory loop of this transcription factor (left panel in Figure 1G). CSF3R expression is lower in CEBPAbi GEP+ specimens only (right panel in Figure 1G).
CSF3R/STAT5 mutations are the most frequent mutations in CEBPAbi AML
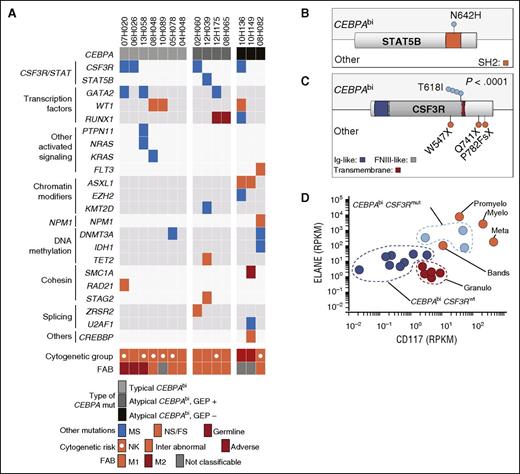
We next investigated the mutations in typical and atypical CEBPAbi cases. A total of 22 additional genes were mutated in this subset: CSF3R (4/14, 29%), WT1, GATA2, and RUNX1 (3/14), DNMT3A and ASXL1 (2/14), STAT5B, FLT3, KRAS, NPM1, IDH1, TET2, PTPN11, NRAS, RAD21, SMC1A, STAG2, U2AF1, ZRSR2, EZH2, CREBBP, and KMT2D (1/14) (Figure 2A; supplemental Table 6). These results are consistent with other targeted mutation analyses also reporting recurrent GATA2 and WT1 mutations in the CEBPAbi AML subgroup (3/14 vs 10/401, P = .007 and 3/14 vs 22/401, P = .045, respectively).19-22 The only STAT5B mutation in this cohort was in the CEBPAbi sample (1/14 vs 0/401) and consisted of an N642H substitution, which is known to increase STAT5 transcriptional activity and phosphorylation (Figure 2B).28 STAT5B mutations have been described in large granular lymphocytic leukemias28 and in other T-cell neoplasms,29,30 but not in AML.
Mutational landscape of CEBPAbi AML. (A) Mutation profile of CEBPAbi AML. Samples are grouped together according to the type of CEBPA mutations: typical CEBPAbi, atypical GEP+ CEBPAbi, and atypical GEP− CEBPAbi, illustrated by shades of gray from light to dark. Each column represents a patient sample. (B-C) Primary structures of STAT5B (B) and CSF3R (C) proteins with corresponding positions of mutations in CEBPAbi and non-CEBPAbi (other) AML. (D) Expression of ELANE and CD117 in GEP+ CEBPAbi compared with normal sorted granulocyte populations from peripheral blood (n = 5, red) and normal marrow precursors (n = 4, orange). FNIII, fibronectin type III; Granulo, granulocytes; Ig, immunoglobulin; Inter, intermediate; Meta, metamyelocytes; Mut, mutated; Myelo, myelocytes; NPM1, nucleophosmin 1; Promyelo, promyelocytes.
Mutational landscape of CEBPAbi AML. (A) Mutation profile of CEBPAbi AML. Samples are grouped together according to the type of CEBPA mutations: typical CEBPAbi, atypical GEP+ CEBPAbi, and atypical GEP− CEBPAbi, illustrated by shades of gray from light to dark. Each column represents a patient sample. (B-C) Primary structures of STAT5B (B) and CSF3R (C) proteins with corresponding positions of mutations in CEBPAbi and non-CEBPAbi (other) AML. (D) Expression of ELANE and CD117 in GEP+ CEBPAbi compared with normal sorted granulocyte populations from peripheral blood (n = 5, red) and normal marrow precursors (n = 4, orange). FNIII, fibronectin type III; Granulo, granulocytes; Ig, immunoglobulin; Inter, intermediate; Meta, metamyelocytes; Mut, mutated; Myelo, myelocytes; NPM1, nucleophosmin 1; Promyelo, promyelocytes.
The most frequent mutations in CEBPAbi subgroup affected CSF3R in 4/14 (29%) (Figure 2A; supplemental Figure 2). CSF3R mutations were not previously reported in CEBPAbi AML and they were strongly associated with this subgroup, as only 3 additional CSF3R mutations were identified in the entire cohort (4/14 vs 3/401, P < .0001; Figure 2C). CSF3R T618I “membrane proximal” mutation characterized all 4 CEBPAbi samples. In contrast, this specific point mutation was not found in the 3 non-CEPBA specimens, which carried nonsense or FS CSF3R mutations (Figure 2C; supplemental Table 7). A single T618I mutation was observed in The Cancer Genome Atlas in a sample with CEBPA monoallelic mutation.7 CSF3R mutations were also found in 1.9% of pediatric AML in an analysis that did not perform CEBPA mutational analysis.8 Rare CSF3R T618I mutations were also identified in AML samples with no CEBPA mutation, indicating that they can also occur at a low frequency in other genetic contexts.31 VAF analysis suggests that co-occurrence of CSF3R and CEBPA mutations were found in the dominant clone (supplemental Table 8).
CSF3RT618I mutated CEBPAbi specimens did not show any distinctive clinical laboratory features (supplemental Table 9) but presented a defined transcriptomic profile (supplemental Figure 3) when compared with their WT CSF3R counterparts. Gene ontology term enrichment analyses showed a marked enrichment in defense and organisms response genes (supplemental Table 10). In particular, these specimens expressed significantly higher levels of genes associated with myeloid maturation, such as ELANE and CD117, which upon analysis of larger patient cohorts may become useful in identifying CSF3R mutated samples (Figure 2D).
Chemical interrogation of CEBPAbi and CSF3R mutated AML
CSF3R signals predominantly through the JAK-STAT pathway.3 Considering the high frequency of activating T618I CSF3R mutations detected in CEBPAbi AML, we conducted a targeted chemical screen employing a collection of compounds enriched for JAK inhibitors (n = 6; supplemental Table 11). For this study, we used our recently described culture system that preserves the integrity of leukemia blasts and leukemia stem cell activity.27 Cytotoxic activity of the selected molecules was measured in dose-response studies on a total of 28 primary AML specimens, including all 14 CEBPAbi AML samples described in the previous sections and 14 randomly selected CEBPAwt NK controls (supplemental Table 12).
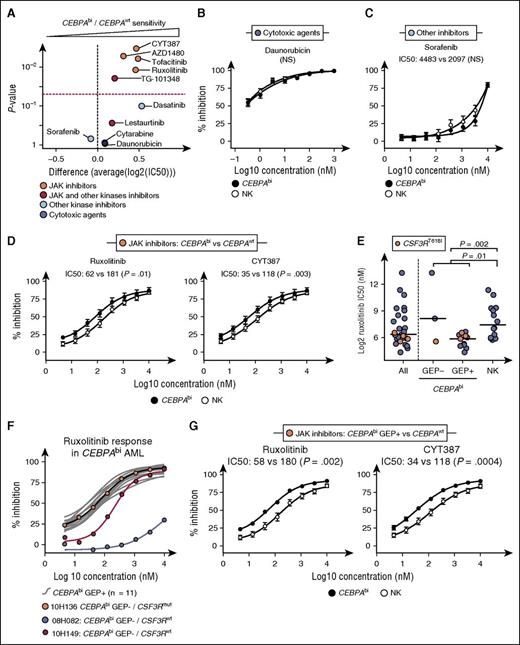
Responses of CEBPAbi specimens (n = 14) and NK CEBPAwt AML (n = 14) to the positive controls cytarabine and daunorubicin were comparable (Figure 3A-B and supplemental Figure 4A, compounds IC50 in supplemental Table 13, and P values in supplemental Table 14). Likewise, these 2 AML subgroups were equally sensitive to the multikinase inhibitors sorafenib and dasatinib reported to lack JAK inhibitory activity (Figure 3A,C; supplemental Figure 4B). In contrast, CEBPAbi AML were statistically more sensitive than control NK AML to inhibitors which more specifically targeted JAK proteins (ruxolitinib, CYT387, tofacitinib, and AZD1480) (Figure 3A,D; supplemental Figure 4C). Although of high potency, compounds such as lestaurtinib and TG-101348 that target JAK and other kinases were less discriminatory (red dots in Figure 3A and supplemental Figure 4D).
CEBPAbi AML is uniformly sensitive to JAK inhibitors. (A) Volcano plot showing the comparative sensitivity of CEBPAbi AML (n = 14) vs CEBPAwt NK AML (n = 14) to selected small molecules. Dashed red line corresponds to P = .05. (B-D) Mean dose-response curves with SEM and IC50-associated statistics comparing CEBPAbi AML (n = 14) to CEBPAwt NK AML (n = 14) for selected compounds as follows: (B) daunorubicin, (C) sorafenib, and (D) ruxolitinib and CYT387. Results for other compounds are shown in supplemental Figure 4. (E) IC50 for ruxolitinib in all samples (n = 28) (left), CEBPAbi GEP− AML (n = 3) and CEBPAbi GEP+ AML (n = 11) (middle), and NK AML (n = 14) (right). CSF3R T618 mutated samples are indicated in orange. Horizontal bars represent medians. (F) Individual dose-response curves (gray) for GEP+ and GEP− (black, red, and blue curves) CEBPAbi AML. Results for other JAK inhibitors are shown in supplemental Figure 6. (G) Mean dose-response curves with SEM and IC50-associated statistics comparing CEBPAbi GEP+ AML (n = 11) to CEBPAwt NK AML (n = 14) for selected JAK inhibitors. Results for other JAK inhibitors are shown in supplemental Figure 7. P values were calculated using a Wilcoxon rank-sum test on IC50 values. NS, not significant; SEM, standard error of the mean.
CEBPAbi AML is uniformly sensitive to JAK inhibitors. (A) Volcano plot showing the comparative sensitivity of CEBPAbi AML (n = 14) vs CEBPAwt NK AML (n = 14) to selected small molecules. Dashed red line corresponds to P = .05. (B-D) Mean dose-response curves with SEM and IC50-associated statistics comparing CEBPAbi AML (n = 14) to CEBPAwt NK AML (n = 14) for selected compounds as follows: (B) daunorubicin, (C) sorafenib, and (D) ruxolitinib and CYT387. Results for other compounds are shown in supplemental Figure 4. (E) IC50 for ruxolitinib in all samples (n = 28) (left), CEBPAbi GEP− AML (n = 3) and CEBPAbi GEP+ AML (n = 11) (middle), and NK AML (n = 14) (right). CSF3R T618 mutated samples are indicated in orange. Horizontal bars represent medians. (F) Individual dose-response curves (gray) for GEP+ and GEP− (black, red, and blue curves) CEBPAbi AML. Results for other JAK inhibitors are shown in supplemental Figure 6. (G) Mean dose-response curves with SEM and IC50-associated statistics comparing CEBPAbi GEP+ AML (n = 11) to CEBPAwt NK AML (n = 14) for selected JAK inhibitors. Results for other JAK inhibitors are shown in supplemental Figure 7. P values were calculated using a Wilcoxon rank-sum test on IC50 values. NS, not significant; SEM, standard error of the mean.
Specimens with activating mutations in CSF3R were among the most sensitive to ruxolitinib (orange dots in Figure 3E) and to the other 3 most specific JAK inhibitors (supplemental Figure 5). Of the 3 CEBPAbi GEP− samples, the highest sensitivity to JAK inhibitors was observed for the CSF3R-mutated specimen (Figure 3E, second column and Figure 3F; see also supplemental Figure 6 for other JAK inhibitors). These results are in agreement with those of Maxson et al,3 who reported a correlation between sensitivity to JAK inhibitors and CSF3RT618I mutation in 2 other related diseases, T acute lymphoblastic leukemia and chronic neutrophilic leukemia.
CEBPAbi GEP+ but not GEP− AML are homogeneously sensitive to JAK inhibitors
The 11 CEBPAbi GEP+ specimens tested, irrespective of their CSF3R mutation status, were equally sensitive to the JAK inhibitors (Figure 3E, third column for ruxolitinib). The uniform response to JAK inhibition in the homogeneous CEBPAbi GEP+ subgroup stands in sharp contrast to the heterogeneous response determined for the NK control group (Figure 3E, fourth column) and the GEP− specimens, and strengthens our hypothesis that the CEBPAbi GEP− and GEP+ are distinct entities (Figure 3G; supplemental Figure 7).
These results suggest that networks or pathways upstream of JAK-STAT are aberrantly activated in the majority of CEBPAbi GEP+ specimens and, less frequently in other NK AML. Activating mutations in CSF3R could account for responses in 3 out of 11 CEBPAbi GEP+, but not for either the remaining 8 CEBPAbi GEP+ specimens and/or the sensitive NK AML specimens (supplemental Table 15). To further investigate this observation, we systematically analyzed all variants present in genes (n = 167) relevant to the JAK network, excluding its proposed downstream targets (supplemental Table 2). In the addition of the CSF3R T618I mutation described above, we detected 2 recurrent variants, JAK2 L383V and EPHB6 S166F, characterized by a 7.4- and >117-fold observed/expected enrichment ratios, respectively (supplemental Table 16). Both variants were confirmed to be germ line. Interestingly, the S166F substitution within the ligand-binding domain of EPHB6 is predicted to affect the ligand binding and consequently activity of this receptor. Moreover, this variant allele detected exclusively in 2 CEBPAbi GEP+ samples has never been described before. We also found a large number of nonrecurrent variants in several cytokine receptors in these specimens (supplemental Table 16), but larger patient cohorts would be required to select potential candidates for functional validation studies.
In conclusion, this study documents for the first time recurrent activating CSF3R mutations in AML with a strict association to the rare CEBPAbi genetic subgroup. Targeted analyses in other cohorts will precise the frequency of CSF3R mutation in CEBPAbi and non-CEBPAbi AML as well. Of interest, Maxson et al recently presented similar findings.32 As might be anticipated, these CSF3R mutated specimens are sensitive to JAK inhibition. Most notably, our study also shows that CEBPAbi GEP+ AMLs are uniformly sensitive to JAK inhibition, raising the possibility that selective genetic pressure resulted in a dependence on the JAK-STAT signaling pathway. An unexpected frequency of variant alleles in JAK-STAT network genes provides a pipeline for future exploration. Considering that most molecules tested herein are available drugs, these studies suggest that JAK inhibitor repositioning could thus represent a true example of therapy targeting a specific well-defined subset of AML patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Muriel Draoui for project coordination, Sophie Corneau for sample coordination, Marianne Arteau and Raphaëlle Lambert at the IRIC genomics platform for sequencing, Jean Duchaine at the IRIC high-throughput screening platform, and Laura Simon. The collaboration of BCLQ coinvestigators and the dedicated work of BCLQ staff, namely Giovanni d’Angelo, Claude Rondeau, and Sylvie Lavallée are also acknowledged, as well as the contribution of Bruno Lamontagne, Guylaine Lépine, and Julie Bergeron in Maisonneuve-Rosemont Hospital Molecular Biology Laboratory.
This work was supported by the Government of Canada through Genome Canada and the Ministère de l’économie, de l’innovation et des exportations du Québec through Génome Québec, with supplementary funds from AmorChem. Support from the Canadian Cancer Society Research Institute to G.S. is also acknowledged. G.S. and J.H. are recipients of research chairs from the Canada Research Chair program and Industrielle-Alliance (Université de Montréal), respectively. BCLQ is supported by grants from the Cancer Research Network of the Fonds de recherche du Québec–Santé. RNA-Seq read mapping and transcript quantification were performed on the supercomputer Briaree from Université de Montréal, managed by Calcul Québec and Compute Canada. The operation of this supercomputer is funded by the Canada Foundation for Innovation, NanoQuébec, Réseau de médecine génétique appliquée, and the Fonds de recherche du Québec-Nature et technologies. V.-P.L. is supported by a fellowship from the Cole Foundation. C.P. was supported by postdoctoral fellowships from German Cancer Aid and the Cole Foundation.
Authorship
Contribution: V.-P.L. contributed to project conception, analyzed NGS and chemical screen data, generated all figures, tables, and supplementary material, and was the main author of this paper; G.S. contributed to project conception and coordination and cowrote the paper; J.H. contributed to project conception, analyzed the cytogenetic and fluorescence in situ hybridization studies, provided all the AML samples, and edited the manuscript; P.G. processed the raw NGS data; G.B. codeveloped the analytical pipeline; S.L. was responsible for supervision of the bioinformatics team and of statistical analyses; I.B. performed data validation and chemical screen; J.K. conceived and performed the chemical screen; A.M. was responsible for the chemistry team as part of the Leucegene project and analyzed the chemical screen data; S.M. contributed to the selection of compounds and interpretation of results with C.J.G; and C.P. processed and sequenced normal peripheral and BM populations.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guy Sauvageau, Institute for Research in Immunology and Cancer, PO Box 6128, Station Centre-Ville, Montreal, QC H3C 3J7, Canada; e-mail: guy.sauvageau@umontreal.ca; and Josée Hébert, Banque de cellules leucémiques du Québec, 5415 L'Assomption Blvd, Montreal, QC H1T 2M4, Canada; e-mail: josee.hebert@umontreal.ca.

![Figure 1. Gene expression signature of CEBPAbi AML reclassifies atypical CEBPAbi AML. (A) Primary structure of CEBPA and positions of mutations in CEBPAbi samples. (B) Comparative analyses of differentially expressed genes in a typical CEBPAbi AML subgroup. Diamonds correspond to the 95 most differentially expressed genes listed in supplemental Table 4. Genes with low expression in both groups, defined by a mean (log10 [(RPKM + 0.0001) × 10 000]) <4 (corresponding approximately to 1 RPKM) were not included in this analysis. (C) PCA performed on the entire cohort (n = 415) using the 95-gene signature that characterizes typical CEBPAbi AML (orange dots). Atypical CEBPAbi samples (light blue dots) that cluster with typical CEBPAbi AML (defined by the dashed line) are grouped under GEP+ CEBPAbi AML, whereas others are termed GEP− CEBPAbi AML. (D) Classification of CEBPAbi AML based on CEBPA mutations and gene expression. (E) Representation of CEBPA mutations in atypical GEP+ or GEP− CEBPAbi AML. (F) HOXA9 expression in GEP+ and GEP− CEBPAbi AML samples. (G) Gene expression of CEBPA (left) and CSF3R (right) in the entire 415 AML sample cohort. C-ter, C-terminal; CEBPAmono, monoallelic CEBPA mutation; IF, inframe; Mid, middle; MS, missense; NS, nonsense; N-ter, N-terminal; PC, principal component.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/24/10.1182_blood-2016-03-705053/4/m_3054f1.jpeg?Expires=1769299823&Signature=rzvAf0W4AUjapSn4bcvDBYfi259V~3DEZIPWwKiui9M~H-NbvhdNrwirs3PmNLWmDfG~7RMdYf03eYY20rdS6fA4-jXFu5-OqL-Mh0G1fz9SOvQLtlM1m9E6xUF1KArCH~7JBFa1ExIBHO9qMWZ2loDpkip82FL42Np3TBXPkEniupcRaPOMT2LcrZrT6vLoswJCLyHDpe1ckPnZ5RgVelHEBj-FzjssWI~H~~MBjNJkbcINkTOb7u7x79M9AGeuuPET5pkfcvgq0d2PHf7bwpLwgY3bG1jfkqmftjpGmUA~Cy5yet-jiHfp5QN-vqTANWDHDcpWvZXL8DWMJyaqdA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)