To the editor:
Childhood cancers represent distinct clinical entities, often with unique genomic alterations and therapeutic responses that differ from cancers arising in adults. Pediatric acute myeloid leukemia (AML) comprises ∼25% of childhood leukemias.1 In contrast to acute lymphoblastic leukemia, which has a 90% survival rate, outcomes for pediatric AML patients remain poor, with an ∼50% relapse rate despite intensive regimens.1 Our limited understanding of the genetic alterations in pediatric AML has hindered development of targeted therapeutic strategies. Given that AML is primarily an adult disease, until recently, our understanding of pediatric AML had been informed by data generated in adults. Yet, some newly discovered mutations in adult AML are rare or entirely lacking in pediatric AML, thus validating critical differences in the pathogenesis. The Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative (https://ocg.cancer.gov/programs/target), a collaboration between the Children’s Oncology Group (COG) and the National Institutes of Health National Cancer Institute, has addressed this issue by large-scale genomic analysis of pediatric AML. For the first time, “pediatric-specific” genomic lesions are being defined that may alter the therapeutic options in childhood AML.
Through the TARGET initiative, comprehensive genomic analysis was performed on 186 cases of pediatric AML, 95 with matched relapse and all with matched remission samples in the discovery phase. Samples were marrow or peripheral blood. Research was approved by the appropriate review board and conducted in accordance with the Declaration of Helsinki. The analysis included whole genome (WGS) and exome sequencing from which a custom capture panel of ∼200 genes of interest was generated. A total of 787 samples were then deep sequenced using this custom capture panel, including samples from the discovery cohort. All sequencing files were deposited to the Database of Genotypes and Phenotypes under TARGET AML substudy ID phs000465 (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000465.v10.p3). Through these efforts, we have identified oncogenic colony-stimulating factor 3 receptor (CSF3R) mutations in pediatric AML.
CSF3R, also known as granulocyte colony-stimulating factor (G-CSF) receptor is a major driver of neutrophil proliferation and differentiation.2 Upon binding to the G-CSF, CSF3R activates a pro-neutrophil program by signaling through downstream cytoplasmic tyrosine kinases, such as JAK3,4 and SRC5-8 family members. Activating CSF3R mutations are found in the majority of patients (∼80%) with chronic neutrophilic leukemia (CNL) and a much smaller fraction of patients with atypical chronic myeloid leukemia.9,10 CNL is a rare myeloproliferative disorder characterized by high levels of neutrophils.11 The mutations fall into 2 classes,9 extracellular point mutations that render the receptor ligand-independent (such as T618I, aka T595I),12,13 and truncations of the cytoplasmic domain that lead to receptor over-expression on the cell surface.14,15 The most common CSF3R mutation, T618I, is sensitive to inhibition of the JAK pathway downstream of the receptor.9 Animal models16 and clinical data from a few patients9,17 have validated JAK inhibition as a potential therapeutic avenue in this disease. A clinical trial is now underway to test whether a JAK inhibitor will be effective against these diseases (#NCT02092324).
CSF3R mutations are rare in adult AML with an estimated frequency of 0.5% to 1%.9,18 However, our efforts through the TARGET AML initiative demonstrated the presence of 19 patients with CSF3R mutations in 787 cases of pediatric AML (2.4%). These somatic mutations include some variants previously identified in adult CNL (T615A, T618I, T640N, and S783fs9 ), as well as a number of additional truncating mutations (Q749X, Q754X, Y767fs, Y787X, and P819/820fs) (Figure 1A). The most common mutation was T618I, which was found in 11/19 CSF3R-mutated cases. The frequency of CSF3R mutations was higher than seen previously in another cohort of pediatric AML.19 This could be because the deep sequencing strategy employed in the TARGET study has a higher sensitivity than the Sanger sequencing employed in the previous study, and also partially because the previous study did not perform sequencing of exon 15, where the CSF3R T640N mutation resides, which was found in 2 cases in our cohort.19 For all of the cases where WGS showed a mutant allele frequency of >10%, we confirmed the existence of the variants by Sanger sequencing (data not shown). One patient had both a T615A and a T618I mutation, which were both at low allele frequency and could therefore be in different minor clones. In CNL, CSF3R truncation mutations often occur alongside an oncogenic point mutation.11 However, in this cohort of pediatric AML, the truncation mutations and point mutations were mutually exclusive. This finding may be indicative of the difference in biological underpinnings of these 2 myeloid malignancies.
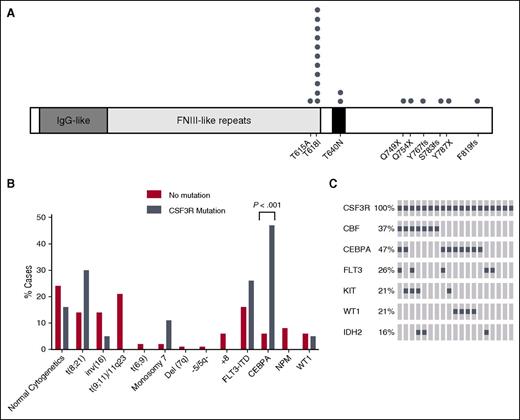
CSF3R mutations co-occur with CEBPA mutations in pediatric AML. (A) Schematic showing the locations of CSF3R mutations found in pediatric AML. One patient had both the T618I and T615A mutations. (B) The percentage of cytogenetic abnormalities (obtained from clinical data) or gene mutations for patients with transforming CSF3R mutations (gray) or CSF3R non-mutated cases (red) are shown. (C) Transforming CSF3R mutation overlap with other gene mutations. The most frequent genomic alterations are shown by OncoPrint analysis. Each column represents a case with a transforming CSF3R mutation. Mutations are represented by a gray square. Some 74% of these cases have either a CBF rearrangement or CEBPA mutation, and there is very minimal overlap between CBF and CEBPA. Custom OncoPrints were generated using the OncoPrinter function of the cBioPortal website (www.cbioportal.org). FNIII, fibronectin type III; IDH2, isocitrate dehydrogenase 2; IgG, immunoglobulin G; KIT, KIT proto-oncogene receptor tyrosine kinase; WT, wild-type.
CSF3R mutations co-occur with CEBPA mutations in pediatric AML. (A) Schematic showing the locations of CSF3R mutations found in pediatric AML. One patient had both the T618I and T615A mutations. (B) The percentage of cytogenetic abnormalities (obtained from clinical data) or gene mutations for patients with transforming CSF3R mutations (gray) or CSF3R non-mutated cases (red) are shown. (C) Transforming CSF3R mutation overlap with other gene mutations. The most frequent genomic alterations are shown by OncoPrint analysis. Each column represents a case with a transforming CSF3R mutation. Mutations are represented by a gray square. Some 74% of these cases have either a CBF rearrangement or CEBPA mutation, and there is very minimal overlap between CBF and CEBPA. Custom OncoPrints were generated using the OncoPrinter function of the cBioPortal website (www.cbioportal.org). FNIII, fibronectin type III; IDH2, isocitrate dehydrogenase 2; IgG, immunoglobulin G; KIT, KIT proto-oncogene receptor tyrosine kinase; WT, wild-type.
Sequencing of the CSF3R exon 14 hotspot (encompassing the T615A and T618I mutations) in diagnostic specimens from 396 adult AML patients enrolled on the Southwest Oncology Group S0106 study did not identify any CSF3R mutations, highlighting the increased prevalence of CSF3R mutations in pediatric over adult AML (data not shown). Clinical characteristics of patients with CSF3R mutations are detailed in supplemental Table 1, available on the Blood Web site.
We next compared the characteristics of patients with CSF3R mutations vs the nonmutated cohort (Table 1). The association of CSF3R variants with CCAAT/enhancer-binding protein alpha (CEBPA) mutations (as detected by a clinical Sanger-based assay) was very strong with frequencies of 47% vs 6% (P < .001), which is approximately eightfold higher than in the pediatric AML patients without CSF3R mutations (Figure 1B). CEBPA is a transcription factor that is expressed in early myeloid precursors with increasing expression during differentiation to mature granulocytes. The mutations in CEBPA occur in either the N-terminal or C-terminal portion of the protein and are dominant negative or inactivating, respectively.20-22 Mutation of CEBPA is thought to block myeloid differentiation and thus promote AML.23
Clinical and genomic characteristics of pediatric AML patients with or without CSF3R mutations
| . | No mutation (N = 768) . | CSF3R mutation (N = 19) . | No mutation vs CSF3R mutation . | ||
|---|---|---|---|---|---|
| Characteristics . | N . | (%) . | N . | (%) . | P . |
| Gender | |||||
| Male | 389 | 51 | 15 | 79 | .015 |
| Female | 379 | 49 | 4 | 21 | — |
| Age (y) | |||||
| Median (range) | 10.08 | (0.02-29.84) | 10.87 | (2.38-22.77) | .133 |
| 0-2 y | 183 | 24 | 1 | 5 | .059 |
| 3-10 y | 229 | 30 | 9 | 47 | .100 |
| +11 y | 356 | 46 | 9 | 47 | .930 |
| FAB | |||||
| M0 | 19 | 3 | 0 | 0 | 1.000 |
| M1 | 84 | 13 | 2 | 13 | 1.000 |
| M2 | 164 | 25 | 7 | 47 | .153 |
| M4 | 166 | 25 | 4 | 27 | 1.000 |
| M5 | 150 | 22 | 0 | 0 | .034 |
| M6 | 12 | 2 | 0 | 0 | 1.000 |
| M7 | 33 | 5 | 0 | 0 | 1.000 |
| Other | 39 | 6 | 2 | 13 | .260 |
| Unknown | 101 | — | 4 | — | — |
| Cytogenetics | |||||
| Normal | 176 | 24 | 3 | 16 | .588 |
| t(8;21) | 108 | 15 | 5 | 26 | .174 |
| inv(16) | 102 | 14 | 1 | 5 | .495 |
| t(9;11)/11q23 | 153 | 21 | 0 | 0 | .034 |
| t(6;9) | 12 | 2 | 0 | 0 | 1.000 |
| Monosomy 7 | 12 | 2 | 2 | 11 | .042 |
| Del(7q) | 5 | 1 | 0 | 0 | 1.000 |
| −5/5q- | 8 | 1 | 0 | 0 | 1.000 |
| +8 | 41 | 6 | 0 | 0 | .618 |
| Other | 117 | 16 | 8 | 42 | .005 |
| Missing | 34 | — | 0 | — | — |
| FLT3/ITD-positive | 118 | 16 | 5 | 26 | .209 |
| FLT3/ITD point mutation | 52 | 7 | 0 | 0 | .632 |
| CEBPA mutation | 44 | 6 | 9 | 47 | < .001 |
| NPM mutation | 63 | 8 | 0 | 0 | .392 |
| Risk group | |||||
| Standard | 353 | 47 | 2 | 11 | .001 |
| Low | 293 | 39 | 12 | 63 | .055 |
| High | 98 | 13 | 5 | 26 | .161 |
| CR1 response | |||||
| CR | 577 | 76 | 14 | 74 | .789 |
| Not in CR | 170 | 22 | 5 | 26 | .780 |
| Death | 12 | 2 | 0 | 0 | 1.000 |
| Unevaluable | 9 | — | 0 | — | — |
| MRD1 | |||||
| No | 443 | 71 | 10 | 71 | 1.000 |
| Yes | 177 | 29 | 4 | 29 | — |
| CR2 response | |||||
| CR | 665 | 88 | 16 | 89 | 1.000 |
| Not in CR | 74 | 10 | 2 | 11 | .695 |
| Death | 15 | 2 | 0 | 0 | 1.000 |
| Unevaluable | 13 | — | 1 | — | — |
| 5-y OS from study entry | 44 | 67 ± 15 | 9 | 89 ± 21 | .146 |
| 5-y EFS from study entry | 768 | 49 ± 4 | 19 | 44 ± 23 | .900 |
| 5yr DFS from end of course 1 (CR patients only) | 577 | 55 ± 4 | 14 | 36 ± 26 | .250 |
| 5-y RR from end of course 1 (CR patients only) | 577 | 40 ± 4 | 14 | 64 ± 27 | .111 |
| WBC × 103 µL: median (range) | 29.8 | (0.2-610) | 39 | (0.9-439.2) | .359 |
| BM blasts %: median (range) | 70.0 | (0-100) | 50.5 | (23-99) | .040 |
| Platelet count (1000/µL): median (range) | 48.0 | (1-1177) | 34 | (8-405) | .240 |
| Hemoglobin: median (range) | 8.1 | (1.8-17.0) | 8.9 | (4.2-12.3) | .347 |
| . | No mutation (N = 768) . | CSF3R mutation (N = 19) . | No mutation vs CSF3R mutation . | ||
|---|---|---|---|---|---|
| Characteristics . | N . | (%) . | N . | (%) . | P . |
| Gender | |||||
| Male | 389 | 51 | 15 | 79 | .015 |
| Female | 379 | 49 | 4 | 21 | — |
| Age (y) | |||||
| Median (range) | 10.08 | (0.02-29.84) | 10.87 | (2.38-22.77) | .133 |
| 0-2 y | 183 | 24 | 1 | 5 | .059 |
| 3-10 y | 229 | 30 | 9 | 47 | .100 |
| +11 y | 356 | 46 | 9 | 47 | .930 |
| FAB | |||||
| M0 | 19 | 3 | 0 | 0 | 1.000 |
| M1 | 84 | 13 | 2 | 13 | 1.000 |
| M2 | 164 | 25 | 7 | 47 | .153 |
| M4 | 166 | 25 | 4 | 27 | 1.000 |
| M5 | 150 | 22 | 0 | 0 | .034 |
| M6 | 12 | 2 | 0 | 0 | 1.000 |
| M7 | 33 | 5 | 0 | 0 | 1.000 |
| Other | 39 | 6 | 2 | 13 | .260 |
| Unknown | 101 | — | 4 | — | — |
| Cytogenetics | |||||
| Normal | 176 | 24 | 3 | 16 | .588 |
| t(8;21) | 108 | 15 | 5 | 26 | .174 |
| inv(16) | 102 | 14 | 1 | 5 | .495 |
| t(9;11)/11q23 | 153 | 21 | 0 | 0 | .034 |
| t(6;9) | 12 | 2 | 0 | 0 | 1.000 |
| Monosomy 7 | 12 | 2 | 2 | 11 | .042 |
| Del(7q) | 5 | 1 | 0 | 0 | 1.000 |
| −5/5q- | 8 | 1 | 0 | 0 | 1.000 |
| +8 | 41 | 6 | 0 | 0 | .618 |
| Other | 117 | 16 | 8 | 42 | .005 |
| Missing | 34 | — | 0 | — | — |
| FLT3/ITD-positive | 118 | 16 | 5 | 26 | .209 |
| FLT3/ITD point mutation | 52 | 7 | 0 | 0 | .632 |
| CEBPA mutation | 44 | 6 | 9 | 47 | < .001 |
| NPM mutation | 63 | 8 | 0 | 0 | .392 |
| Risk group | |||||
| Standard | 353 | 47 | 2 | 11 | .001 |
| Low | 293 | 39 | 12 | 63 | .055 |
| High | 98 | 13 | 5 | 26 | .161 |
| CR1 response | |||||
| CR | 577 | 76 | 14 | 74 | .789 |
| Not in CR | 170 | 22 | 5 | 26 | .780 |
| Death | 12 | 2 | 0 | 0 | 1.000 |
| Unevaluable | 9 | — | 0 | — | — |
| MRD1 | |||||
| No | 443 | 71 | 10 | 71 | 1.000 |
| Yes | 177 | 29 | 4 | 29 | — |
| CR2 response | |||||
| CR | 665 | 88 | 16 | 89 | 1.000 |
| Not in CR | 74 | 10 | 2 | 11 | .695 |
| Death | 15 | 2 | 0 | 0 | 1.000 |
| Unevaluable | 13 | — | 1 | — | — |
| 5-y OS from study entry | 44 | 67 ± 15 | 9 | 89 ± 21 | .146 |
| 5-y EFS from study entry | 768 | 49 ± 4 | 19 | 44 ± 23 | .900 |
| 5yr DFS from end of course 1 (CR patients only) | 577 | 55 ± 4 | 14 | 36 ± 26 | .250 |
| 5-y RR from end of course 1 (CR patients only) | 577 | 40 ± 4 | 14 | 64 ± 27 | .111 |
| WBC × 103 µL: median (range) | 29.8 | (0.2-610) | 39 | (0.9-439.2) | .359 |
| BM blasts %: median (range) | 70.0 | (0-100) | 50.5 | (23-99) | .040 |
| Platelet count (1000/µL): median (range) | 48.0 | (1-1177) | 34 | (8-405) | .240 |
| Hemoglobin: median (range) | 8.1 | (1.8-17.0) | 8.9 | (4.2-12.3) | .347 |
The data from AAML0531, AAML03P1, and CCG-2961 were acquired from the TARGET project, current as of June 30, 2015. The χ2 test was used to test the significance of observed differences in patients’ characteristics, and Fisher’s exact test was used when data were sparse. Median differences were compared by the Mann–Whitney or Wilcoxon signed-rank test as appropriate. A P value < .05 was considered statistically significant. The Kaplan–Meier method was used to estimate OS (defined as time from study entry to death) and EFS (time from study entry until failure to achieve CR during induction, relapse, or death). RR was calculated by cumulative incidence methods defined as time from the end of induction I for patients in CR to relapse or death, where deaths without a relapse were considered competing events. Patients who withdrew from therapy due to relapse, persistent central nervous system disease, or refractory disease with >20% BM blasts by the end of induction I were defined as induction I failures. The significance of predictor variables was tested with the log-rank statistic for OS, EFS, and with Gray’s statistic for RR. All estimates are reported with 2× the Greenwood standard errors. Children lost to follow-up were censored at their date of last known contact.
BM, bone marrow; CR, complete remission; EFS, event-free survival; FAB, French-American-British; FLT3, FMS-like tyrosine kinase; ITD, internal tandem duplications; MRD, minimal residual disease; NPM, nucleophosmin; OS, overall survival; RR, relapse risk.
Another common genetic alteration among patients with CSF3R mutations were the core binding factor (CBF) abnormalities, which occurred in 37% of patients with CSF3R mutations. CBF-AML is comprised of RUNX1/RUNX1T1 t(8;21) translocations and inversion 16, which form fusion complexes involving CBF components. CBF abnormalities block differentiation and promote the formation of immature AML blasts. Interestingly, CEBPA mutations and CBF abnormalities were often mutually exclusive in patients with transforming CSF3R mutations (Figure 1C), together comprising 74% of CSF3R-mutated cases. This suggests that perhaps a differentiation-blocking signal from either CBF or a CEBPA mutation, alongside the CSF3R mutation, drives the production of immature myeloid cells in pediatric AML, whereas isolated CSF3R mutations can drive a mature neutrophil phenotype in CNL. Testing these possibilities will be the subject of future studies.
Likely due to the association with CEBPA, transforming CSF3R mutations had a trend toward low-risk disease (63% vs 39%; P = .055). Risk groups were defined using cytogenetics and the presence of FLT3-ITD, CEBPA, and NPM mutations. Actuarial OS at 5 years for those with CSF3R mutations vs no CSF3R mutations was 83% vs 65% (P = .196). EFS was 44% with CSF3R mutations vs 49% (P = .900), and relapse risk was 64% vs 40% (P = .111).
This study identifies a distinct molecular subtype of pediatric AML defined by CSF3R mutations. The CSF3R mutations found in pediatric AML are either the same point mutations or similar truncation mutations that are observed in adult CNL, suggesting that other cooperating mutations contribute to the distinct pathobiology of these 2 diseases. The majority of patients with CSF3R mutations have either a mutation in CEBPA or CBF translocations. Strikingly, CEPBA mutations are approximately eightfold more frequent in patients with transforming CSF3R mutations than in those patients without transforming CSF3R mutations. In future studies we will investigate how cooperating mutations alter the disease phenotype of myeloid malignancies driven by CSF3R mutations.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors gratefully acknowledge the important contributions of the late Robert Arceci to the AML TARGET initiative.
The TARGET initiative is supported by a grant from the National Institutes of Health National Cancer Institute (U10 CA98543). Work performed under contracts from the National Cancer Institute (HHSN261200800001E) include: specimen processing (COG Biopathology Center) and whole genomic sequencing (Complete Genomics, Inc.). This work is also supported by the JJ’s Angels St. Baldrick’s research grant (#360525) (J.E.M. and S.M.). J.E.M. is supported by a K99/R00 award from the National Cancer Institute (CA190605-02); Y.-C.W. is supported by the Andrew McDonough B+ Foundation; S.M. is supported by a grant from the National Cancer Institute (5R01CA114563), a St. Baldrick’s Foundation Consortium Research Grant (#280920), TARGET (3U01CA098543), and a COG chair’s grant (3U10CA180886); J.E.F. is supported by the Arkansas Biosciences Institute; and T.A.A. is supported by the COG Statistics and Data Center (1U10CA180899) and COG chair’s grant (5U10CA098543). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Contribution: J.E.M., R.E.R., S.L.T., S.B.H., M.A.M., Y.M., Z.Z., and A.J.M. performed the experiments; J.E.M., R.E.R., M.A.M., Y.M., Z.Z., R.M., W.L., and S.M. analyzed the data; Y.-C.W., R.B.G., and T.A.A. performed statistical analysis; J.E.M., J.M.G.A., P.G., T.M.D., L.C.H., J.E.F., J.P.R., E.A.K., and S.M. conceived and designed the experiments; J.E.M., Y.-C.W., T.A.A., and S.M. wrote the paper; and M.A.S., D.S.G., A.S.G., T.A.A., E.A.K., and S.M. supervised the research.
Conflict-of- interest disclosure: J.P.R. is a consultant for Incyte Corporation. The remaining authors declare no competing financial interests.
Correspondence: Soheil Meshinchi, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, D5-380, Seattle, WA 98109; e-mail: smeshinc@fredhutch.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal