Key Points
Clonal PCs in AL have similar phenotypic and CNA profiles as those in MM, but their transcriptome is similar to that of normal PCs.
First-ever WES in AL amyloidosis reveals potential lack of a unifying mutation.
Abstract
Immunoglobulin light-chain amyloidosis (AL) and multiple myeloma (MM) are 2 distinct monoclonal gammopathies that involve the same cellular compartment: clonal plasma cells (PCs). Despite the fact that knowledge about MM PC biology has significantly increased in the last decade, the same does not apply for AL. Here, we used an integrative phenotypic, molecular, and genomic approach to study clonal PCs from 24 newly diagnosed patients with AL. Through principal-component-analysis, we demonstrated highly overlapping phenotypic profiles between AL and both monoclonal gammopathy of undetermined significance and MM PCs. However, in contrast to MM, highly purified fluorescence-activated cell–sorted clonal PCs from AL (n = 9) showed almost normal transcriptome, with only 38 deregulated genes vs normal PCs; these included a few tumor-suppressor (CDH1, RCAN) and proapoptotic (GLIPR1, FAS) genes. Notwithstanding, clonal PCs in AL (n = 11) were genomically unstable, with a median of 9 copy number alterations (CNAs) per case, many of such CNAs being similar to those found in MM. Whole-exome sequencing (WES) performed in 5 AL patients revealed a median of 15 nonrecurrent mutations per case. Altogether, our results show that in the absence of a unifying mutation by WES, clonal PCs in AL display phenotypic and CNA profiles similar to MM, but their transcriptome is remarkably similar to that of normal PCs.
Introduction
Light-chain amyloidosis (AL) and multiple myeloma (MM) are 2 distinct monoclonal gammopathies that involve the same cellular compartment: clonal plasma cells (PCs). Despite the fact that knowledge about the biology of MM PCs has greatly increased in recent years,1 the same does not apply to AL. Thus, the cytogenetic profile of patients with AL has only been reported in a few studies2-7 that investigated a limited number of alterations (eg, IGH translocations or 13q and 17p deletion). Apparently, t(11;14) is the most frequent cytogenetic alteration in AL and confers a poor outcome,4,8 whereas MM-related high-risk cytogenetic abnormalities (ie, [t(4;14), t(14;16) and/or del(17p)]) are prognostically neutral in AL.8 Conversely, no data have been reported on copy number alterations (CNAs) and potential recurrent mutations present in clonal PCs from AL patients.9 Of note, the only study in which the gene expression profile (GEP) of (total) bone marrow (BM) PCs was investigated showed that deregulated genes in AL have intermediate expression between MM and normal PCs,10 although this observation may result from the systematic coexistence of clonal and normal PCs in the BM of patients with AL.11 In fact, the low incidence of AL and its typically low tumor burden, often masked by a polyclonal PC background,11 most probably account for the limited information on AL tumor cell biology, particularly when compared with MM. Accordingly, in this study, we characterized the phenotypic, transcriptomic, and genetic profile of highly-purified clonal PCs from patients with newly diagnosed AL.
Methods
Overall, 24 patients with confirmed diagnosis of AL based on the presence of amyloid-related systemic syndrome, positive amyloid tissue staining with Congo red, and evidence of PC clonality were studied. Patients’ demographics and clinical characteristics are described in supplemental Table 1, available on the Blood Web site. Samples were collected after informed consent was given, in accordance with the local ethics committee guidelines and the Declaration of Helsinki.
Approximately 200 μL of EDTA-anticoagulated BM aspirate of sample aliquots was immunophenotyped using 2 different 8-color combinations of monoclonal antibodies, and a direct immunofluorescence stain-and-then-lyse technique (PacB/PO/FITC/PE/ PerCP-Cy5.5/ PE-Cy7/APC/APC-H7): CD38/CD45/β2M/CD56/CD138/CD19/cyKappa/cyLambda and CD38/CD45/CD27/CD28/CD138/CD19/CD117/CD81. Data acquisition was performed in a FACS Canto II flow cytometer (Becton Dickinson Biosciences [BD], San Jose, CA) using the FACSDiva 6.1 software (BD). Generation of immunophenotypic protein expression profiles (iPEP) based on the 12 different markers evaluated at the single-cell level was performed as described elsewhere12 using the Infinicyt software (Cytognos SL, Salamanca, Spain).
GEP was performed in 9 of 24 AL cases with adequate RNA extracted from fluorescence-activated cell sorted (FACS)-purified clonal PCs according to patient-specific aberrant phenotypes (≥95% purity), and compared with that of normal PCs from 5 healthy individuals (FACS Aria IIb, BD). RNA was retrotranscribed into cDNA and hybridized to the Human Gene 1.0 ST Array (Affymetrix, Santa Clara, CA); normalization of the measured data was done using the expression console (Affymetrix) and the RMA algorithm, which includes background correction, normalization, and calculation of expression (log2) values.12 Differentially expressed genes between classes were identified using the Significant Analysis of Microarrays algorithm (http://www.statweb.standford.edu/∼tibs/SAM) and the lowest q-value (<10−5).12
Genome-wide detection of CNAs and copy number–neutral (CNN) loss-of-heterozygosity (LOH) were investigated using the Cytoscan 750K platform (Affymetrix) in 11 of 24 cases, with adequate DNA extracted from FACS-sorted clonal PCs. Given that matched normal DNA was only available in a subset of cases (n = 7/11), an unpaired analysis was performed using 240 Hapmap files. In those cases with paired normal DNA (peripheral blood T lymphocytes), the latter was also used to eliminate patient-specific CNAs. The AGCC and ChAS software programs (Affymetrix) were used for data analysis, as described elsewhere.12,13 Full microarray data are available at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/; accession number GSE73042). Fluorescence in-situ hybridization (FISH) investigating IGH translocations and del(17p13) was also performed; del(13q14) was tested in selected cases.
Whole-exome sequencing (WES) was performed in FACS-sorted clonal PCs and paired T lymphocytes from 5 of 24 patients with AL. Exonic sequences were enriched using the Ion Ampliseq Exome kit (Life Technologies, Carlsbad, CA), sequenced with a 150× depth coverage using semiconductor technology (Ion Proton, Carlsbad, CA), and analyzed with the Ion Reporter and Torrent Suite Software (Life Technologies). Single-nucleotide variants (SNVs) were considered to be positive when called by ≥10% variant reads. Full WES data are available at the Sequence Read Archive under the study accession number SRP070988 (http://www.ncbi.nlm.nih.gov/bioproject/313508).
Results and discussion
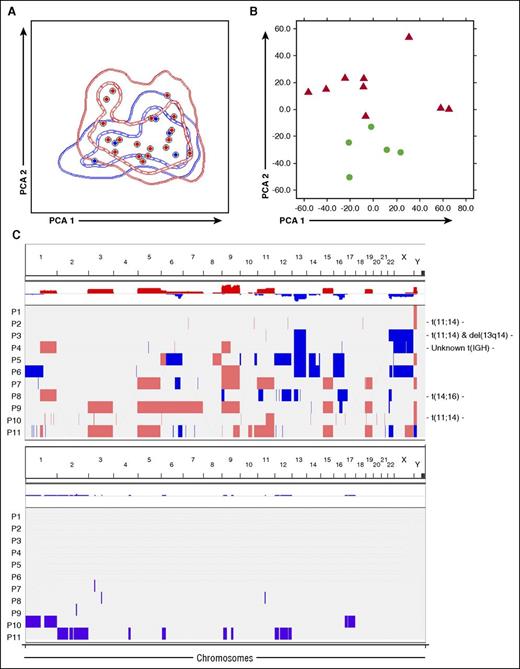
Clonal PCs were detectable in all 24 (100%) patients with AL (median, 0.81%; range, 0.01%-30%). The median distribution between clonal/normal PCs within the BM PC compartment was of 85%/15%, and only in 8 of 24 (33%) cases, the majority (≥95%) of PCs were phenotypically clonal. Aberrant PCs were identified based on downregulation of CD19 (100% of cases), CD27 (67%), CD38 (42%), CD45 (53%), and CD81 (48%), and overexpression of CD117 (29%), CD28 (50%), and CD56 (61%). β2M was strongly expressed on all clonal PCs, similar to normal PCs. Overall, the iPEP of clonal PCs in AL overlapped with that of monoclonal gammopathy of undetermined significance (MGUS)/MM (Figure 1A), and, similarly to the former, transitioning from 4-color to 8-color multiparameter flow cytometry (MFC) based on the EuroFlow approach improved the detection rate of PC clonality in AL from 97%11 to 100% of patients. Hence, despite lower BM PC percentages compared with conventional morphology (partly explained because samples evaluated by the latter contain cells associated with lipid-enriched spicules, whereas MFC is performed on the BM fluid, which is depleted in the lipid-adhesive PC),2,4,8,11 MFC represents a highly sensitive tool to demonstrate clonality in AL.
Phenotypic, transcriptomic, and genomic features of clonal plasma cells in AL. (A) Immunophenotypic protein expression profiles of clonal plasma cells from patients with AL (red) and MGUS plus MM (blue). These latter patients were initially screened for bone marrow clonality using the same immunophenotypic approach because of suspected AL, but were finally diagnosed as having MGUS (n = 3) and MM (n = 3). In the principal component analysis (PCA) graphic view, every patient is represented by a single dot, and disease reference groups by 1 (dashed lines) and 2 (solid lines) standard deviation curves. (B) Unsupervised PCA shows differences in gene expression profiling between clonal PCs from patients with AL (red) and normal PCs from healthy donors (green). (C) Overview of CNAs and copy number neutral loss of heterozygosity (CNN-LOH) detected in clonal PCs from patients with newly diagnosed AL. Samples are distributed in the y-axis and chromosome location in the x-axis. Red shading indicates the presence of copy number gains, and blue indicates copy number losses. CNAs were reported when the 3 following criteria were met: ≥25 consecutive imbalanced markers per segment, ≥100 Kb minimum genomic size, and <50% overlap with paired control DNA and/or genomic variants of Toronto DB (DGV). Those cytogenetics alterations detected by FISH in each individual patient are detailed at the beginning of the corresponding row. CNN-LOH are represented by violet bars. Only CNN-LOH >5 Mb, with ≥25 consecutive imbalanced markers per segment, and <50% overlap with patient-paired CNAs were considered.
Phenotypic, transcriptomic, and genomic features of clonal plasma cells in AL. (A) Immunophenotypic protein expression profiles of clonal plasma cells from patients with AL (red) and MGUS plus MM (blue). These latter patients were initially screened for bone marrow clonality using the same immunophenotypic approach because of suspected AL, but were finally diagnosed as having MGUS (n = 3) and MM (n = 3). In the principal component analysis (PCA) graphic view, every patient is represented by a single dot, and disease reference groups by 1 (dashed lines) and 2 (solid lines) standard deviation curves. (B) Unsupervised PCA shows differences in gene expression profiling between clonal PCs from patients with AL (red) and normal PCs from healthy donors (green). (C) Overview of CNAs and copy number neutral loss of heterozygosity (CNN-LOH) detected in clonal PCs from patients with newly diagnosed AL. Samples are distributed in the y-axis and chromosome location in the x-axis. Red shading indicates the presence of copy number gains, and blue indicates copy number losses. CNAs were reported when the 3 following criteria were met: ≥25 consecutive imbalanced markers per segment, ≥100 Kb minimum genomic size, and <50% overlap with paired control DNA and/or genomic variants of Toronto DB (DGV). Those cytogenetics alterations detected by FISH in each individual patient are detailed at the beginning of the corresponding row. CNN-LOH are represented by violet bars. Only CNN-LOH >5 Mb, with ≥25 consecutive imbalanced markers per segment, and <50% overlap with patient-paired CNAs were considered.
After identification and sensitive FACS-purification of clonal PCs, we performed GEP to identify uniquely deregulated genes in AL vs normal PCs. Even though multidimensional scaling analysis showed differences between both groups of PCs (Figure 1B), there were only 38 genes significantly deregulated (3 upregulated and 35 downregulated) in AL PCs (excluding 22 immunoglobulin coding genes, which are physiologically upregulated in normal PCs and mainly reflect differences between clonal vs polyclonal PCs; supplemental Excel File 1). Interestingly, the CDH1 and RCAN tumor-suppressor genes and the GLIPR1 and FAS proapoptotic genes14-18 were all significantly downregulated in AL; in contrast, IFITM1, which has been associated with more aggressive solid tumors,19 was overexpressed in clonal PCs. It is worth noting that CD19 and CD81 mRNAs were significantly decreased in AL, confirming the correlation between the iPEP and the GEP of clonal vs normal PCs. Overall, these results indicate that, although clonal PCs in AL and MM share similarly altered iPEP, clonal PCs in AL show a much less altered GEP than MM, where ∼400 genes have been reported to be deregulated vs normal PCs.20
The lack of significant transcriptomic deregulation in clonal PCs from AL could be related to a lower genetic instability; in fact, Bochtler et al21 have previously reported that hyperdiploid karyotypes are significantly less frequent in AL than in MGUS or MM. Here, by performing high-throughput genetic profiling of purified clonal PCs (Table 1; supplemental Table 2), CNAs were detected in all cases tested with a median number of 9 CNAs per patient (range, 1-23) (Figure 1C), similar to what has been described for MM (median of 12 CNAs/patient)13 ; furthermore, the pattern of CNA observed in AL was also similar to that observed in MM.13 Interestingly, the 3 cases with t(11;14) displayed fewer or small-sized chromosomal alterations; although the series is small, these findings are consistent with those reported by Bryce et al4 using a reduced number of FISH probes. CNN-LOH were also detected in approximately half of the cases (5/11; mean, 1.6 CNN-LOH/patient) (Figure 1C). Because of the lack of data on the mutation landscape of AL, we then investigated whether AL could potentially harbor a unifying mutation as occurs in Waldenström macroglobulinemia.22 Overall, 94 SNVs were detected in 93 genes among 5 AL-sequenced patients (median, 15 SNVs/case). Although 2 different SNVs in the XKR8 gene were noted, no recurrent mutations were identified (Table 1). In contrast to the recently reported similarities between AL and MM in their inherited susceptibility,23 all but 1 (KRAS c.35G>T) SNV identified herein have not been previously reported as significantly frequent in MM (supplemental Excel File 2)24,25 ; however, approximately two-thirds (62/93) of the mutated genes in AL are also mutated in MM.24,25 Further studies are warranted to confirm these findings in larger series of patients.
CNAs and mutations detected in clonal PCs from patients with newly-diagnosed AL amyloidosis
| . | Most frequent alteration . | 2nd most frequent alteration . | 3rd most frequent alteration . | Less frequent alterations . |
|---|---|---|---|---|
| Chromosomal imbalances | ||||
| Gains | +9* | +19* | +5* | +3, +6, +7, +11 |
| Losses | −X | −16 | — | −12, −14 |
| Chromosomal arms | ||||
| Gains | +15q | +1q | — | +8q, +9p, +11q |
| Losses | −Yp | −13q | −22q | −1p, −6q, 12q, −16q |
| Mutations† | XKR8‡ | AHNAK, ALKBH4, AP3B2, ASCC3, ASIC4, BIRC6, BSN, BTBD17, C14orf39, C22orf39, C4orf3, CACNA1I, CATSPERD, CCDC17, CCDC39, CELSR1, CPXCR1, CSF3R, CTNNAL1, CTNND2, DCAF12L2, DIS3, DNAH5, DNAH9, DSEL, EML6, ENO4, FAM208A, FBXL21, FBXO15, FKBP3, FYCO1, GEMIN2, GFRAL, HECW1, HIST1H3D, HIST1H3H, HPN, IKZF1, ITSN1, KANSL1L, KCNT1, KRAS, KRT28, LAMA3, LMX1A, LYG1, MIA2, PTCD2, MYO18B, NAALAD2, NBEAL1, NBR1, NFIB, OCA2, ORC4, PCLO, PDE8B, PKD1, PLCB4, PRKD2, PRPF4B, PSCA, PVRL3, QPCT, RAD51D, RAPGEF4, RBP3, RETN, RGS7BP, RPL19, SALL2, SERPINA5, SFMBT2, SI, SLX4, SPATA31D1, SPOCK1, SPTAN1, STPG2, SWSAP1, TMEM200B, TRADD, TTLL4, TTN, TTR, TXNRD3, USP54, XKR8,, YTHDF2, ZFYVE1, ZNF519, ZNF729 |
| . | Most frequent alteration . | 2nd most frequent alteration . | 3rd most frequent alteration . | Less frequent alterations . |
|---|---|---|---|---|
| Chromosomal imbalances | ||||
| Gains | +9* | +19* | +5* | +3, +6, +7, +11 |
| Losses | −X | −16 | — | −12, −14 |
| Chromosomal arms | ||||
| Gains | +15q | +1q | — | +8q, +9p, +11q |
| Losses | −Yp | −13q | −22q | −1p, −6q, 12q, −16q |
| Mutations† | XKR8‡ | AHNAK, ALKBH4, AP3B2, ASCC3, ASIC4, BIRC6, BSN, BTBD17, C14orf39, C22orf39, C4orf3, CACNA1I, CATSPERD, CCDC17, CCDC39, CELSR1, CPXCR1, CSF3R, CTNNAL1, CTNND2, DCAF12L2, DIS3, DNAH5, DNAH9, DSEL, EML6, ENO4, FAM208A, FBXL21, FBXO15, FKBP3, FYCO1, GEMIN2, GFRAL, HECW1, HIST1H3D, HIST1H3H, HPN, IKZF1, ITSN1, KANSL1L, KCNT1, KRAS, KRT28, LAMA3, LMX1A, LYG1, MIA2, PTCD2, MYO18B, NAALAD2, NBEAL1, NBR1, NFIB, OCA2, ORC4, PCLO, PDE8B, PKD1, PLCB4, PRKD2, PRPF4B, PSCA, PVRL3, QPCT, RAD51D, RAPGEF4, RBP3, RETN, RGS7BP, RPL19, SALL2, SERPINA5, SFMBT2, SI, SLX4, SPATA31D1, SPOCK1, SPTAN1, STPG2, SWSAP1, TMEM200B, TRADD, TTLL4, TTN, TTR, TXNRD3, USP54, XKR8,, YTHDF2, ZFYVE1, ZNF519, ZNF729 |
Interstitial CNAs are detailed in supplemental Table 1.
All 3 chromosomal gains were detected in 3 of 11 patients with AL.
None of each SNV was simultaneously found in ≥2 patients.
Two different SNVs found in the same gene (c.445G>A and c.86C>T).
In summary, in this study, based on the analysis of highly purified clonal PCs from patients with AL, we showed that although they display similar iPEP to that of MM, their GEP is remarkably similar to that of normal PCs; such stable transcriptomes could not be explained by a lower genetic instability. First-ever WES revealed potential lack of a unifying gene mutation in AL, and therefore further studies are warranted to better understand the pathogenesis and predisposition of AL clonal PCs to produce amyloid light-chain fibrils.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by the Cooperative Research Thematic Network grants RD12/0036/0058, RD12/0036/0061, and RD12/0036/0048 of the Red de Cancer (Cancer Network of Excellence); Instituto de Salud Carlos III/Subdirección General de Investigación Sanitaria (FIS: PI13/02196); Consejería de Sanidad, Junta de Castilla y León, Valladolid, Spain (557/A/10); and the International Myeloma Foundation.
Authorship
Contribution: B.P., J.M.-L., F.P., J.-J.L., and J.F.S.M. conceived and designed the study; B.P., J.M.-L., L.A.C., I.R., N.P., B.S.-V., S.B., M.-L.S., D.A., A.O., M.-B.V., and N.C.G. developed the study methodology: B.P., J.M.-L., L.A.C., I.R., N.P., B.S.-V., S.B., M.-L.S., D.A., M.L., A.G.d.C., E.P., A.O., M.-E.G.G., F.E., T.J.G.-L., L.P., J.A., F.P., A.O., M.-B.V., M.-V.M., J.-J.L., N.C.G., and J.F.S.M. acquired the data; B.P., J.M.-L., L.A.C., I.R., J.-J.L., and J.F.S.M. analyzed and interpreted the data; B.P. and J.F.S.M. wrote, reviewed, and revised the manuscript; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bruno Paiva, Clinica Universidad de Navarra; Centro de Investigacion Médica Aplicada (CIMA), Av Pio XII 36, 31008 Pamplona, Spain; e-mail: bpaiva@unav.es.