Key Points
NR3C1 haploinsufficiency is found in patients with a plasmacytoid dendritic cell neoplasm characterized by very poor clinical outcome.
Overexpression of lincRNA-3q is a consistent feature of malignant cells in these patients and can be abrogated by BET protein inhibition.
Abstract
Blastic plasmacytoid dendritic cell neoplasm (BPDCN) is a rare and highly aggressive leukemia for which knowledge on disease mechanisms and effective therapies are currently lacking. Only a handful of recurring genetic mutations have been identified and none is specific to BPDCN. In this study, through molecular cloning in an index case that presented a balanced t(3;5)(q21;q31) and molecular cytogenetic analyses in a further 46 cases, we identify monoallelic deletion of NR3C1 (5q31), encoding the glucocorticoid receptor (GCR), in 13 of 47 (28%) BPDCN patients. Targeted deep sequencing in 36 BPDCN cases, including 10 with NR3C1 deletion, did not reveal NR3C1 point mutations or indels. Haploinsufficiency for NR3C1 defined a subset of BPDCN with lowered GCR expression and extremely poor overall survival (P = .0006). Consistent with a role for GCR in tumor suppression, functional analyses coupled with gene expression profiling identified corticoresistance and loss-of-EZH2 function as major downstream consequences of NR3C1 deletion in BPDCN. Subsequently, more detailed analyses of the t(3;5)(q21;q31) revealed fusion of NR3C1 to a long noncoding RNA (lncRNA) gene (lincRNA-3q) that encodes a novel, nuclear, noncoding RNA involved in the regulation of leukemia stem cell programs and G1/S transition, via E2F. Overexpression of lincRNA-3q was a consistent feature of malignant cells and could be abrogated by bromodomain and extraterminal domain (BET) protein inhibition. Taken together, this work points to NR3C1 as a haploinsufficient tumor suppressor in a subset of BPDCN and identifies BET inhibition, acting at least partially via lncRNA blockade, as a novel treatment option in BPDCN.
Introduction
Blastic plasmacytoid dendritic cell neoplasm (BPDCN) is a rare and clinically aggressive disorder that shows dismal prognosis whatever the treatment.1 Median overall survival is less than 2 years, even with high-dose chemotherapy, although longer-term, albeit short-lived, remissions have been observed in allotransplanted patients.2-4
BPDCN derives from malignant transformation of plasmacytoid dendritic cell (pDC) precursors5-7 and is currently classified with acute myeloid leukemia (AML) and related precursor neoplasms in the 2008 World Health Organization classification of hematologic malignancies.1 Tumor cells infiltrate skin, bone marrow, peripheral blood, and lymph nodes and show the characteristic immunophenotypic profile CD4+ CD56+ HLA-DRhi CD123+ lineage (Lin)−, although atypical profiles are reported.8,9
BPDCN presents heterogeneous genetic features characterized by chromosomal losses and deletions10,11 and a mutational landscape that overlaps with other hematologic malignancies without evidence of unique, disease-specific, driver genetic lesions.12-14 As in myeloid and lymphoid malignancies, mutations in key epigenetic modifier-encoding genes, such as TET2, ASXL1, and EZH2, have been described in a proportion of BPDCN cases, thus supporting a role for deregulation of epigenetic signaling in disease pathogenesis.12-14 Gene expression profiling (GEP), although performed in relatively few cases, has shown a distinctive transcriptional program in BPDCN characterized by predominant expression of genes that are typical of the pDC lineage15,16 and evidence of aberrant activation of the NF-κB pathway.16
Deletion of chromosome 5q, commonly seen in myelodysplastic syndrome (MDS) and AML, is frequent in BPDCN.10,11 Deletion 5q (outside of the 5q− syndrome) is associated with increased risk of leukemic transformation in MDS17 and with very poor prognosis in AML.18,19 This is attributed to haploinsufficiency for critical 5q gene(s), which in cooperation with additional signaling networks, drives malignant transformation and clonal evolution in these disorders.20-22 In keeping with this, treatments that are effective in 5q-deleted myeloid malignancies have been shown to directly target critical haploinsufficient 5q genes and their signaling networks.20
Based on the latter findings,20-22 we hypothesized that similar 5q gene haploinsufficiency mechanisms are likely to operate in BPDCN and that identification of the relevant 5q target genes might lead to new insight into disease biology and therapy. In this setting, we have used molecular cloning coupled with molecular cytogenetics and next-generation sequencing to characterize 5q alterations in BPDCN. Our work points to NR3C1, encoding the glucocorticoid receptor (GCR), as a haploinsufficient tumor suppressor in this disorder. NR3C1 loss defines a subset of highly aggressive BPDCN characterized by a loss-of-EZH2 function gene expression signature. In addition, we extend previous observations that identified a potential role for epigenetic modifier gene mutations in BPDCN pathogenesis by providing the first evidence of a key role for nuclear long noncoding RNA (lncRNA) deregulation in the pathogenesis of this disorder.
Methods
BPDCN patients and cell lines
BPDCN patients investigated in this study were recruited retrospectively between 2008 and 2014 through 2 French study groups, the Groupe Francophone de Cytogénétique Hématologique and the French BPDCN network (identified as cohorts A and B, respectively, in supplemental Table 1, available on the Blood Web site). After centralized review of clinical and biological criteria for BPDCN diagnosis,8 and on the basis of available cytogenetic/molecular cytogenetic data, 47 patients (median age, 66 years; range, 7-82 years) were enrolled in the current study (supplemental Tables 1-4). All patient data were obtained at diagnosis. All patients provided written informed consent. The study was approved by the institutional review boards of the participating centers. For 2 patients, derived cell lines that displayed the same cytogenetic characteristics as the original patient blasts were used for analyses (unique patient number 1 [UPN 1]: GEN2.2 and UPN 2: CAL-1).23,24 BPDCN cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum.23,24 Murine stromal cell support was provided for GEN2.2 cells, as previously described.23
Cytogenetics, FISH, molecular analyses, and aCGH
R-banded karyotyping, fluorescence in situ hybridization (FISH) analyses, and array comparative genomic hybridization (aCGH) were performed by standard methods, as previously described.10,25 All cytogenetic and aCGH data were centrally reviewed by the Groupe Francophone de Cytogénétique Hématologique and the French BPDCN network. Karyotypes were described according to the International System for Human Cytogenetic Nomenclature. Bacterial artificial chromosome and fosmid probes for FISH mapping are listed in supplemental Table 5. Additional molecular analyses (see below) used reagents given in supplemental Tables 6-12.
NR3C1, EZH2, and ASXL1 mutation screening
For mutation screening of the coding regions of NR3C1, a custom, targeted next-generation sequencing panel covering all 9 exons of the NR3C1 gene (total panel size, 3.3 kb; 31 amplicons) was designed using the Ampliseq Designer software (Thermo Fisher Scientific). Ion Ampliseq DNA libraries were prepared using 10 ng of amplified genomic DNA (for a list of the cases studied and the tissue source of DNA, see supplemental Table 2). Libraries were submitted to emulsion polymerase chain reaction (PCR) with the Ion PGM Hi-Q OneTouch 2 template kit v2. The generated ion sphere particles were enriched with the Ion OneTouch Enrichment System, loaded, and sequenced with the Ion PGM Hi-Q Sequencing 200 Kit on Ion 316 v2 chips (Thermo Fisher Scientific). Torrent Suite version 5.0 software (Thermo Fisher Scientific) was used to perform primary analysis, including signal processing, base calling, sequence alignment to the reference genome (hg19), and generation of binary alignment/map files. Binary alignment/map files were used by Torrent Suite Variant Caller to detect point mutations and short indels using the PGM Somatic Low Stringency profile. Variant Call Format files generated by Variant Caller were annotated using Variant Effect Predictor.26 Samples were considered of sufficient quality when >90% of targeted bases were read with a base call accuracy of at least Q20 (success rate, 100%). The average coverage was 1800× (range, 400-4000×). Variants present in single nucleotide polymorphism databases and absent in COSMIC (version 64) were discarded, as were variants with a predictive SIFT (sorting intolerant from tolerant) score >0.05.26
EZH2 (all coding exons) and ASXL1 (exon 12) mutation screening was performed in a subset of BPDCN patients (those undergoing GEP; supplemental Table 2) by using primers and PCR cycling conditions, as previously described.27,28 Purified PCR products were directly sequenced in both directions, using BigDye Terminator Mix (Applied Biosystems), on the Applied Biosystems 3130xl Genetic Analyzer. Data were analyzed with the SeqScanner software version 1.0 (Applied Biosystems).
LD-PCR and fusion transcript cloning
Long-distance PCR (LD-PCR) cloning of the t(3;5)(q21;q31) genomic breakpoint was performed using the Expand Long Template PCR system (Sigma) and LD-PCR primers, TA cloning of PCR products, and sequencing (supplemental Table 6). For RNA analysis, total RNA was prepared by TRIzol reagent, according to the manufacturer’s instructions. Rapid amplification of complementary DNA (cDNA) ends (RACE)-PCR (5′ and 3′) was used to clone the t(3;5)-encoded fusion transcript and was performed using the SMARTer RACE cDNA Amplification Kit (Clontech), according to the manufacturer’s instructions. RACE primers are listed in supplemental Table 7. For functional studies, GCR fusion protein (GCR-FP) cDNA was cloned into pcDNA3.1 and pEGFP-C1 and subcloned as a green fluorescent protein (GFP)-tagged isoform into a modified pHIV-SFFV-mRFP-WPRE lentiviral vector, using primers described in supplemental Table 8. CAL-1 cell lines stably expressing GFP alone or GFP-tagged GCR-FP were obtained by lentiviral transduction and cell sorting (BD FACSAria). Successful transduction was confirmed by flow cytometry and western blotting.
RT-qPCR, Northern blotting, and GEP
Total RNA was obtained from whole cells or from subcellular fractions (nuclear and cytoplasmic), reverse transcribed (RT) by using the SuperScript III First-Strand Synthesis SuperMix (Invitrogen), and subjected to quantitative PCR (qPCR) by using SYBR Green PCR Master Mix (Applied Biosystems) and the primers shown in supplemental Table 7, according to the manufacturers’ instructions. The ABL gene was used as a control for normalization of gene expression data. qPCR was performed on an MX3000P machine (Stratagene). Northern blotting used NR3C1 cDNA probe (see supplemental Methods and supplemental Table 9 for details). Affymetrix GEP was performed as described previously by using total RNA and Affymetrix U133 2.0 chips.29 GEP data normalization and bioinformatics analysis were performed as described previously (see supplemental Methods for details).29
IHC and western blotting
For GCR expression analysis in formalin-fixed paraffin-embedded tissue, sections of 3-μm thickness were stained with an anti-human polyclonal GCR antibody (SC-1003; Santa Cruz Biotechnology; polyclonal E20), at an optimized dilution of 1:800. Immunohistochemistry (IHC) was performed on a Benchmark XT autostainer (Ventana Medical Systems) using standard antigen retrieval (1 hour at 98°C) in Cell Conditioning Solution (CC1; Ventana Medical Systems). Primary antibody was incubated for 1 hour at room temperature, and the signal was visualized by diaminobenzidine with the OptiView DAB IHC Detection Kit (Ventana Medical Systems). Sections were counterstained with hematoxylin. The H-Score method was used for evaluation of the immunostaining by multiplying the intensity of the staining (0, no staining; 1, weak; 2, moderate; and 3, strong staining) by the percentage of the tumor stained. The minimum score was 0 and the maximum was 300.
For western blotting, whole cell lysates, obtained by sonication of cell pellets in Laemmli buffer, were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes for blotting with anti-N-terminal GCR (polyclonal E-20), anti-GFP (clone B2; Santa Cruz Biotechnology), anti-H3K27me3 (Millipore), anti-histone H3 (Upstate), anti-E2F1 (clone KH95; Santa Cruz Biotechnology), and anti-ACTIN (Sigma) antibodies. After treatment with blocking solution (1× phosphate-buffered saline, 8% skimmed milk, and 0.1% Tween 20), membranes were incubated with primary antibody (anti-GCR [0.4 mg/mL] and anti-H3K27me3, anti-histone H3, and anti-ACTIN [0.5 mg/mL] in phosphate-buffered saline, 3% skimmed milk, and 0.1% Tween 20), washed, and incubated with anti-rabbit or anti-mouse immunoglobulin G–horseradish peroxidase (Thermo Fisher Scientific) before incubation in enhanced chemiluminescent SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific). Images were captured using autoradiography films.
Gene silencing
Nontargeting (control; Ctrl), GCR-targeting, and lincRNA-3q-targeting short hairpin RNA (shRNA) sequences were designed using the DSIR (designer of small interfering RNA [siRNA]) algorithm (http://biodev.cea.fr/DSIR/DSIR.html).30 Hairpin sequences are provided in supplemental Table 10. Short hairpin sequences were cloned into the pLKO-1 lentiviral vector (Addgene) and packaged, as described previously.31 Cell lines stably expressing shCtrl, shGCR, and shlincRNA-3q were established under puromycin selection.
Glucocorticoid response element and E2F reporter assays and RNA interference
Transient transfection assays using the glucocorticoid response element luciferase reporter plasmid (gift from J. Cidlowski, National Institute of Environmental Health Sciences, National Institutes of Health, Durham, NC) were performed in COS7 cells using Lipofectamine (Invitrogen), according to the manufacturer’s instructions. Transient transfection assays with the E2F reporter system were performed in H1299 cells using X-tremeGENE (Roche) following the manufacturer’s instructions. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega). siRNAs were ordered from Eurogentec (supplemental Table 10) and transfected using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions.
Evaluation of drug sensitivity
For drug sensitivity testing, 5 × 105 cells per milliliter were treated for the desired time and drug dose (dexamethasone at 100 nM or 10 μM, etoposide at 10 μM, and JQ1 at 1 μM). Dexamethasone was provided by the pharmacy at Grenoble University Hospital. Etoposide was purchased from Sigma. JQ1 was synthesized, as described previously.29 Cell viability was evaluated by AnnexinV/propidium iodide (Beckman Coulter) or propidium iodide staining and flow cytometry analysis (BD LSR II). Specific cell death was calculated as (% drug-induced cell death − % control cell death) ÷ (100 − % control cell death) × 100, as described previously.32
ChIP
Chromatin immunoprecipitation (ChIP) experiments were performed as described by Fournier et al25 (native chromatin) or using the Magna ChIP kit (Millipore; cross-linked chromatin), following the manufacturer’s instructions. Briefly, all ChIP assays were performed in triplicate in at least 2 independent chromatin preparations. For qPCR analyses of immunoprecipitated chromatin fractions at sequences of interest, a SYBR Green PCR Kit (Applied Biosystems) was used with primer pairs, as listed in supplemental Table 11. The qPCR data obtained on immunoprecipitated fractions were normalized to input chromatin (% Input = 2ΔCt = 2[Ct(Immunoprecipitation)−Ct(Input)]). Background precipitation was evaluated with a mock immunoprecipitation.
Clonogenicity and xenotransplantation assays
Clonogenic cell frequencies were estimated by a method33 checking that the limiting dilution data do not include false-negative outcomes and conform to the single-hit Poisson model. Statistical differences of clonogenic cell frequency estimates between assays were analyzed by a likelihood ratio test developed for limiting dilution assays.34 Animal studies are described in supplemental Methods.
Statistics
Excel 2010 (Microsoft), JMP 10.0.0 (SAS Institute), Stata/IC 10.1 (StataCorp), and GraphPad (Prism) software programs were used for graphical representations and to perform statistical analysis across all experiments. Survival curves were built according to the Kaplan-Meier method. Comparison of Kaplan-Meier survival estimates was performed using the log-rank test. Univariate and multivariate survival analyses were performed using Cox proportional hazards regression to identify statistically significant predictors of overall survival and to obtain hazard ratio estimates as well as corresponding 95% confidence intervals. Multivariate models were constructed by including all variables with P < .2 in univariate analysis. The Wald test was used to assess the significance of variables in models. The Wilcoxon nonparametric method or the 2-sided Student t test were used for statistical analysis, as indicated. Histograms represent the average for each group, and error bars represent standard deviation or standard error on the mean, as indicated.
Results
5q Anomalies in BPDCN confer adverse clinical outcome
To uncover 5q-linked disease mechanisms in BPDCN, we screened 47 BPDCN patients at diagnosis for the presence of 5q alterations/chromosome 5 loss, by conventional metaphase karyotyping (39/47 cases), coupled with aCGH (25/47 cases), and/or locus-specific FISH (23/47 cases) when material was available (supplemental Tables 1 and 2; Figure 1). Overall, 5q alterations, including a single case of balanced 5q translocation [UPN 1; t(3;5)(q21;q31); supplemental Figure 1A] and 1 case with chromosome 5 loss, were observed in 17 of 47 patients (36%) (Figure 1A). In agreement with our previous findings,10,11 a majority of 5q abnormalities occurred in the context of complex karyotypes, in either founder clones (13 cases) or subclones at diagnosis (4 cases) (supplemental Table 2). Consistent with a role in leukemia initiation and/or clonal evolution, and as seen in other myeloid cancers, the presence of 5q anomalies in BPDCN was associated with very poor prognosis (P = .0063) (Figure 1B, top).
Targeting of the NR3C1 gene by 5q deletion/translocation in BPDCN. (A) Schematic representation of 5q alterations in BPDCN patients (n = 17/47), as indicated (left). aCGH image of case UPN 33 showing focal deletion of the NR3C1 gene (right). (B) Kaplan-Meier cumulative survival curves in BPDCN according to 5q alteration status (for simplicity, this is labeled del(5q) [UPNs 1, 2, 8, 12, 14, 21, 22, 24, 26, 27, 29, 31, 35, and 45] and non-del(5q) [UPNs 3, 9, 10, 16-19, 25, 27, 36, 37, and 40-42]; upper) and to NR3C1 deletion status (UPNs 1, 8, 14, 22, 24, 26, 27, 31, 35, and 45) (lower), determined by FISH or aCGH; ΔNR3C1). P values were determined by log-rank test. (C) FISH analysis of the t(3;5)(q21;q31) in the BPDCN cell line GEN2.2 (UPN 1) with chromosome 5q probes CTB-88J14 (green) and RP11-278J6 (red) (supplemental Table 2). Note a split signal for RP11-278J6 (NR3C1) on the der(3) and der(5) chromosomes. (D) Affymetrix-derived NR3C1 expression in BPDCN patients presenting del(5q) targeting NR3C1 or not. P value was determined by Wilcoxon test. (E) aCGH image showing 5q/NR3C1 deletion in DNA from a skin biopsy sample with leukemic infiltrate (UPN 22) compared with control DNA (Ctrl), and analysis in 2 BPDCN cases without 5q/NR3C1 deletion (UPNs 19 and 21). (F) Overview of immunostaining (diaminobenzidine and hematoxylin) of GCR on BPDCN dermal localization in a del(5q) case: NR3C1(+) (left) and a del(5q)/NR3C1 case: NR3C1(−) (upper right). Epidermal squamous cells showing positive nuclear staining are marked with thin gray arrows; leukemia infiltrate cells are marked with thick black arrows. A higher magnification of the clonal infiltrate in dermis of the same case as the upper right image shows mainly negative tumor cells (red arrow) and some positive cells (thick black arrow) (lower right). BM, bone marrow.
Targeting of the NR3C1 gene by 5q deletion/translocation in BPDCN. (A) Schematic representation of 5q alterations in BPDCN patients (n = 17/47), as indicated (left). aCGH image of case UPN 33 showing focal deletion of the NR3C1 gene (right). (B) Kaplan-Meier cumulative survival curves in BPDCN according to 5q alteration status (for simplicity, this is labeled del(5q) [UPNs 1, 2, 8, 12, 14, 21, 22, 24, 26, 27, 29, 31, 35, and 45] and non-del(5q) [UPNs 3, 9, 10, 16-19, 25, 27, 36, 37, and 40-42]; upper) and to NR3C1 deletion status (UPNs 1, 8, 14, 22, 24, 26, 27, 31, 35, and 45) (lower), determined by FISH or aCGH; ΔNR3C1). P values were determined by log-rank test. (C) FISH analysis of the t(3;5)(q21;q31) in the BPDCN cell line GEN2.2 (UPN 1) with chromosome 5q probes CTB-88J14 (green) and RP11-278J6 (red) (supplemental Table 2). Note a split signal for RP11-278J6 (NR3C1) on the der(3) and der(5) chromosomes. (D) Affymetrix-derived NR3C1 expression in BPDCN patients presenting del(5q) targeting NR3C1 or not. P value was determined by Wilcoxon test. (E) aCGH image showing 5q/NR3C1 deletion in DNA from a skin biopsy sample with leukemic infiltrate (UPN 22) compared with control DNA (Ctrl), and analysis in 2 BPDCN cases without 5q/NR3C1 deletion (UPNs 19 and 21). (F) Overview of immunostaining (diaminobenzidine and hematoxylin) of GCR on BPDCN dermal localization in a del(5q) case: NR3C1(+) (left) and a del(5q)/NR3C1 case: NR3C1(−) (upper right). Epidermal squamous cells showing positive nuclear staining are marked with thin gray arrows; leukemia infiltrate cells are marked with thick black arrows. A higher magnification of the clonal infiltrate in dermis of the same case as the upper right image shows mainly negative tumor cells (red arrow) and some positive cells (thick black arrow) (lower right). BM, bone marrow.
Targeting of NR3C1 by 5q anomalies in BPDCN
As an entry point to identification of pathogenically relevant 5q genes in BPDCN, we next focused our attention on molecular cytogenetic characterization of the balanced t(3;5)(q21;q31) identified in UPN 1 (supplemental Table 2; supplemental Figure 1A).11 Involvement of chromosomal arm 3q was of added interest because 3q aberrations are known to confer extremely poor prognosis, at least in AML and MDS.17-19,35 Remarkably, FISH mapping of the t(3;5)(q21;q31) in UPN 1 metaphases identified the NR3C1 gene, encoding the GCR, a ligand-dependent transcription factor of the nuclear hormone receptor family,36 as the 5q target gene. Subsequently, aCGH and/or FISH analysis with an NR3C1-specific probe in our cohort of BPDCN showed NR3C1 deletion to occur in 13 of 17 5q-rearranged cases (76%), of which 4 occurred in subclones detected at diagnosis (Figure 1A; supplemental Tables 1 and 2). Strikingly, 1 case with normal cytogenetics (UPN 33) presented a focal, morphologically cryptic NR3C1 deletion (chromosome 5:141,529,340 to 143,696,321), thus defining a minimally deleted 5q region in our BPDCN patient series (Figure 1A, right). It is of note that focal deletions of NR3C1 have previously been implicated in B-cell acute lymphoblastic leukemia (B-ALL),37,38 but not in myeloid malignancies. No further NR3C1 translocations were identified. Targeted deep sequencing in 36 cases, including 10 of the 13 cases with NR3C1 translocation/deletion (UPNs 1, 8, 14, 22, 24, 26, 27, 31, 32, 33), did not detect NR3C1 point mutations or indels (see supplemental Table 12 for NR3C1 amplicon primers). This finding suggested that haploinsufficiency for NR3C1 is a pathogenically relevant event in BPDCN with 5q/NR3C1 alteration. In keeping with this, GEP in bone marrow and skin biopsy tissue from cases with NR3C1 deletion showed lower NR3C1 expression in a subset of NR3C1-deleted BPDCN cases compared with nondeleted cases (Figure 1D).
We next examined GCR protein expression levels according to NR3C1 deletion status in leukemia-infiltrated skin biopsy tissue from 2 BPDCN cases, 1 with NR3C1 deletion (UPN 22) and 1 without (UPN 21) (Figure 1E-F). Material was not available for GCR protein expression analysis in any other cases. Quite strikingly, IHC with the anti-GCR antibody (clone A20) revealed markedly diminished GCR protein levels in the NR3C1-deleted case (UPN 22) compared with the nondeleted case (UPN 21; H-Score, 300) (Figure 1F). In the latter, GCR protein expression was high and localized to the nucleus of the leukemia cells, indicating engagement of GCR signaling. BPDCN cells in the NR3C1 haploinsufficient case appeared mostly negative for GCR immunostaining (H-Score, 0), although we suspect that IHC may not be sufficiently sensitive to detect very low level GCR expression. In the few GCR-positive leukemic cells seen in the NR3C1-deleted case (UPN 22), GCR expression was nuclear, indicative of active, nuclear GCR signaling.
Taken together, these results show that NR3C1 deletion impacts NR3C1 expression at the RNA and protein levels in BPDCN. Nuclear GCR signaling is engaged in BPDCN and appears attenuated in NR3C1-deleted BPDCN.
Haploinsufficiency for NR3C1 defines a very high risk subset of BPDCN
We next wished to determine the clinical impact of monoallelic NR3C1 deletion in BPDCN. We first analyzed the impact on overall survival of variables, including 5q/NR3C1 deletion, in univariate and multivariate Cox proportional hazards regression models (supplemental Table 3). By univariate analysis, patient age (>66 years) (P = .03), lack of allogeneic bone marrow transplant (allo-BMT) (P = .008), presence of del(5q) (P = .013), and NR3C1 gene alterations (P = .004) were significant hazards for death. Age was significantly linked to allo-BMT (P < .0001 by standard χ2 test), and del(5q) significantly linked to NR3C1 alterations (P < .0001), suggesting that allo-BMT and NR3C1 alterations, rather than age and del(5q), were the main factors of interest. Accordingly, allo-BMT and NR3C1 alteration were investigated for their impact on survival by multivariate analysis. Although allo-BMT did not reach statistical significance (P = .08), the analysis suggests that lack of allo-BMT and monoallelic loss of NR3C1 (P = .022) are independent factors that negatively impact survival in BPDCN (supplemental Table 3). Thus, haploinsufficiency for NR3C1 appears to define a very high risk subgroup of BPDCN patients. Of note, none of the 3 chemotherapy regimens was statistically significantly related to favorable or unfavorable outcome in univariate analysis (supplemental Table 3).
Molecular characterization of a novel t(3;5)(q21;q31) in BPDCN reveals fusion of the NR3C1 gene to a lncRNA-encoding gene in 3q21
To further explore the pathological relevance of NR3C1 rearrangement in BPDCN, we performed detailed molecular and functional characterization of the t(3;5)(q21;q31) observed in the primary cells and in the derived cell line GEN2.2 of patient UPN 1 (Figure 1C; supplemental Figure 1A; supplemental Table 2). RACE-PCR and sequencing revealed the t(3;5)(q21;q31) to encode a single chimeric transcript comprising exons 1 and 2 of the NR3C1 gene fused to an antisense sequence deriving from a novel lincRNA gene located in 3q21 (named here, lincRNA-3q) (Figure 2A; supplemental Figure 1C-D). LD-PCR genomic breakpoint cloning identified this transcript to result from fusion of NR3C1 intron 2 (chromosome 5:142,732,115) to lincRNA-3q intron 3 on the der(3) chromosome (chromosome 3:129,837,689) (supplemental Figure 1D). No further abnormal transcripts were detectable by Northern blotting across the NR3C1 gene, thus ruling out NR3C1 reciprocal fusion transcripts deriving from the der(5) chromosome (supplemental Figure 1B). Sequencing predicted the NR3C1-lincRNA-3q fusion gene transcript to encode a GCR fusion protein comprising the GCR N-terminal activation domain AF1 (amino acids 1-393) fused to 18 amino acids derived from transcriptional read-through into the lincRNA-3q locus (Figure 2B, left). In keeping with this, western blotting in t(3;5)-positive GEN2.2 cells, using an anti-N-terminal GCR antibody (clone E20), detected a single abnormal GCR protein isoform (∼52 kDa, hereafter referred to as GCR-FP), in addition to the wild-type GCR (90 kDa) (Figure 2B, right).
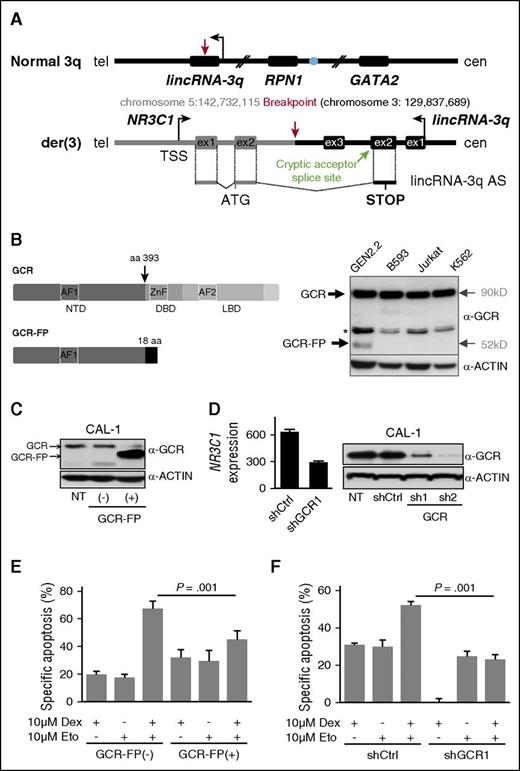
The t(3;5)-encoded GCR-lincRNA-3q fusion protein is associated with glucocorticoid resistance. (A) Schematic representation of the genomic organization surrounding the normal 3q and der(3) breakpoint regions, indicating the resulting fusion transcript NR3C1-lincRNA-3q. The blue filled circle denotes the GATA2 superenhancer. In black, 3q sequences. In gray, 5q sequences. The NR3C1-lincRNA-3q fusion transcript likely derives from a splicing event between NR3C1 exon 2 and a cryptic splice acceptor site located in the antisense orientation in lincRNA-3q exon 2 (chromosome 3:129,832,478). (B) Schematic representation of the structure of wild-type GCR (top left) and predicted GCR-FP (bottom left). GCR and ACTIN western blot analyses showing expression of an abnormal GCR isoform (GCR-FP) exclusively in GEN2.2 cells compared with t(3;5)-negative cell lines, as indicated (right); * denotes nonspecific band. (C) Western blot using anti-GCR and anti-ACTIN antibodies in the parent CAL-1 cell line (nontransduced; NT) compared with CAL-1 cells transduced with empty GFP vector (−) or with the GFP-tagged GCR-FP expression vector (+), as indicated (n = 2). (D) Western blot using anti-GCR and anti-ACTIN antibodies in the parent CAL-1 cell line (NT) compared with CAL-1 cells transduced with shCtrl or shGCR (n = 2 independent hairpins, shGCR1 and 2), as indicated (right). Affymetrix NR3C1 messenger RNA expression in shGCR1 compared with shCtrl-transduced cells (experimental haploinsufficiency, n = 3) (left). (E) Evaluation of drug sensitivity of CAL-1 cells overexpressing GCR-FP compared with control CAL-1 cells after 16 hours of treatment with 10 μM etoposide (Eto), 72 hours of treatment with 10 μM dexamethasone (Dex), or a combination of both treatments. Specific apoptosis was measured by AnnexinV/propidium iodide staining and flow cytometry, and calculated as follows: (% drug-induced cell death − % control cell death) ÷ (100 − % control cell death) × 100. P value determined by Wilcoxon test; n = 3. (F) Evaluation of drug sensitivity of CAL-1 cells shGFP compared with shCtrl as described in panel E. aa, amino acid; AF, activation function domain; cen, centromere; DBD, DNA binding domain; LBD, ligand binding domain; ex, exon; NTD, N-terminal domain; tel, telomere; TSS, transcription start site; ZnF, zinc finger domain.
The t(3;5)-encoded GCR-lincRNA-3q fusion protein is associated with glucocorticoid resistance. (A) Schematic representation of the genomic organization surrounding the normal 3q and der(3) breakpoint regions, indicating the resulting fusion transcript NR3C1-lincRNA-3q. The blue filled circle denotes the GATA2 superenhancer. In black, 3q sequences. In gray, 5q sequences. The NR3C1-lincRNA-3q fusion transcript likely derives from a splicing event between NR3C1 exon 2 and a cryptic splice acceptor site located in the antisense orientation in lincRNA-3q exon 2 (chromosome 3:129,832,478). (B) Schematic representation of the structure of wild-type GCR (top left) and predicted GCR-FP (bottom left). GCR and ACTIN western blot analyses showing expression of an abnormal GCR isoform (GCR-FP) exclusively in GEN2.2 cells compared with t(3;5)-negative cell lines, as indicated (right); * denotes nonspecific band. (C) Western blot using anti-GCR and anti-ACTIN antibodies in the parent CAL-1 cell line (nontransduced; NT) compared with CAL-1 cells transduced with empty GFP vector (−) or with the GFP-tagged GCR-FP expression vector (+), as indicated (n = 2). (D) Western blot using anti-GCR and anti-ACTIN antibodies in the parent CAL-1 cell line (NT) compared with CAL-1 cells transduced with shCtrl or shGCR (n = 2 independent hairpins, shGCR1 and 2), as indicated (right). Affymetrix NR3C1 messenger RNA expression in shGCR1 compared with shCtrl-transduced cells (experimental haploinsufficiency, n = 3) (left). (E) Evaluation of drug sensitivity of CAL-1 cells overexpressing GCR-FP compared with control CAL-1 cells after 16 hours of treatment with 10 μM etoposide (Eto), 72 hours of treatment with 10 μM dexamethasone (Dex), or a combination of both treatments. Specific apoptosis was measured by AnnexinV/propidium iodide staining and flow cytometry, and calculated as follows: (% drug-induced cell death − % control cell death) ÷ (100 − % control cell death) × 100. P value determined by Wilcoxon test; n = 3. (F) Evaluation of drug sensitivity of CAL-1 cells shGFP compared with shCtrl as described in panel E. aa, amino acid; AF, activation function domain; cen, centromere; DBD, DNA binding domain; LBD, ligand binding domain; ex, exon; NTD, N-terminal domain; tel, telomere; TSS, transcription start site; ZnF, zinc finger domain.
NR3C1 translocation/deletion drives altered GCR signaling and drug resistance in BPDCN
NR3C1 gene alterations have been described in a subset of relapsing B-ALL, suggesting a role in therapy resistance.37,38 We thus investigated responses to drug therapy in CAL-1 cells after stable overexpression of a GFP-tagged isoform of GCR-FP (CAL-1 GCR-FP[+]; Figure 2C; supplemental Figure 2A-B) or after stable GCR knockdown (CAL-1 shGCR1 and 2; Figure 2D) designed to mimic haploinsufficiency for NR3C1 as seen in patients compared with control cells (ie, CAL-1 cells expressing GFP alone [CAL1 GCR-FP(−)] or CAL-1 shCtrl cells, respectively). Strikingly, responses to combination therapy with high-dose synthetic GCR ligand (dexamethasone at 10 μM) and etoposide (10 μM) were markedly reduced in CAL-1 GCR-FP-overexpressing cells compared with controls (Figure 2E). Cell death after dexamethasone (10 μM) or etoposide (10 μM) treatment alone was unchanged, at least under these conditions (Figure 2E). Similarly, GCR depletion by short hairpin markedly reduced cell death responses to single-agent dexamethasone or to combination therapy by etoposide–dexamethasone compared with controls, but did not affect cell death response to etoposide alone (Figure 2F).
The next step was to analyze more globally the potential impact of GCR haploinsufficiency on ligand-dependent GCR transcriptional regulatory activity in BPDCN. We first used luciferase reporter assays to test the impact of GCR-FP on wild-type GCR activity in the absence (dimethyl sulfoxide) or presence of dexamethasone (Figure 3A). As expected, this revealed GCR-FP dose-dependent attenuation of GCR transcriptional transactivation activity (Figure 3A). We next performed GEP in GCR-FP-overexpressing or shGCR-treated CAL-1 cells (designed to mimic haploinsufficiency; Figure 2D), in the absence (dimethyl sulfoxide) or presence of dexamethasone (100 nM, 6 hours) (Figure 3B). Low-dose, as opposed to high-dose (therapeutic-level) dexamethasone, was used in order to simulate the conditions in which NR3C1 alterations might be expected to undergo clonal selection in treatment-naive leukemia cells in vivo (ie, physiological glucocorticoid concentrations). Strikingly, this analysis revealed specific, hormone-dependent deregulation of 2332 (1205 down; 1127 up) and 3205 genes (1456 down; 1749 up) in CAL-1 shGCR and CAL-1 GCR-FP dexamethasone-treated cells, respectively, compared with CAL-1 control cells. Further analysis revealed a common subset of 264 deregulated genes (139 down; 125 up) (Figure 3B; supplemental Table 13), which included known GCR target genes such as PFKB2 (encoding 6-phosphofructo-2-kinase) and genes encoding nuclear factors that have been implicated in normal (IKZF2/Helios) or leukemic (ZEB2) T lymphoid39,40 or dendritic cell lineage development (SPIB).41 Supporting the clinical relevance of the latter findings, deregulated expression of IKZF2, ZEB2, and SPIB was also observed by GEP in primary patient cells with NR3C1 deletion compared with nondeleted control cases (supplemental Figure 3A).
Misregulated GCR and EZH2 signaling as a consequence of NR3C1 alterations in BPDCN. (A) Luciferase reporter assay for glucocorticoid transcriptional transactivation activity in COS7 cells transduced with empty pcDNA3.1 vector, pcDNA3.1-GCR, and pcDNA3.1-GCR-FP, as indicated, after a 6-hour treatment with either dimethyl sulfoxide (DMSO) or 100 nM dexamethasone (n = 3). (B) Heatmap representation of the common differentially expressed genes among CAL-1-GCR-FP (GCR-FP[+]), CAL-1 control cells (GCR-FP[−]), and shGCR-transduced cells (shGCR[+]) compared with controls (shGCR[−]) after treatment with 100 nM dexamethasone for 6 hours (n = 3 for each group). Black arrows indicate genes cited in the text. (C) GSEA plots showing gene regulatory circuits that are differentially expressed between dexamethasone-treated CAL-1-GCR-FP or shGCR compared with respective control CAL-1 cells (CAL-1-GFP or CAL-1 shCtrl). (D) Western blot analysis and quantification of global H3K27me3 levels in CAL-1-GCR-FP (GCR-FP[+]) and CAL-1 shGCR (upper) and respective CAL-1 control cells (CAL-1 GFP [GCR-FP(−)] and CAL-1 shCtrl) (lower) upon 6-hour or 24-hour treatment with 100 nM dexamethasone (n = 3). (E) ChIP for H3K27me3 followed by qPCR analysis for enrichment on HOXA gene promoters, as indicated, in CAL-1-GCR-FP (GCR-FP[+]), CAL-1 shGCR, and respective CAL-1 control cells (CAL-1 GFP [GCR-FP(−)] and CAL-1 shCtrl) treated for 24 hours with 100 nM dexamethasone (n = 3). (F) GSEA plots showing gene regulatory circuits that are differentially expressed between BPDCN patients presenting, or not, del(5q) abnormalities that target NR3C1 (bone marrow or skin, as indicated; unmutated EZH2 and ASXL1 cases only). (G) Tumor bioluminescence (7 and 14 days postinjection) for xenotransplanted nude mice bearing tumors derived from CAL-1 shCtrl and shGCR cell lines (left). P value determined by Wilcoxon test; n = 6 for each group. Bioluminescence imaging for representative mouse (right). D, day; GSEA, gene set enrichment analysis.
Misregulated GCR and EZH2 signaling as a consequence of NR3C1 alterations in BPDCN. (A) Luciferase reporter assay for glucocorticoid transcriptional transactivation activity in COS7 cells transduced with empty pcDNA3.1 vector, pcDNA3.1-GCR, and pcDNA3.1-GCR-FP, as indicated, after a 6-hour treatment with either dimethyl sulfoxide (DMSO) or 100 nM dexamethasone (n = 3). (B) Heatmap representation of the common differentially expressed genes among CAL-1-GCR-FP (GCR-FP[+]), CAL-1 control cells (GCR-FP[−]), and shGCR-transduced cells (shGCR[+]) compared with controls (shGCR[−]) after treatment with 100 nM dexamethasone for 6 hours (n = 3 for each group). Black arrows indicate genes cited in the text. (C) GSEA plots showing gene regulatory circuits that are differentially expressed between dexamethasone-treated CAL-1-GCR-FP or shGCR compared with respective control CAL-1 cells (CAL-1-GFP or CAL-1 shCtrl). (D) Western blot analysis and quantification of global H3K27me3 levels in CAL-1-GCR-FP (GCR-FP[+]) and CAL-1 shGCR (upper) and respective CAL-1 control cells (CAL-1 GFP [GCR-FP(−)] and CAL-1 shCtrl) (lower) upon 6-hour or 24-hour treatment with 100 nM dexamethasone (n = 3). (E) ChIP for H3K27me3 followed by qPCR analysis for enrichment on HOXA gene promoters, as indicated, in CAL-1-GCR-FP (GCR-FP[+]), CAL-1 shGCR, and respective CAL-1 control cells (CAL-1 GFP [GCR-FP(−)] and CAL-1 shCtrl) treated for 24 hours with 100 nM dexamethasone (n = 3). (F) GSEA plots showing gene regulatory circuits that are differentially expressed between BPDCN patients presenting, or not, del(5q) abnormalities that target NR3C1 (bone marrow or skin, as indicated; unmutated EZH2 and ASXL1 cases only). (G) Tumor bioluminescence (7 and 14 days postinjection) for xenotransplanted nude mice bearing tumors derived from CAL-1 shCtrl and shGCR cell lines (left). P value determined by Wilcoxon test; n = 6 for each group. Bioluminescence imaging for representative mouse (right). D, day; GSEA, gene set enrichment analysis.
Loss-of-EZH2 function is a hallmark of 5q alterations that target NR3C1 in BPDCN
To further explore the pathological relevance of 5q alterations targeting NR3C1 and related gene expression changes in BPDCN, we used GSEA to identify, in an unbiased manner, common dexamethasone-dependent gene signatures that are abnormally modulated upon overexpression of the t(3;5)-encoded GCR-FP or experimental NR3C1 haploinsufficiency in CAL-1 BPDCN cells. Consistent with our functional studies (Figure 2E-F), this analysis revealed enrichment of prednisone resistance gene signatures that have previously been described in B-ALL42,43 in dexamethasone-treated CAL-1-GCR-FP-expressing cells and CAL-1 shGCR cells compared with controls (supplemental Figure 2C). Of note, 1 of these resistance signatures43 was also enriched in NR3C1-deleted primary BPDCN cells compared with nondeleted cases, thus further reinforcing the notion that NR3C1 deletion contributes to treatment failure in BPDCN, as in B-ALL37,38 (supplemental Figure 2C). Strikingly, this analysis also revealed marked enrichment for signatures related to loss-of-EZH2 function in cancer cells, including gene expression programs that are functionally linked to HOXA locus derepression in myeloid leukemia44,45 (Figure 3C). This prompted us to examine the impact of attenuated GCR signaling on global H3K27me3 levels in BPDCN. For this, we used CAL-1 cells overexpressing GCR-FP or undergoing GCR knockdown to assess global H3K27me3 levels after low-dose dexamethasone treatment compared with control conditions. Consistent with our GEP experiments, western blot analyses revealed global loss of H3K27me3 under attenuated GCR signaling (Figure 3D; supplemental Figure 3D). Further analysis by site-specific anti-H3K27me3 ChIP confirmed reduced H3K27me3 levels at the HOXA locus, a major downstream target of the polycomb-repressive complex 2 (PRC2) complex in the hematopoietic lineage, specifically at the HOXA5, HOXA9, and HOXA11B gene promoters, in dexamethasone-treated GCR-FP overexpressing CAL-1 cells and in CAL-1 shGCR cells compared with controls (Figure 3E). In keeping with the clinical pertinence of these findings, GEP in primary NR3C1-deleted BPDCN leukemia cells compared with nondeleted cases from bone marrow (UPNs 32-42) or skin (UPNs 43-47) also identified EZH2 loss-of-function signatures (Figure 3F; supplemental Figure 3B). Genomic DNA sequencing was performed to check whether ASXL1 and EZH2 mutations might be responsible for the observed loss-of-EZH2 function signatures. This is important because mutations in these genes have been described in BPDCN.14 We screened the 2 BPDCN cell lines (UPNs 1 and 2) and the bone marrow or blood-derived DNA from the 12 patients for which GEP was used for GSEA (UPNs 31-42, including 4 cases with NR3C1 monoallelic loss [UPNs 31-33 and UPN 35]). Sequencing identified 2 BPDCN cases with deleterious ASXL1 exon 12 mutations (UPNs 34 and 40) (supplemental Table 4; supplemental Figure 3C) and 1 case (UPN 36) with a heterozygous EZH2 mutation. UPN 40 showed 2 different ASXL1 mutations (supplemental Table 4). In 5 cases (ie, 3 non-del(5q) [UPNs 38, 39, and 41] and 2 5q/NR3C1 deletion [UPNs 32 and 33]), the region spanning the G-rich region chromosome 20:31,022,184 to 31,022,744 of ASXL1 exon 12 could not be fully sequenced because of insufficient DNA. Of note however, mutations in this region appear rare in BPDCN (1/25 reported cases).14 When GSEA was performed, without the GEP data from the EZH2 and ASXL1 mutant cases (UPN 36 and UPNs 34 and 40, respectively), NR3C1 deletion remained significantly associated with multiple loss-of-EZH2 function signatures (Figure 3F; bone marrow, top; skin, bottom). Although this does not exclude other, yet-to-be-determined factors, in the emergence of the loss-of-EZH2 function signature in our NR3C1-deleted BPDCN cases, gene mutations affecting ASXL1 and EZH2 are unlikely to be responsible for the observed phenotype. In keeping with the relevance of these findings to BPDCN pathogenesis, knockdown of GCR significantly accelerates leukemia growth in a xenotransplant assay utilizing CAL-1 BPDCN cells (Figure 3G; supplemental Figure 3E-F). Of note, the loss-of-EZH2 function GEP signatures did not include PRC2 factor or H3K27me3 demethylase encoding genes, suggesting that altered PRC2 activity, rather than deregulated expression of individual PRC2 components per se, is responsible for the observed phenotypes. Taken together, these findings point to the existence of a novel cellular mechanism by which GCR and EZH2 signaling collaborate to reprogram EZH2 targets in leukemia.
LincRNA-3q is overexpressed in BPDCN and AML and encodes a nuclear noncoding RNA that regulates cell proliferation and leukemic stem cell gene expression programs
We next wished to assess the pathological relevance of the lncRNA gene identified at the 3q21 breakpoint in the t(3;5)(q21;q31) observed in BPDCN case UPN 1 (Figure 2A). For this, we first measured expression levels of lincRNA-3q in normal pDC, BPDCN cell lines, and primary BPDCN leukemia samples by RT-qPCR (Figure 4A). This revealed evidence of illegitimate lincRNA-3q activation in primary BPDCN cases compared with normal pDC. Deregulation of lincRNA-3q was also evident in AML (Figure 4A), B-cell lymphoma, and solid cancers (supplemental Figure 4A-B). In normal tissues, lincRNA-3q was predominantly expressed in fetal brain, bone marrow, and testis (supplemental Figure 4A). We next assessed the subcellular localization of lincRNA-3q by cell fractionation and qRT-PCR analysis in CAL-1 and U937 AML cells (Figure 4B). lincRNA-3q was found to be predominantly nuclear, suggestive of a role in chromatin signaling.46 Quite strikingly, lincRNA-3q knockdown in CAL-1 and U937 cells induced a G1/S arrest (Figure 4C and supplemental Figure 4C, respectively), without inducing cell death (supplemental Figure 4D). In view of this phenotype, GEP, followed by GSEA, was performed in the same cells (Figure 4D and supplemental Figure 4E, respectively). Short hairpin-mediated lincRNA-3q depletion was associated with differential expression of 371 (174 up; 197 down) and 904 genes (428 up; 476 down) in CAL-1 and U937 cells, respectively, thus suggestive of a cell context–dependent role for lincRNA-3q in both gene activation and repression. Among the lincRNA-3q regulated genes in BPDCN CAL-1 cells were TCL1B,47 HOXB4,48 and TP53INP1, a tumor suppressor gene49 (Figure 4D). Similarly, E2F family members (E2F7 and E2F8) known to negatively regulate E2F transcriptional activity were modulated by lincRNA-3q knockdown in U937 cells50 (supplemental Figure 4E). Consistent with G1/S arrest in lincRNA-3q-depleted leukemia cells, E2F-regulated gene expression signatures were markedly reduced upon lincRNA-3q depletion in both CAL-1 and U937 cells compared with controls (Figure 4D and supplemental Figure 4E, respectively), thus suggesting a role for lincRNA-3q in the regulation of E2F transcriptional transactivation functions. In keeping with this, lincRNA-3q depletion by siRNA transient transfection in H1299 cells led to G1/S arrest coincident to markedly reduced E2F activity, as measured by a cyclin E–luciferase reporter assay, in the same cells compared with control conditions (Figure 4E; supplemental Figure 5A-E). Knockdown of lincRNA-3q in CAL-1 and U937 AML cells was found to downregulate normal and/or leukemic hematopoietic stem cell gene expression programs48,51 (Figure 4D and supplemental Figure 4E, respectively), suggestive of a role in the normal hematopoietic/leukemic stem cell compartment, which is consistent with high-level lincRNA-3q expression in normal bone marrow samples (Figure 4A). This was borne out by in vitro limiting dilution clonogenicity assays in CAL-1 cells, which showed dramatically reduced clonogenic potential upon lincRNA-3q depletion compared with controls (Figure 4F). Concordant with this, lincRNA-3q knockdown markedly impaired CAL-1 leukemia growth in an in vivo xenotransplantation assay (Figure 5A; supplemental Figure 4F). Thus, lincRNA-3q displays features of a bona fide leukemic driver that warrants assessment for therapeutic intervention.
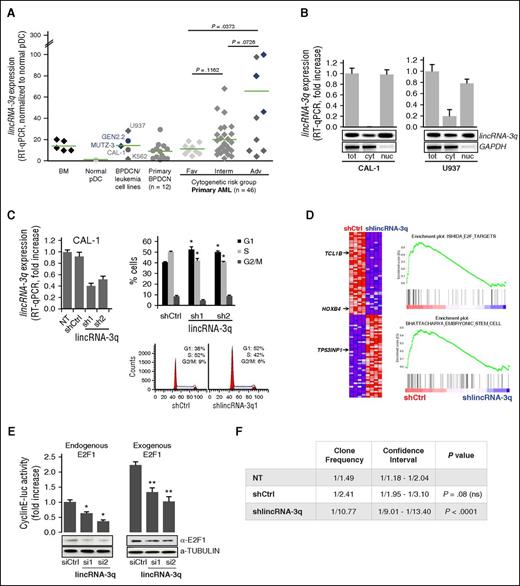
lincRNA-3q overexpression in BPDCN and AML drives G1/S and leukemia-driver gene expression programs. (A) RT-qPCR-derived lincRNA-3q expression profiles in normal bone marrow; normal pDC; BPDCN (CAL-1, GEN2.2), AML (U937, MUTZ-3), and chronic myeloid leukemia cell lines (K562); and in BPDCN and AML patient samples. AML patients are classified according to cytogenetic risk group. Cases presenting chromosome 3q abnormalities are identified in blue. (B) RT-qPCR and RT-PCR analysis of the subcellular localization of lincRNA-3q in CAL-1 BPDCN cells and U937 AML cells. RNA extracted from whole-cell and subcellular fractions (n = 2). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control. (C) RT-qPCR-derived lincRNA-3q expression in CAL-1 cells transduced with control (shCtrl) or lincRNA-3q-targeting short hairpin constructs (sh1 and 2) (left). Cell cycle analysis in CAL-1 cells transduced with control (shCtrl) or lincRNA-3q sh1 and 2 constructs (upper right). *P < .05 by Wilcoxon test; n = 6. Representative histogram representation of percentage of cells in cell cycle phases (n = 4) (lower right). (D) GSEA plots obtained by comparing gene expression profiles of CAL-1 cells transduced with control (shCtrl) or lincRNA-3q-targeting short hairpin (shlincRNA-3q) (right) and associated heatmap (n = 4 for each group) (left). Genes mentioned in the text are marked with an arrow. (E) E2F cyclin E–luciferase (luc) reporter assay in H1299 cells transiently transfected with control or siRNA targeting lincRNA-3q with or without addition of exogenous E2F1 (100 ng) (upper). *P < .05, **P < .01 by Wilcoxon test; n = 6. Western blot using anti-E2F1 antibody in H1299 cells transduced with siCtrl or silincRNA-3q, as indicated (n = 2) (lower). (F) Table presenting clone frequency and associated statistics derived from in vitro limiting dilution clonogenicity for CAL-1 either nontransduced (NT) or transduced with control (shCtrl) or lincRNA-3q-targeting (shlincRNA-3q) shRNA. Adv, adverse; cyt, cytosolic fraction; Fav, favorable; Interm, intermediate; nuc, nuclear fraction; tot, whole cell extract.
lincRNA-3q overexpression in BPDCN and AML drives G1/S and leukemia-driver gene expression programs. (A) RT-qPCR-derived lincRNA-3q expression profiles in normal bone marrow; normal pDC; BPDCN (CAL-1, GEN2.2), AML (U937, MUTZ-3), and chronic myeloid leukemia cell lines (K562); and in BPDCN and AML patient samples. AML patients are classified according to cytogenetic risk group. Cases presenting chromosome 3q abnormalities are identified in blue. (B) RT-qPCR and RT-PCR analysis of the subcellular localization of lincRNA-3q in CAL-1 BPDCN cells and U937 AML cells. RNA extracted from whole-cell and subcellular fractions (n = 2). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control. (C) RT-qPCR-derived lincRNA-3q expression in CAL-1 cells transduced with control (shCtrl) or lincRNA-3q-targeting short hairpin constructs (sh1 and 2) (left). Cell cycle analysis in CAL-1 cells transduced with control (shCtrl) or lincRNA-3q sh1 and 2 constructs (upper right). *P < .05 by Wilcoxon test; n = 6. Representative histogram representation of percentage of cells in cell cycle phases (n = 4) (lower right). (D) GSEA plots obtained by comparing gene expression profiles of CAL-1 cells transduced with control (shCtrl) or lincRNA-3q-targeting short hairpin (shlincRNA-3q) (right) and associated heatmap (n = 4 for each group) (left). Genes mentioned in the text are marked with an arrow. (E) E2F cyclin E–luciferase (luc) reporter assay in H1299 cells transiently transfected with control or siRNA targeting lincRNA-3q with or without addition of exogenous E2F1 (100 ng) (upper). *P < .05, **P < .01 by Wilcoxon test; n = 6. Western blot using anti-E2F1 antibody in H1299 cells transduced with siCtrl or silincRNA-3q, as indicated (n = 2) (lower). (F) Table presenting clone frequency and associated statistics derived from in vitro limiting dilution clonogenicity for CAL-1 either nontransduced (NT) or transduced with control (shCtrl) or lincRNA-3q-targeting (shlincRNA-3q) shRNA. Adv, adverse; cyt, cytosolic fraction; Fav, favorable; Interm, intermediate; nuc, nuclear fraction; tot, whole cell extract.
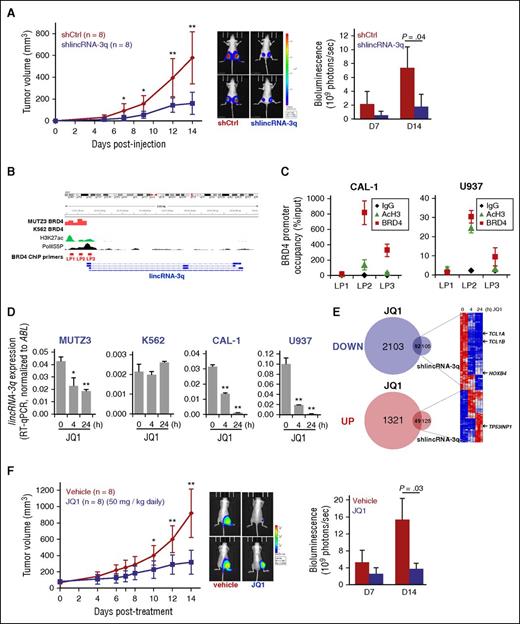
Abrogation of lincRNA-3q activity by BET inhibition in BPDCN and AML leukemia cells. (A) Tumor growth kinetics measured in nude mice engrafted with shCtrl or shlincRNA-3q-transduced CAL-1 cells (left); bioluminescence imaging for 2 representative mice (day 14) (middle); and quantification of tumor bioluminescence (7 and 14 days postinjection) for each group (right). *P < .05, **P < .01 by Student t test; n = 8 for each group. (B) Genomic organization of the lincRNA-3q locus and BRD4 ChIP-sequencing analysis (ArrayExpress accession number ERP004614) showing promoter occupancy by BRD4 in MUTZ-3 cell line (3q rearrranged AML) compared with K562 cell line (no 3q rearrangement). ENCODE data showing positions of active chromatin marks and phospho-RNA polymerase II (Pol II) binding, as indicated. (C) Anti-BRD4 and anti-AcH3 ChIP at the promoter region of lincRNA-3q in CAL-1 and U937 cells, as indicated. (D) RT-qPCR-derived lincRNA-3q expression in MUTZ3, K562, U937, and CAL-1 cells treated with 1 μM JQ1 at the time indicated. *P < .05, **P < .01 by Wilcoxon test; n = 3. (E) Venn diagrams (left) and heatmap (right) showing time course of differential gene expression of lincRNA-3q targets upon JQ1 treatment in CAL-1, as indicated. (F) Tumor growth kinetics measured in nude mice bearing tumors derived from CAL-1 cell lines after control or JQ1 treatment (left); bioluminescence imaging for 2 representative mice (day 14) (middle); and quantification of tumor bioluminescence (7 and 14 days posttreatment) for each group (right). *P < .05, **P < .01 by Student t test; n = 6 for each group. AcH3, acetylated histone H3; BET, bromodomain and extraterminal domain; Ig, immunoglobulin; LP, lincRNA ChIP Primer.
Abrogation of lincRNA-3q activity by BET inhibition in BPDCN and AML leukemia cells. (A) Tumor growth kinetics measured in nude mice engrafted with shCtrl or shlincRNA-3q-transduced CAL-1 cells (left); bioluminescence imaging for 2 representative mice (day 14) (middle); and quantification of tumor bioluminescence (7 and 14 days postinjection) for each group (right). *P < .05, **P < .01 by Student t test; n = 8 for each group. (B) Genomic organization of the lincRNA-3q locus and BRD4 ChIP-sequencing analysis (ArrayExpress accession number ERP004614) showing promoter occupancy by BRD4 in MUTZ-3 cell line (3q rearrranged AML) compared with K562 cell line (no 3q rearrangement). ENCODE data showing positions of active chromatin marks and phospho-RNA polymerase II (Pol II) binding, as indicated. (C) Anti-BRD4 and anti-AcH3 ChIP at the promoter region of lincRNA-3q in CAL-1 and U937 cells, as indicated. (D) RT-qPCR-derived lincRNA-3q expression in MUTZ3, K562, U937, and CAL-1 cells treated with 1 μM JQ1 at the time indicated. *P < .05, **P < .01 by Wilcoxon test; n = 3. (E) Venn diagrams (left) and heatmap (right) showing time course of differential gene expression of lincRNA-3q targets upon JQ1 treatment in CAL-1, as indicated. (F) Tumor growth kinetics measured in nude mice bearing tumors derived from CAL-1 cell lines after control or JQ1 treatment (left); bioluminescence imaging for 2 representative mice (day 14) (middle); and quantification of tumor bioluminescence (7 and 14 days posttreatment) for each group (right). *P < .05, **P < .01 by Student t test; n = 6 for each group. AcH3, acetylated histone H3; BET, bromodomain and extraterminal domain; Ig, immunoglobulin; LP, lincRNA ChIP Primer.
BET inhibition abrogates lincRNA-3q overexpression and signaling in BPDCN
Numerous cancer genes,29,52,53 including the cancer-related lncRNA gene HOTAIR,54 have been shown to critically depend on proteins of the BET domain family for their expression. Because high-level overexpression of lincRNA-3q was detected in 3q-rearranged AML in which BET proteins (bromodomain-containing protein 4 [BRD4]) have been implicated in deregulation of a critical 3q oncogene (EVI1), we asked whether BRD4 might also play a role in the regulation of lincRNA-3q. To address this question, we interrogated BRD4 ChIP sequencing data sets52 derived from MUTZ3 (3q-rearranged AML cell line) compared with K562 (no 3q rearrangement) leukemia cells (ArrayExpress accession number ERP004614). This revealed marked enrichment of BRD4 across the promoter region of the linc-RNA-3q gene in MUTZ3 cells, but not K562 cells (Figure 5B). Site-specific anti-BRD4 ChIP subsequently revealed BRD4 occupancy of the lincRNA-3q promoter in CAL-1 and U937 cells (Figure 5C). In aggregate, this indicated that BRD4 directly regulates linc-RNA-3q in CAL-1, U937, and MUTZ-3 cells, but not K562 cells (no detectable occupancy of the lincRNA-3q promoter by BRD4) and that BET inhibition might constitute a means to reverse abnormal lincRNA-3q expression in leukemia cells. In keeping with this, treatment with one such BET inhibitor (JQ1) led to suppression of lincRNA-3q levels in a time-dependent manner in MUTZ-3, U937, and CAL-1 BPDCN cells, but not in K562 cells (Figure 5D). In view of this, we next assessed the consequences on global gene expression profiles of JQ1 treatment BET-sensitive lincRNA-3q-overexpressing CAL-1 cells compared to those obtained upon depletion of lincRNA-3q in the same cells. Quite strikingly, JQ1 treatment in CAL-1 cells modulated a common subset of lincRNA-3q targets (92 down; 49 up), including downregulation of known leukemic driver genes (eg, TCL1A, TCL1B, and HOXB4, of which the former 2 are of relevance to BPDCN), as well as enhanced expression of candidate tumor suppressor genes such as TP53INP1 (Figures 4D and 5E, arrows; supplemental Table 14). As expected for BET inhibition, numerous additional genes not coregulated by lincRNA-3q were also identified (Figure 5E). Thus, BET inhibition would be expected to counteract at least some of the oncogenic signaling properties of lincRNA-3q in BPDCN and AML. To test this in vivo, we performed a JQ1 treatment protocol in a CAL-1 xenotransplantation assay, which revealed significant suppression of CAL-1 leukemia growth in the treatment (50 mg/kg daily for 14 days) compared with the control arm (vehicle alone) (Figure 5F) at kinetics that closely mirrored those seen in lincRNA-3q-depleted compared with control tumors (Figure 5A; supplemental Figure 4F), indicating that BET inhibition is a potent therapeutic strategy in BPDCN.
Discussion
A remarkable finding from our focused genetic screen was the identification of the GCR-encoding gene, NR3C1, as a candidate 5q-linked haploinsufficient tumor suppressor in BPDCN. Based on functional studies coupled with GEP and survival analyses in BPDCN patients, we provide novel evidence of a role for NR3C1 haploinsufficiency in corticoresistance and functional antagonism of gene silencing via EZH2 in a subset of BPDCN. In addition, this report contains the first description of an NR3C1 fusion gene translocation in human leukemia, thereby raising the necessity for screening for balanced structural aberrations at this locus in BPDCN.
That corticoresistance could be driven by haploinsufficiency for GCR expression in BPDCN is in line with recent findings that identify diminished GCR protein levels as a major determinant of de novo corticoresistance in B-ALL.55 NR3C1 haploinsufficiency in BPDCN is associated with gene expression programs that characterize corticoresistance in B-ALL, suggestive of the existence of common cellular mechanisms for resistance to glucocorticoid-induced apoptosis in BPDCN and B-ALL.42,43 In the specific case of BPDCN, most cases initially respond well clinically to corticotherapy-based regimens, suggesting that glucocorticoid resistance may arise initially at the subclinical level as a consequence of the persistence of residual resistant cells that are haploinsufficient for NR3C1. Larger prospective studies will be required to address the relationship, if any, between NR3C1 haploinsufficiency, residual disease, and the presence of central nervous system involvement at diagnosis which has been implicated in rapid disease recurrence in BPDCN patients.56 As proposed recently,55 restoring normal GCR levels from the intact NR3C1 allele might represent a route to overcoming glucocorticoid-based therapy failure in NR3C1-haploinsufficient BPDCN. Of note in this context is the finding that thalidomide appears to specifically benefit patients presenting with multiple myeloma and low GCR expression.57
The present study has revealed a previously unsuspected connection between GCR signaling and EZH2 activity in human leukemia. EZH2 activity seems to be acutely sensitive to GCR signaling thresholds. Attenuated GCR signaling was associated with profound reductions in H3K27me3, including at the HOXA locus, which is a target for derepression in leukemia. We speculate that under conditions in which GCR is limiting, such as in haploinsufficiency for NR3C1, GCR-ligand binding elicits dynamic GCR recruitment to high affinity sites in chromatin where either transcriptional activation or repression, depending on the regulatory context of nearby promoters or enhancers, occurs.58,59 Enhanced GCR recruitment to high affinity sites would occur at the expense of GCR recruitment to lower-affinity sites across the genome, including at loci that may encode hormone-sensitive, developmentally regulated, and/or lineage-specific transcriptional regulatory factors that are direct PRC2 targets or that control deployment of PRC2 to other critical downstream targets such as the HOXA locus in BPDCN. In an alternative, but not mutually exclusive scenario, GCR binding to chromatin increases active marks such as H3K4me2/3 and H3K27ac,60 which are known to antagonize H3K27me3 by PRC2.61,62
We propose that hormone-dependent, epigenetic reprogramming is the ultimate consequence of NR3C1 haploinsufficiency in BPDCN (Figure 6). This hypothesis is supported by our clinical and functional analysis in vitro and in vivo and by reports in other cancers showing that low NR3C1/GCR expression is linked to poor prognosis57,63,64 and tumor progression.65 This postulate is also consistent with analyses showing increased tumor incidence in Nr3c1 haploinsufficient mice.64
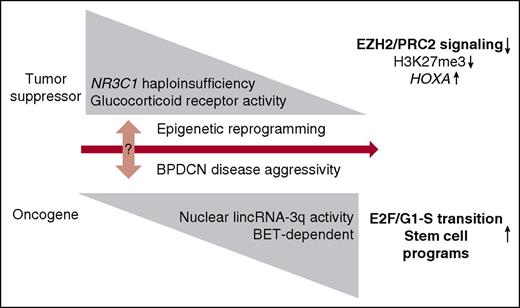
A model illustrating how NR3C1 haploinsufficiency and lincRNA-3q misregulation contribute to BPDCN pathogenesis. We postulate that attenuated GCR signaling and lincRNA-3q malfunction drive BPDCN disease pathogenesis through epigenetic reprogramming. This is proposed to favor emergence of clinically aggressive disease and predicted to occur progressively by 2 routes. In the first, altered GCR signaling drives a loss-of-EZH2 phenotype that rewires key downstream PRC2 targets (eg, the HOXA locus) and drives deregulation of pDC differentiation pathways and treatment resistance in BPDCN. In the second, BET-dependent desilencing of oncogenic lncRNA genes (lincRNA-3q) may occur as “collateral damage” downstream of altered GCR and EZH2 activity or through other mechanisms. Abnormal activity of the affected nuclear lncRNA (in this case, lincRNA-3q) would engage further rounds of epigenetic reprogramming, leading to misregulation of E2F activity and activation of leukemia stem cell programs.
A model illustrating how NR3C1 haploinsufficiency and lincRNA-3q misregulation contribute to BPDCN pathogenesis. We postulate that attenuated GCR signaling and lincRNA-3q malfunction drive BPDCN disease pathogenesis through epigenetic reprogramming. This is proposed to favor emergence of clinically aggressive disease and predicted to occur progressively by 2 routes. In the first, altered GCR signaling drives a loss-of-EZH2 phenotype that rewires key downstream PRC2 targets (eg, the HOXA locus) and drives deregulation of pDC differentiation pathways and treatment resistance in BPDCN. In the second, BET-dependent desilencing of oncogenic lncRNA genes (lincRNA-3q) may occur as “collateral damage” downstream of altered GCR and EZH2 activity or through other mechanisms. Abnormal activity of the affected nuclear lncRNA (in this case, lincRNA-3q) would engage further rounds of epigenetic reprogramming, leading to misregulation of E2F activity and activation of leukemia stem cell programs.
A second striking finding in this work resulted from the detailed characterization of the NR3C1 fusion partner in our index case presenting the balanced t(3;5)(q21;q31). Molecular cloning of this case identified a novel nuclear lncRNA-encoding gene as the partner gene in 3q21. Significantly, ectopic expression of this lncRNA was linked to enrichment of the BET protein BRD4 to the promoter region. BET-inhibitor treatment rapidly abrogated lincRNA-3q expression only in leukemia cells showing BRD4 binding to the lincRNA-3q promoter, suggestive of a critical role for BET proteins in the maintenance of high-level lincRNA-3q expression in BPDCN and AML. We speculate that BRD4 recruitment to the lincRNA-3q promoter may depend on the activity of upstream (oncogenic) transcription factors, including possibly the GCR itself, that direct BRD4 to the lincRNA-3q promoter region or to neighboring “superenhancers.” The latter are known to mediate long-range dysregulation of key oncogenes in hematologic cancers52 and to coordinate activity of key hematopoietic transcription factors in leukemia.66 Alternatively, disruption of BRD4 interaction with transcription and elongation factors (eg, polymerase II, positive transcription elongation factor b, Mediator, and Activator) may be part of the mechanism by which JQ1 inhibits lincRNA-3q expression. Finally, intrinsic histone acetylase activity of BRD4 itself may play a role.67 Our findings indicate that lncRNA function constitutes a novel BET-inhibitor target in human leukemia.
In conclusion, our focused, functional genomics analysis has identified attenuated GCR signaling and unscheduled activation of a novel nuclear lncRNA as important drivers of deregulated epigenetic signaling, treatment resistance, and malignant reprogramming in the dendritic cell lineage (Figure 6). Further studies will be required to fully leverage these findings for treatment innovation in BPDCN and potentially other myeloid and lymphoid cancers that share biological features with this disease. One avenue of investigation is BET protein inhibition.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank members of the Groupe Francophone de Cytogénétique Hématologique for enrollment of BPDCN cases and for centralized review of karyotype and FISH data; Veronique Pantesco at the Microarray Core Facility of the Institute of Research on Biotherapy, Centre Hospitalier Régional Universitaire de Montpellier, INSERM Université Montpellier 1, for performing Affymetrix microarrays; Amandine Chatagnon for advice on next-generation sequencing (NGS), NGS core facility Grenoble University Hospital; Nicolas Lemaitre of the Plateforme de Pathologie Moléculaire in situ, Grenoble University Hospital, for IHC analysis; Céline Villenet, Frédéric Leprêtre, and Martin Figeac of the Plateforme de génomique fonctionnelle et structurale, Université de Lille, for performing and analyzing aCGH; Mylene Pezet and Alexandra Debernardi for expert advice on flow cytometry and bioinformatics, respectively (Flow cytometry and EpiMed core facilities, INSERM U1209, CNRS 5309, Université Grenoble Alpes); Beatrice Eymin for providing the E2F activity reporter system; Patricia Betton-Fraisse and staff in the cytogenetics and molecular genetics laboratory and at the Centre de Resources Biologiques–Grenoble University Hospital for additional technical assistance; André Verdel, Ramin Shiekhattar, Saadi Khochbin, Carlo Petosa, Alexandra Traverse-Glehen, and Tony Petrella for helpful discussions during the course of this work.
This study was supported by the Ligue Nationale Contre le Cancer (comités de l’Isère et de la Haute Savoie) (M.B.C.) and by the French Ministry for Higher Education and Research and the Société Française d’Hématologie (N.H.; doctoral funding). Additional funding in the M.B.C. laboratory is from the Institut National du Cancer “Epigénétique et Cancer Programme.” NGS diagnostics in oncology/hematology at Grenoble University Hospital is funded through the Institut National du Cancer “NGS à visé diagnostique” program via the “Plateforme de génétique moléculaire des cancers ” of Grenoble Hospital (P. Hainaut).
Authorship
Contributions: A.E. analyzed the data and jointly coordinated the research; N.H., S.D., A.H., S.B., S.H., M.C., and J.B.-C. performed all molecular and cell biology experiments and analyzed the data; C.S.-M. performed the targeted deep sequencing; E.B. interpreted the IHC and pathology; V.J. performed the bioluminescence imaging in vivo and analyzed the data; E.V. and F.-L.C. provided advice on the lentiviral constructs, supplied all lentiviral stocks, and analyzed the data; T.B. coordinated the flow cytometry and the limiting dilution analysis and performed the patient survival analysis; S.R. performed the bioinformatics analysis; P.B., C.R., A.R., J.C., I.T., T.M., L.C., J.P., M.-C.J., S.P., R.G., C.P.T., C.M., E.M., D.L., F.N.-K., I.L., D.P., C.B., F.J., C.L., and F.G. provided the patient samples, clinical and molecular cytogenetic/transcriptome data, and expert analysis on BPDCN (through the French BPDCN network and Groupe Francophone de Cytogénétique Hématologique); and M.B.C. conceived and coordinated the research, analyzed the data, and wrote the paper. All authors contributed to manuscript preparation, writing, and final approval.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mary B. Callanan, Centre de Recherche, INSERM U1209, CNRS UMR 5309, Université Grenoble Alpes, Site Santé, IAB, BP 170, La Tronche, 38042 Grenoble Cedex 9, France; e-mail: mary.callanan@univ-grenoble-alpes.fr.
References
Author notes
A.E., N.H., and S.D. contributed equally to this study.
F.G. and M.B.C. are co-senior authors.

![Figure 1. Targeting of the NR3C1 gene by 5q deletion/translocation in BPDCN. (A) Schematic representation of 5q alterations in BPDCN patients (n = 17/47), as indicated (left). aCGH image of case UPN 33 showing focal deletion of the NR3C1 gene (right). (B) Kaplan-Meier cumulative survival curves in BPDCN according to 5q alteration status (for simplicity, this is labeled del(5q) [UPNs 1, 2, 8, 12, 14, 21, 22, 24, 26, 27, 29, 31, 35, and 45] and non-del(5q) [UPNs 3, 9, 10, 16-19, 25, 27, 36, 37, and 40-42]; upper) and to NR3C1 deletion status (UPNs 1, 8, 14, 22, 24, 26, 27, 31, 35, and 45) (lower), determined by FISH or aCGH; ΔNR3C1). P values were determined by log-rank test. (C) FISH analysis of the t(3;5)(q21;q31) in the BPDCN cell line GEN2.2 (UPN 1) with chromosome 5q probes CTB-88J14 (green) and RP11-278J6 (red) (supplemental Table 2). Note a split signal for RP11-278J6 (NR3C1) on the der(3) and der(5) chromosomes. (D) Affymetrix-derived NR3C1 expression in BPDCN patients presenting del(5q) targeting NR3C1 or not. P value was determined by Wilcoxon test. (E) aCGH image showing 5q/NR3C1 deletion in DNA from a skin biopsy sample with leukemic infiltrate (UPN 22) compared with control DNA (Ctrl), and analysis in 2 BPDCN cases without 5q/NR3C1 deletion (UPNs 19 and 21). (F) Overview of immunostaining (diaminobenzidine and hematoxylin) of GCR on BPDCN dermal localization in a del(5q) case: NR3C1(+) (left) and a del(5q)/NR3C1 case: NR3C1(−) (upper right). Epidermal squamous cells showing positive nuclear staining are marked with thin gray arrows; leukemia infiltrate cells are marked with thick black arrows. A higher magnification of the clonal infiltrate in dermis of the same case as the upper right image shows mainly negative tumor cells (red arrow) and some positive cells (thick black arrow) (lower right). BM, bone marrow.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/24/10.1182_blood-2015-09-671040/4/m_3040f1.jpeg?Expires=1769147487&Signature=SWoG1P7IkCjM4I~R~Z49xTd0~kXP9~K3IBxZHStLV4qGCijMOblFLbY8Eykzy1HE~WFq-M7U6W1GLbIhejIcRLojiJAFh9~4ChvzCUshS38nd59v2mhPoeFo0e6qeVvJ7C61OUO5ZrM4F3KqDs2RqdZ4dgdjwra0sT-915GGjVVMdAN3WCxyml-48ecMfXGntN~b8bpuUsz8jBava4r41JDnkGrdYNC5KVPyY5nxV9sApe3F0EOn0F9GR9d9AEUjJ-bI00HymVTVtME~ZNM7lhPcGwvtvqxOuZmvRH2CpgETNzdJiCy1TaxjEc8q6eLOYOW7TC0on3miA1wOw~Kw0A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Misregulated GCR and EZH2 signaling as a consequence of NR3C1 alterations in BPDCN. (A) Luciferase reporter assay for glucocorticoid transcriptional transactivation activity in COS7 cells transduced with empty pcDNA3.1 vector, pcDNA3.1-GCR, and pcDNA3.1-GCR-FP, as indicated, after a 6-hour treatment with either dimethyl sulfoxide (DMSO) or 100 nM dexamethasone (n = 3). (B) Heatmap representation of the common differentially expressed genes among CAL-1-GCR-FP (GCR-FP[+]), CAL-1 control cells (GCR-FP[−]), and shGCR-transduced cells (shGCR[+]) compared with controls (shGCR[−]) after treatment with 100 nM dexamethasone for 6 hours (n = 3 for each group). Black arrows indicate genes cited in the text. (C) GSEA plots showing gene regulatory circuits that are differentially expressed between dexamethasone-treated CAL-1-GCR-FP or shGCR compared with respective control CAL-1 cells (CAL-1-GFP or CAL-1 shCtrl). (D) Western blot analysis and quantification of global H3K27me3 levels in CAL-1-GCR-FP (GCR-FP[+]) and CAL-1 shGCR (upper) and respective CAL-1 control cells (CAL-1 GFP [GCR-FP(−)] and CAL-1 shCtrl) (lower) upon 6-hour or 24-hour treatment with 100 nM dexamethasone (n = 3). (E) ChIP for H3K27me3 followed by qPCR analysis for enrichment on HOXA gene promoters, as indicated, in CAL-1-GCR-FP (GCR-FP[+]), CAL-1 shGCR, and respective CAL-1 control cells (CAL-1 GFP [GCR-FP(−)] and CAL-1 shCtrl) treated for 24 hours with 100 nM dexamethasone (n = 3). (F) GSEA plots showing gene regulatory circuits that are differentially expressed between BPDCN patients presenting, or not, del(5q) abnormalities that target NR3C1 (bone marrow or skin, as indicated; unmutated EZH2 and ASXL1 cases only). (G) Tumor bioluminescence (7 and 14 days postinjection) for xenotransplanted nude mice bearing tumors derived from CAL-1 shCtrl and shGCR cell lines (left). P value determined by Wilcoxon test; n = 6 for each group. Bioluminescence imaging for representative mouse (right). D, day; GSEA, gene set enrichment analysis.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/24/10.1182_blood-2015-09-671040/4/m_3040f3.jpeg?Expires=1769147487&Signature=ML3fmLB2CU42~KEbmTCg8yw~cxTXQXtJIdDAdGPrnMpTjnPddJZoQFhtwFTGF94-SRik7KxMXfWstwHeG1Ak907yMTab8sXNvwiq8U95FQBEAw2Hze0WbqQ9vOMAm~oubHmilxFpFFa6Fdgz-ALxSS9yBTjJZLp-g56-eUuXsMYnrCVdy0M6M8Y~XlnoYUKYdhlaLMaWVnTXi9sOx9DLl-qFyFk7ajEtJyKr-gnxYE9-~cCp~ShXhPQAdtlnH6N7PNjPz0ygAcvf-jTk~9rIRifNkAhEVn8v0x36aJ3~ocYF5p5TjR9QDeqWMy2py2qYClhVc7uExknn-Ql4cVs-Eg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)