Key Points
miR-486-5p is expressed in megakaryocyte-erythroid progenitors and regulates growth and survival by regulating FOXO1 and AKT.
miR-486-5p is overexpressed in CML progenitors and enhances their growth, survival, and response to tyrosine kinase inhibitors.
Abstract
MicroRNAs (miRNAs) are key regulators of hematopoietic cell differentiation and may contribute to altered growth of leukemic stem cells. Using microarray-based miRNA profiling, we found that miRNA 486 (miR-486) is significantly upregulated in chronic myeloid leukemia (CML) compared with normal CD34+ cells, particularly in the megakaryocyte-erythroid progenitor population. miR-486-5p expression increased during erythroid differentiation of both CML and normal CD34+ cells. Ectopic miR-486-5p expression enhanced in vitro erythroid differentiation of normal CD34+ cells, whereas miR-486-5p inhibition suppressed normal CD34+ cell growth in vitro and in vivo and inhibited erythroid differentiation and erythroid cell survival. The effects of miR-486-5p on hematopoietic cell growth and survival are mediated at least in part via regulation of AKT signaling and FOXO1 expression. Using gene expression and bionformatics analysis, together with functional screening, we identified several novel miR-486-5p target genes that may modulate erythroid differentiation. We further show that increased miR-486-5p expression in CML progenitors is related to both kinase-dependent and kinase-independent mechanisms. Inhibition of miR-486-5p reduced CML progenitor growth and enhanced apoptosis following imatinib treatment. In conclusion, our studies reveal a novel role for miR-486-5p in regulating normal hematopoiesis and of BCR-ABL–induced miR-486-5p overexpression in modulating CML progenitor growth, survival, and drug sensitivity.
Introduction
MicroRNAs (miRNAs) are small noncoding RNAs that represent an important mechanism for control of gene expression in addition to transcription factors.1 miRNAs bind to 3′ untranslated regions (3′ UTRs) of messenger RNAs (mRNAs) to induce translational repression or RNA destabilization.2 Over 2000 miRNAs are reported in humans.3 Sets of combinatorially expressed miRNAs can precisely delineate specific cell types and play an important role in determining the differentiated state.4,5 Changes in miRNA expression are observed during hematopoietic stem cell (HSC) differentiation along specific lineages.6 Analysis of miRNA function has uncovered regulatory circuits where miRNAs modulate expression of transcription factors and are activated by transcription factors to fine-tune or maintain differentiation and function.1 Mice deficient in or overexpressing specific miRNAs demonstrate a critical role for miRNAs in B- and T-lymphocyte development, erythropoiesis, megakaryocytopoiesis, monocytopoiesis, and granulopoiesis.7,8 The importance of miRNAs is further supported by reports of deregulated expression of several miRNAs in hematologic malignancies.9-11 However, functional analysis of miRNA in human as opposed to murine hematopoiesis has been challenging and is less well described.
Chronic myeloid leukemia (CML) is a lethal hematologic malignancy resulting from transformation of a primitive hematopoietic cell by the BCR-ABL tyrosine kinase.12 The cancer-associated miRNA 17-92 (miR-17-92) cluster was reported to be aberrantly expressed in CML CD34+ cells in a BCR-ABL– and c-MYC–dependent manner.13 On the other hand, miRNA 10a, 150, and 151 were downregulated in CML CD34+ cells.14 Loss of miRNA 328 was identified in blast crisis CML leading to loss of function as an RNA decoy modulating hnRNPE2 regulation of mRNA translation.15 miRNA 203, a tumor-suppressor miRNA targeting BCR-ABL and ABL kinases, is epigenetically silenced in human Ph-positive leukemic cell lines.16,17 Other miRNAs are associated with resistance to the BCR-ABL tyrosine kinase inhibitor (TKI) imatinib mesylate (IM) and identified as a possible predictor for IM resistance.18 However, the role of miRNAs in regulating CML leukemia stem cell growth remains poorly understood.
In this study, we evaluated global miRNA expression in CML compared with normal CD34+ cells and identified miRNA 486-5p (miR-486-5p) as significantly upregulated in CML CD34+ cells. We evaluated the role of miR-486-5p in normal hematopoiesis and in modulating CML progenitor growth and identified target genes that mediate these effects. Our studies identify a novel miRNA regulatory network that regulates normal hematopoietic development and contributes to the transformed phenotype of CML progenitors and modulates their response to IM treatment.
Materials and methods
Cell lines
Human embryonic kidney 293T cells were maintained in Dulbecco’s modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum (HyClone Laboratories, Logan, UT). Human leukemia cell lines TF-1 and TF-1-BA were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal calf serum and 2 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF).
Patient samples and CD34+ cell isolation
Human cord blood (CB) and CML bone marrow (BM) samples were obtained under protocols approved by the institutional review board at City of Hope, in accordance with assurances filed with the Department of Health and Human Services, and meeting all requirements of the Declaration of Helsinki. CML patients were in chronic phase and had not received IM treatment. Mononuclear cells were prepared by Ficoll-Hypaque (Sigma-Aldrich, St. Louis, MO) separation. CD34+ cells were isolated as previously described.19 Sorting of CD34+ fractions was performed based on expression of the following cell-surface markers: granulocyte-macrophage progenitors (GMPs), Lin−CD34+CD38+CD123lowCD45RA+; megakaryocyte-erythrocyte progenitors (MEPs), Lin−CD34+CD38+CD123−CD45RA−; common myeloid progenitors (CMPs), Lin−CD34+CD38+CD123+CD45RA−; HSC, Lin−CD34+CD38−.
miRNA array, mRNA array, and TaqMan miRNA assay
Total RNAs including miRNA were extracted from cells using the miRNeasy Mini Kit (Qiagen, Valencia, CA). Integrity of RNA samples was analyzed using an Agilent bioanalyzer. Microarray analysis of miRNA was performed using G4470B human miRNA microarrays 8x15K version 2 and for mRNA using Agilent Human Gene 1.0 ST arrays. Analyses were performed by the City of Hope integrated genomics core. miRNA expression was log2 transformed after quantile normalization. mRNA expression was log2 transformed after RMA normalization using the Affymetrix gene expression console. Differential miRNA and mRNA expression between peripheral blood stem cell (PBSC) and CML samples was identified by 2-sample t test using the LIMMA package in R and Bioconductor. Multiple testing was adjusted for by calculating the false discovery rate. For quantitative PCR (qPCR) analysis, complementary DNAs specific for miR-486-5p, miR-486-3p, pri-miR-486, and RNU44 (internal control) were transcribed and amplified using gene-specific primer sets and the TaqMan microRNA assay protocol (Applied Biosystems, Foster City, CA). Microarray data have been deposited to the Gene Expression Omnibus under accession number GSE64011.
Retrovirus and lentivirus vector production
The MIG-R1 and MIG-p210 retroviral vectors were kind gifts from Dr Warren Pear, University of Pennsylvania. Replication incompetent retroviruses were obtained by transient transfection of 293 cells with retroviral plasmids and the pCL-ampho plasmid.20 To express miRNA, the EF1 promoter and human β-globin intron from pEGP-miR vector (Cell Biolabs, San Diego, CA) were inserted into the pHIV7 lentivirus vector. The multiple cloning sites from the pcDNA3 vector were introduced into the human β-globin intron to generate a pHIV7-EF1-miR vector suitable for miRNA cloning. The pre-miR-486 construct (kindly provided by Dr X. F. Zheng, Beijing Institute of Radiation Medicine) was ligated into pHIV7-EF1-miR to generate the pHIV7-EGP-miR-486 vector. miRZip-anti-miR-486-5p and control vectors were purchased from System Biosciences. The cytomegalovirus (CMV) promoter was replaced with the EF1 promoter. The lentivirus pLKO.1-Foxo1 short hairpin RNA (shRNA) and control vectors were purchased from Addgene, and a spleen focus forming virus promoter and red fluorescent protein (RFP) gene construct was inserted. Lentivirus particles were produced by cotransfecting 293T cells with lentivirus vectors and pCMV-gp, pCMV-rev, and pCMV-VSV-G packaging plasmids. Supernatants containing virus particles were collected, filtered, and concentrated using PEG-it (System Biosciences).
Transduction and transfection of CD34+ cells
CD34+ cells isolated from CB or CML BM were cultured on Retronectin-coated (Pan Vera, Madison, WI) plates in serum-free medium (Stem Cell Technologies, Vancouver, BC, Canada) containing growth factors (high growth factors [HGF], IL-3 [25 ng/mL], interleukin-6 (10 ng/mL), Flt-3 ligand (100 ng/mL), stem cell factor [SCF; 50 ng/mL], and thrombopoietin [100 ng/mL]) at 37°C in 5% CO2 for 48 hours and exposed to lentivirus (multiplicity of infection [MOI] = 5, 2 exposures 24 hours apart). Cells were labeled 48 hours later with anti-CD34-APC antibodies (Becton Dickinson, San Jose, CA) and CD34+GFP+ cells selected using flow cytometry (Dako-Cytomation, Fort Collins, CO). For dual transduction, CBCD34+ cells were transduced with retroviral vectors expressing BCR-ABL (MIG-p210), KI (MIG-p210-KI) or green fluorescent protein (GFP) alone (MIG-R1) at MOI of 10. Selected CD34+GFP+ cells were transduced with miRZip-EF1-scramble and miRZip-EF1-anti-miR486-5p at MOI of 10, selected with 2 µg/mL puromycin for 48 hours, and analyzed.
For small interfering RNA (siRNA) transfection, CB CD34+ cells were suspended in Nucleofector Solution with siRNA (2 µM) and nucleofection performed using the Nucleofector 96-well Shuttle System. The CD34+ cells were transferred to GEMM medium (Iscove modified Dulbecco medium, 30% fetal bovine serum [Stem Cell Technologies], β-mercaptoethanol [63 μM], erythropoietin [3 U/mL], interleukin-3 [IL-3; 5 ng/mL], SCF [5 ng/mL], granulocyte CSF [G-CSF; 20 ng/mL], and GM-CSF [20 ng/mL]) to assay differentiation.
Cell proliferation, differentiation, cell cycle, and apoptosis assays
Cells were labeled with Dye670 (Invitrogen) in Iscove modified Dulbecco medium at 37°C for 30 minutes, incubated overnight, and cultured in medium with HGF as used for transduction. After 3 days, Dye670 fluorescence was assessed by flow cytometry. The position of the parent generation was set based on a cell aliquot treated with paraformaldehyde after Dye670 labeling. The proliferation index was calculated using ModFit software.
To assess differentiation, CD34+ cells were cultured in GEMM medium. On day 3 or day 6, cells were labeled with antibodies to CD33, CD11b, CD14, GPA, and CD45 and analyzed by flow cytometry. Cells were also cultured in myeloid (5 ng/mL SCF, 5 ng/mL IL-3, 20 ng/mL G-CSF, and 20 ng/mL GM-CSF) or erythroid differentiation conditions (5 ng/mL SCF, 5 ng/mL IL-3, 3 U/mL erythropoietin [EPO]).
For cell-cycle analysis, CD34+ cells were cultured in serum-free medium with HGF for 48 hours. Hoechst 33342 was added at a concentration of 2 μg/mL. After 1 hour, the DNA content was detected using fluorescence-activated cell sorter (FACS) and cell-cycle phase analyzed using ModFit software.
To assess apoptosis CD34+ cells were cultured in the serum-free expansion medium (SFEM) with low concentrations of GF similar to those present in long-term BM culture stroma-conditioned medium (GM-CSF [200 pg/mL], leukemia inhibitory factor [50 pg/mL], G-CSF [1 ng/mL], SCF [200 pg/mL], MIP-1α [200 pg/mL], and interleukin-6 [1 ng/mL]) for 48 hours, labeled with annexin V-Cy5 and 4,6 diamidino-2-phenylindole, and assessed by flow cytometry.
Luciferase reporter constructs and luciferase assays
The 3-UTR fragments containing predicted miRNA binding sites of target genes were PCR amplified using the HepG2 cell genomic DNA as a template. The primers used for PCR are shown in supplemental Table 1 (available on the Blood Web site). The fragments were cloned downstream of the firefly luciferase–coding region of pMIR-REPORT luciferase plasmid (Applied Biosystems). The authenticity and orientation of the inserts were confirmed by sequencing. Mixtures of 100 ng firefly luciferase reporter plasmid, 20 ng β-galactosidase control plasmid (Applied Biosystems), and 400 ng of miR-486 expression vector were transfected into 293T (5 × 104) cells using lipofectamine 2000 transfection reagent (Invitrogen). Luciferase activity was measured 48 hours after transfection using a luciferase reporter assay system (Promega).
Western blotting
Protein extracts were prepared as previously described.21 Protein were resolved on 4% to 20% sodium dodecyl sulfate polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membranes. Membranes were blocked with 5% nonfat milk in phosphate-buffered saline and 0.1% Tween-20 and labeled with primary antibodies (anti-actin [AC-15; Sigma, St. Louis, MO], anti-FoxO1 [Cell Signaling Danvers, MA], anti-PTEN, and anti-p-AKT [Ser 308, Cell Signaling]), followed by horseradish peroxidase–conjugated secondary anti-mouse and anti-rabbit antibodies (1:8000; Jackson, West Grove, PA). Antibody detection was performed using enhanced chemiluminescence (Superfemto kit; Pierce Biotechnology, Rockford, IL).
Immunodeficient mouse engraftment studies
CB CD34+ cells were transduced with miRZip-scramble and miRZip-anti-miR-486-5p respectively. For transduction, CB CD34+ cells (2 × 105/mouse) were cultured in medium supplemented with Flt-3 ligand (100 ng/mL), SCF (50 ng/mL), and thrombopoietin (100 ng/mL) for 16 hours, followed by 2 exposures to virus (MOI = 10) via spinoculation. Cells harvested 8 hours after the second exposure were injected IV into irradiated (300 cGy) 8-week-old NOD-SCID IL2-rγ-null (NSG) mice. Human cell engraftment in blood assayed at 3, 8, and 12 weeks. Recipients were sacrificed at either 3 or 12 weeks posttransplantation, and engraftment in BM was analyzed. Cells expressing human CD45+ cell and CD45+ subsets (CD34, CD33, CD14, CD19, and CD3) and GFP were analyzed by flow cytometry.
Statistics
Data obtained from multiple experiments were reported as the mean ± standard error of the mean (SEM). Significance levels were determined by Student t test and analysis of variance analysis.
Results
Increased expression of miR-486-5p in human CML CD34+ cells
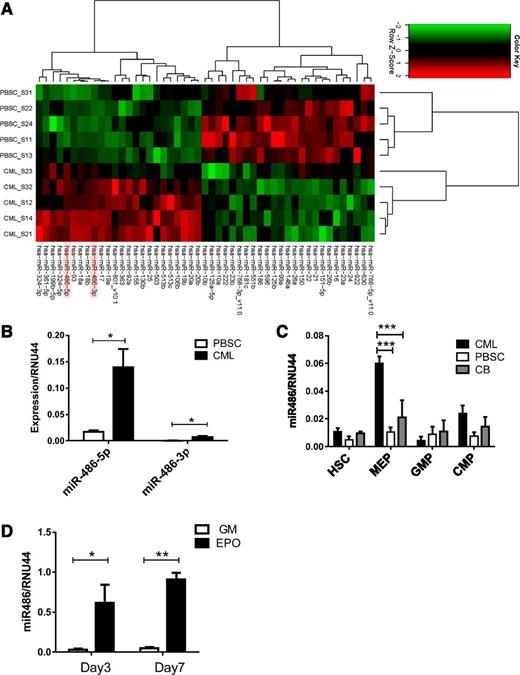
We analyzed miRNA expression in CML and normal PBSC CD34+ cells using Agilent microarrays. The miRNAs exhibiting significantly altered expression in CML compared with normal CD34+ cells are shown in Figure 1A and Table 1. The miRNAs most significantly upregulated in CML CD34+ cells included miR-486-5p and miR-486-3p and the miR-17-92 and miRNA 106b-25 (miR-106b-25) clusters. We confirmed previous observations of downregulation of miRNAs 10a, 150, and 151 in CML cells. Because miR-486 overexpression has not been previously studied in leukemia, we further evaluated its function in normal and malignant hematopoiesis. Although both miR-486-5p and miR-486-3p were differentially overexpressed in CML compared with normal CD34+ cells, we focused on miR-486-5p because its absolute expression was considerably higher than that of miR-486-3p, as confirmed using quantitative reverse-transcription polymerase chain reaction (qRT-PCR) (Figure 1B).
Increased miR-486-5p levels in CML progenitor cells. (A) Heatmap showing hierarchical clustering of differentially expressed miRNA in CML and normal PBSC CD34+ cells (n = 5 each). (B) miR-486-5p and miR-486-3p expression by CML and PBSC CD34+ cells (n = 8 each) evaluated by qRT- PCR. (C) HSC, MEP, GMP, and CMP subpopulations selected from CML, normal PBSCs, and CB CD34+ cells were evaluated for miR-486-5p expression by qRT-PCR. (D) CD34+ cells cultured in myeloid (5 ng/mL SCF, 5 ng/mL IL-3 , 20 ng/mL G-CSF, and 20 ng/mL GM-CSF) and erythroid differentiation conditions (5 ng/mL SCF, 5 ng/mL IL-3, 3 U/mL EPO) were analyzed for miR-486-5p expression by qRT-PCR at days 3 and 7. Cumulative results represent the mean ± SEM. *P < .05, ***P < .001; n = 5.
Increased miR-486-5p levels in CML progenitor cells. (A) Heatmap showing hierarchical clustering of differentially expressed miRNA in CML and normal PBSC CD34+ cells (n = 5 each). (B) miR-486-5p and miR-486-3p expression by CML and PBSC CD34+ cells (n = 8 each) evaluated by qRT- PCR. (C) HSC, MEP, GMP, and CMP subpopulations selected from CML, normal PBSCs, and CB CD34+ cells were evaluated for miR-486-5p expression by qRT-PCR. (D) CD34+ cells cultured in myeloid (5 ng/mL SCF, 5 ng/mL IL-3 , 20 ng/mL G-CSF, and 20 ng/mL GM-CSF) and erythroid differentiation conditions (5 ng/mL SCF, 5 ng/mL IL-3, 3 U/mL EPO) were analyzed for miR-486-5p expression by qRT-PCR at days 3 and 7. Cumulative results represent the mean ± SEM. *P < .05, ***P < .001; n = 5.
Altered miRNA expression in CML CD34+ cells
| Upregulated in CML . | Downregulated in CML . | ||||
|---|---|---|---|---|---|
| miRNA . | logFC . | Adjusted P . | miRNA . | logFC . | Adjusted P . |
| hsa-miR-486-3p | 2.3196694 | 6.99E-06 | hsa-miR-146a | −1.781099 | 4.84E-05 |
| hsa-miR-18b | 2.1646587 | 4.84E-05 | hsa-miR-551b | −2.470109 | .0003938 |
| hsa-miR-19a† | 1.7367785 | 4.84E-05 | hsa-miR-125b | −1.260167 | .0026857 |
| hsa-miR-486-5p | 2.1636705 | .0001093 | hsa-miR-768-3p | −1.216743 | .009664 |
| hsa-miR-18a† | 2.5785991 | .0001093 | hsa-miR-10a | −1.686725 | .009664 |
| hsa-miR-17† | 2.0330101 | .0001185 | hsa-miR-21 | −2.005972 | .010417 |
| hsa-miR-20b | 1.8770134 | .0008293 | hsa-miR-630 | −3.466621 | .0236791 |
| hsa-miR-93‡ | 1.2873623 | .0013853 | hsa-miR-26a | −0.739312 | .0236791 |
| hsa-miR-20a† | 1.6312808 | .010417 | hsa-miR-99a | −0.881689 | .0241247 |
| hsa-miR-106b‡ | 0.869845 | .0241247 | hsa-miR-181c | −0.907926 | .0241247 |
| hsa-miR-17* | 1.8963968 | .0353565 | hsa-miR-23a | −1.049268 | .0391509 |
| hsa-miR-363 | 1.0551691 | .0393167 | hsa-miR-151-5p | −1.505794 | .0531846 |
| hsa-miR-199b-5p | 0.5604006 | .0393167 | hsa-miR-16 | −0.564943 | .0531846 |
| hsa-miR-361-5p | 0.2939439 | .0531846 | hsa-miR-24 | −0.80473 | .0989177 |
| hsa-miR-324-5p | 0.5038724 | .0531846 | hsa-miR-222 | −0.893684 | .1025662 |
| hsa-miR-324-3p | 0.6832275 | .0535795 | hsa-miR-26b | −0.751628 | .1095676 |
| hsa-miR-92a | 0.9787604 | .056494 | hsa-miR-768-5p | −1.554897 | .1121387 |
| hsa-miR-130b | 0.8568465 | .0734194 | hsa-miR-23b | −0.674667 | .1334663 |
| hsa-miR-503 | 0.8823222 | .0917508 | hsa-miR-22 | −2.852556 | .1334663 |
| hsa-miR-19b | 1.6196121 | .1025662 | hsa-miR-29c* | −0.376277 | .1587193 |
| hsa-miR-19b-1*† | 1.0021163 | .1025662 | hsa-miR-10b | −0.961031 | .1768722 |
| hsa-miR-340* | 0.7749786 | .1095676 | hsa-miR-922 | −0.337589 | .1768722 |
| hsa-miR-155 | 1.057247 | .1334663 | hsa-miR-596 | −0.302939 | .2051711 |
| hsa-miR-181a* | 0.7631197 | .1587193 | hsa-miR-150 | −3.752058 | .2321378 |
| hsa-miR-513b | 1.14356 | .1941551 | |||
| hsa-miR-513c | 0.6830905 | .2016059 | |||
| hsa-miR-340 | 0.9628093 | .2321378 | |||
| hsa-miR-92a-1*† | 0.4735888 | .2321378 | |||
| hsa-miR-25‡ | 0.5208347 | .2321378 | |||
| Upregulated in CML . | Downregulated in CML . | ||||
|---|---|---|---|---|---|
| miRNA . | logFC . | Adjusted P . | miRNA . | logFC . | Adjusted P . |
| hsa-miR-486-3p | 2.3196694 | 6.99E-06 | hsa-miR-146a | −1.781099 | 4.84E-05 |
| hsa-miR-18b | 2.1646587 | 4.84E-05 | hsa-miR-551b | −2.470109 | .0003938 |
| hsa-miR-19a† | 1.7367785 | 4.84E-05 | hsa-miR-125b | −1.260167 | .0026857 |
| hsa-miR-486-5p | 2.1636705 | .0001093 | hsa-miR-768-3p | −1.216743 | .009664 |
| hsa-miR-18a† | 2.5785991 | .0001093 | hsa-miR-10a | −1.686725 | .009664 |
| hsa-miR-17† | 2.0330101 | .0001185 | hsa-miR-21 | −2.005972 | .010417 |
| hsa-miR-20b | 1.8770134 | .0008293 | hsa-miR-630 | −3.466621 | .0236791 |
| hsa-miR-93‡ | 1.2873623 | .0013853 | hsa-miR-26a | −0.739312 | .0236791 |
| hsa-miR-20a† | 1.6312808 | .010417 | hsa-miR-99a | −0.881689 | .0241247 |
| hsa-miR-106b‡ | 0.869845 | .0241247 | hsa-miR-181c | −0.907926 | .0241247 |
| hsa-miR-17* | 1.8963968 | .0353565 | hsa-miR-23a | −1.049268 | .0391509 |
| hsa-miR-363 | 1.0551691 | .0393167 | hsa-miR-151-5p | −1.505794 | .0531846 |
| hsa-miR-199b-5p | 0.5604006 | .0393167 | hsa-miR-16 | −0.564943 | .0531846 |
| hsa-miR-361-5p | 0.2939439 | .0531846 | hsa-miR-24 | −0.80473 | .0989177 |
| hsa-miR-324-5p | 0.5038724 | .0531846 | hsa-miR-222 | −0.893684 | .1025662 |
| hsa-miR-324-3p | 0.6832275 | .0535795 | hsa-miR-26b | −0.751628 | .1095676 |
| hsa-miR-92a | 0.9787604 | .056494 | hsa-miR-768-5p | −1.554897 | .1121387 |
| hsa-miR-130b | 0.8568465 | .0734194 | hsa-miR-23b | −0.674667 | .1334663 |
| hsa-miR-503 | 0.8823222 | .0917508 | hsa-miR-22 | −2.852556 | .1334663 |
| hsa-miR-19b | 1.6196121 | .1025662 | hsa-miR-29c* | −0.376277 | .1587193 |
| hsa-miR-19b-1*† | 1.0021163 | .1025662 | hsa-miR-10b | −0.961031 | .1768722 |
| hsa-miR-340* | 0.7749786 | .1095676 | hsa-miR-922 | −0.337589 | .1768722 |
| hsa-miR-155 | 1.057247 | .1334663 | hsa-miR-596 | −0.302939 | .2051711 |
| hsa-miR-181a* | 0.7631197 | .1587193 | hsa-miR-150 | −3.752058 | .2321378 |
| hsa-miR-513b | 1.14356 | .1941551 | |||
| hsa-miR-513c | 0.6830905 | .2016059 | |||
| hsa-miR-340 | 0.9628093 | .2321378 | |||
| hsa-miR-92a-1*† | 0.4735888 | .2321378 | |||
| hsa-miR-25‡ | 0.5208347 | .2321378 | |||
logFC, log fold change (CML: normal).
miR-17-92 cluster.
miR-106b-25 cluster.
Because CD34+ cells are heterogeneous, we evaluated miR-486-5p expression in purified HSCs, CMPs, GMPs, and MEPs from CML, normal PBSCs, and CB CD34+ cells (supplemental Figure 1 A-B). miR-486-5p expression was increased 6-fold in CML compared with normal PBSC MEPs and 3-fold compared with CB populations (P < .001) (Figure 1C). miR-486-5p expression was upregulated following erythroid differentiation of CB CD34+ cells with EPO, but not after myeloid differentiation with G-CSF and GM-CSF (Figure 1D). Erythroid differentiation was confirmed by glycophorin A (GPA) (CD235a) expression and presence of hemoglobinized cells (supplemental Figure 1C-D). These results indicate that miR-486-5p expression increases with differentiation toward the erythroid lineage.
Effect of miR-486-5p on growth and erythroid differentiation of CB CD34+ cells
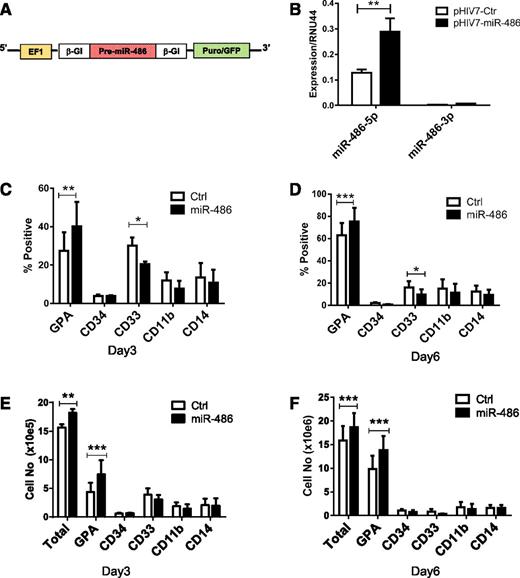
To explore the function of miR-486-5p in regulating hematopoietic progenitor cells, we constructed a pHIV7-EF1-miR-486-EGFP lentivirus vector to overexpress hsa-pre-miR-486 (Figure 2A). Transduction with pHIV7-EF1-miR-486 resulted in a significant increase of miR-486-5p expression in CB CD34+ cells (Figure 2B). The increase in miR-486-3p was not statistically significant. We analyzed the number and type of cells generated after 6 days culture of transduced CD34+ cells (>95% CD34+ cells at start of culture). Overexpression of miR-486-5p modestly increased the numbers of cells generated in culture compared with control vectors (Figure 2E-F). Increased proportions of GPA-expressing erythroid cells and reduced proportions of CD33+ myeloid cells were generated from miR-486-5p–expressing cells (Figure 2C-D). The total number of GPA+ cells was also significantly increased (Figure 2E-F). These results suggest that miR-486-5p enhances growth and erythroid differentiation of CB CD34+ cells.
miR-486-5p overexpression enhances the growth and erythroid differentiation of CB CD34+ cells. (A) The EF1-miR-486 expression cassette was cloned into lentivirus vector pHIV7-EGFP. (B) miR-486-5p and miR-486-3p expression in CB CD34+ cells transduced with pHIV7-EF1-miR-486 and control vector was determined by qRT-PCR. (C-F) miR-486-5p and control vector–transduced cells (Ctrl) (n = 5) were cultured in GEMM medium and GPA, CD34, CD33, CD11b, and CD14 expression analyzed by flow cytometry. The percentage of cells expressing these marker at day 3 (C) and day 6 (D) and the total number of cells expressing these markers at day 3 (E) and day 6 (F) are shown. Cumulative results represent the mean ± SEM. *P < .05, **P < .01, ***P < .001.
miR-486-5p overexpression enhances the growth and erythroid differentiation of CB CD34+ cells. (A) The EF1-miR-486 expression cassette was cloned into lentivirus vector pHIV7-EGFP. (B) miR-486-5p and miR-486-3p expression in CB CD34+ cells transduced with pHIV7-EF1-miR-486 and control vector was determined by qRT-PCR. (C-F) miR-486-5p and control vector–transduced cells (Ctrl) (n = 5) were cultured in GEMM medium and GPA, CD34, CD33, CD11b, and CD14 expression analyzed by flow cytometry. The percentage of cells expressing these marker at day 3 (C) and day 6 (D) and the total number of cells expressing these markers at day 3 (E) and day 6 (F) are shown. Cumulative results represent the mean ± SEM. *P < .05, **P < .01, ***P < .001.
We further investigated the role of miR-486-5p in growth and differentiation of CB CD34+ cells by inhibiting miR-486-5p using a miRZip lentivirus expressed anti-miR-486-5p sequence (Figure 3A). Transduction of miRZip-anti-miR-486-5p vector reduces levels of mature miR-486-5p in CB CD34+ cells (supplemental Figure 2A). Anti-miR-486-5p–expressing CB CD34+ cells generated a significantly reduced percentage of GPA+ cells and increased CD33+ cells compared with scrambled control vector–transduced cells in culture (Figure 3B-D), together with reduced total cell numbers and the number of GPA+ erythroid cells (Figure 3E-F) and reduced burst-forming unit-erythroid colony formation (P < .005; Figure 3G). Expression of anti-miR-486-5p reduced cell division in CB CD34+ cells as assessed by Dye670 labeling (Figure 3H) and reduced the percentage of CB CD34+ cells in S phase (P < .05; Figure 3I and supplemental Figure 2B). Anti-miR-486–expressing CD34+ cells induced toward erythroid or myeloid differentiation demonstrated significantly enhanced apoptosis compared with controls (supplemental Figure 2C). Similarly, GPA (CD235a)+ erythroid cells expressing anti-miR-486 demonstrated significantly enhanced apoptosis compared with scrambled controls (supplemental Figure 2D). Anti-miR-486–expressing CD34+ cells also generated reduced numbers of CD41/CD42+ cells in megakaryocytic differentiation culture (supplemental Figure 2E). However, the percentage of CD41/CD42+ cells was not significantly reduced (supplemental Figure 2F), suggesting that reduced output of megakaryocytic cells may relate to overall reduction in hematopoietic cell numbers rather than selective reduction in this population. These results further support a role for miR-486-5p in regulating proliferation, erythroid differentiation, and survival of CB CD34+ cells.
miR-486-5p inhibition blocks erythroid differentiation and reduces growth of CB CD34+ cells. (A) The lentivirus vector miRZip-anti-miR-486-5p is shown. (B-F) CD34+ cells transduced with anti-miR-486-5p and control vector (n = 3) were selected by flow cytometry and cultured in GEMM medium for 6 days and the percentage of GPA, CD34, CD11b, CD33, and CD14 cells were analyzed by flow cytometry. Representative results of flow cytometry analysis of GPA and CD11b expression are shown in (B). The percentage of cells expressing these markers at day 3 (C) and day 6 (D) and the total cell number of expressing these markers at day 3 (E) and day 6 (F) are shown. (G) The number of CFU-GM and burst-forming unit-erythroid colonies generated from selected CD34+GFP+ cells after culture in methylcellulose progenitor assays. (H) CD34+GFP+ cells were labeled with Dye670 and cultured in HGF of SFEM medium for 3 days. Cell division was evaluated on the basis of reduction of Dye670 fluorescence intensity. A proliferation index was calculated using ModFit software. (I) CD34+GFP+ cells were cultured for SFEM medium with HGF combination or 48 hours and treated with Hoechst 33342 (2 µg/mL) for 1 hour. DNA content was analyzed by flow cytometry and cell-cycle distribution calculated using ModFit software. Cumulative results represent the mean ± SEM. *P < .05, **P < .01, ***P < .001.
miR-486-5p inhibition blocks erythroid differentiation and reduces growth of CB CD34+ cells. (A) The lentivirus vector miRZip-anti-miR-486-5p is shown. (B-F) CD34+ cells transduced with anti-miR-486-5p and control vector (n = 3) were selected by flow cytometry and cultured in GEMM medium for 6 days and the percentage of GPA, CD34, CD11b, CD33, and CD14 cells were analyzed by flow cytometry. Representative results of flow cytometry analysis of GPA and CD11b expression are shown in (B). The percentage of cells expressing these markers at day 3 (C) and day 6 (D) and the total cell number of expressing these markers at day 3 (E) and day 6 (F) are shown. (G) The number of CFU-GM and burst-forming unit-erythroid colonies generated from selected CD34+GFP+ cells after culture in methylcellulose progenitor assays. (H) CD34+GFP+ cells were labeled with Dye670 and cultured in HGF of SFEM medium for 3 days. Cell division was evaluated on the basis of reduction of Dye670 fluorescence intensity. A proliferation index was calculated using ModFit software. (I) CD34+GFP+ cells were cultured for SFEM medium with HGF combination or 48 hours and treated with Hoechst 33342 (2 µg/mL) for 1 hour. DNA content was analyzed by flow cytometry and cell-cycle distribution calculated using ModFit software. Cumulative results represent the mean ± SEM. *P < .05, **P < .01, ***P < .001.
CB CD34+ cells transduced with anti-miR-486-5p or scrambled control sequences were transplanted into sublethally irradiated NSG mice. Aliquots of cells used for transplantation were also cultured for 48 hours and assessed for GFP expression. Both control scramble– and anti-miR-486–expressing cells showed similar percentage GFP expression. The percentage of human CD45+GFP+ cells in peripheral blood at 12 weeks were significantly reduced in mice receiving anti-miR-486-5p compared with scrambled sequence–transduced cells (Figure 4A). Analysis for engraftment of erythroid cells (GPA+CD45−) was performed at week 3, because previous reports have indicated that this is the optimal time to analyze for erythroid engraftment in xenografts. The percentage and number of GFP+ erythroid cells were significantly reduced in mice receiving anti-miR-486-5p–transduced CB CD34+ cells (P < .05) (Figure 4B-C). Evaluation at 12 weeks posttransplant did not reveal consistent alteration in the percentage of CD34-, CD33-, CD14-, CD11b-, and CD19-expressing cells, indicating that the absolute numbers of each of these populations were reduced following miR-486 inhibition (Figure 4D-E). These results indicate that anti-miR-486-5p inhibits in vivo growth and erythroid differentiation of CB CD34+ cells.
Effect of miR-486-5p inhibition on growth of CD34+ cells in vivo in NSG mice. (A-E) A total of 2 × 105 CB CD34+ cells were transduced with miRZip-scramble and miRZip-anti-486-5p and transplanted into sublethally irradiated NSG mice by tail vein injection. (A) The percentage of human CD45+ cells in peripheral blood. (B) The percentage of GPA+GFP+ cells in BM at 3 weeks posttransplantation. (C) The total numbers GPA+GFP+ cells in BM at 3 weeks posttransplantation. (D) The percentage of CD34-, CD11b-, CD14-, CD33-, and CD19-expressing cells within the CD45+GFP+ population in BM at 3 weeks posttransplantation. (E) The percentage of CD34-, CD11b-, CD14-, CD33-, and CD19-expressing cells within the CD45+GFP+ population in BM at 12 weeks posttransplantation. Cumulative results represent the mean ± SEM. *P < .05, ***P < .001.
Effect of miR-486-5p inhibition on growth of CD34+ cells in vivo in NSG mice. (A-E) A total of 2 × 105 CB CD34+ cells were transduced with miRZip-scramble and miRZip-anti-486-5p and transplanted into sublethally irradiated NSG mice by tail vein injection. (A) The percentage of human CD45+ cells in peripheral blood. (B) The percentage of GPA+GFP+ cells in BM at 3 weeks posttransplantation. (C) The total numbers GPA+GFP+ cells in BM at 3 weeks posttransplantation. (D) The percentage of CD34-, CD11b-, CD14-, CD33-, and CD19-expressing cells within the CD45+GFP+ population in BM at 3 weeks posttransplantation. (E) The percentage of CD34-, CD11b-, CD14-, CD33-, and CD19-expressing cells within the CD45+GFP+ population in BM at 12 weeks posttransplantation. Cumulative results represent the mean ± SEM. *P < .05, ***P < .001.
Identification of miR-486-5p targets in CD34+ cells
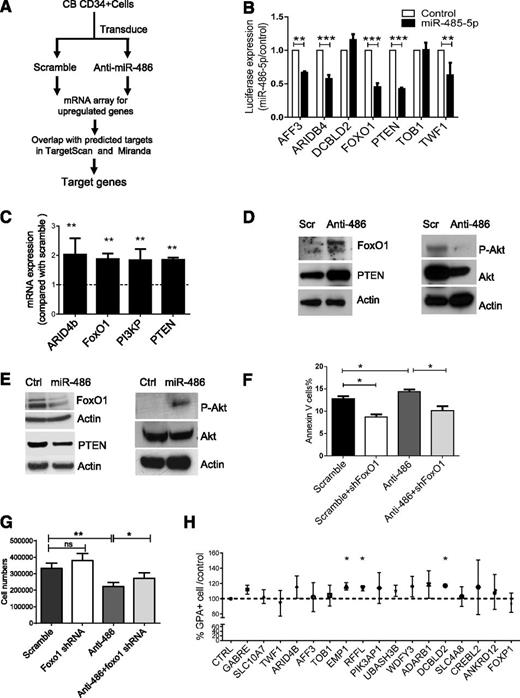
We reasoned that miR-486-5p–targeted genes would be upregulated following miR-486-5p inhibition. We evaluated gene expression in CB CD34+ cells expressing anti-miR-486-5p or a scramble control sequence using microarray analysis (Figure 5A). We then compared genes upregulated after anti-miR-486-5p expression with potential miR-486-5p target genes in the TargetScan and miRanDa databases and identified 19 potential miR-486-5p target genes (Table 2). Using 3′-UTR reporter assays, we confirmed that miR-486-5p targeted several of these genes, including FoxO1, PTEN, ARID4b, AFF3, and TWF1 (Figure 5B). We also validated upregulation of FoxO1, PTEN, ARID4B, and PI3KP expression in anti-miR-486-5p–transduced CB CD34+ cells using qRT-PCR (Figure 5C).
Identification of miR-486-5p target genes in CB CD34+ cells. (A) Strategy for identification of miR-486-5p targets in CBCD34+ cells. (B) Effect of miR-486-5p expression on luciferase activity in 293T cells transfected with reporter plasmids containing miR-486-5p binding sequences in 3′ UTRs of potential target genes. The data show inhibition of luciferase activity in cells expressing miR-486-5p compared with control vectors. (C) Gene expression in anti-miR-486-5p compared with scramble sequence–expressing cells determined by qPCR. (D) CB CD34+ cells were transduced with miRZip-anti-486-5p and scramble vector and cultured in GEMM medium for 3 days. FoxO1, PTEN, p-AKT, and AKT expression was detected by western blotting. (E) TF1 cells were transduced with pHIV7-EF1-miR-486or control vector. FoxO1, PTEN, p-AKT, and AKT expression was detected by western blotting. (F-G) CB CD34+ cells were cotransduced with miRZip-scramble-GFP or miRZip-anti-486-5p vectors (expressing GFP) and pLKO.1-FoxO1 shRNA and pLKO.1-scramble control vectors (expressing RFP). The CD34+RFP+GFP+ cells were sorted and cultured in GEMM medium for 3 days. The number of annexin V+ cells (F) and total cell numbers (G) were determined (n = 3). (H) CD34+ cells were transfected with siRNAs to potential miR-486-5p target genes, cultured in GEMM medium for 3 days and GPA+ cells detected by FACS. The percentage of GPA+ cells compared with control siRNA were determined (n = 3). Cumulative results represent the mean ± SEM. *P < .05, **P < .01, ***P < .001.
Identification of miR-486-5p target genes in CB CD34+ cells. (A) Strategy for identification of miR-486-5p targets in CBCD34+ cells. (B) Effect of miR-486-5p expression on luciferase activity in 293T cells transfected with reporter plasmids containing miR-486-5p binding sequences in 3′ UTRs of potential target genes. The data show inhibition of luciferase activity in cells expressing miR-486-5p compared with control vectors. (C) Gene expression in anti-miR-486-5p compared with scramble sequence–expressing cells determined by qPCR. (D) CB CD34+ cells were transduced with miRZip-anti-486-5p and scramble vector and cultured in GEMM medium for 3 days. FoxO1, PTEN, p-AKT, and AKT expression was detected by western blotting. (E) TF1 cells were transduced with pHIV7-EF1-miR-486or control vector. FoxO1, PTEN, p-AKT, and AKT expression was detected by western blotting. (F-G) CB CD34+ cells were cotransduced with miRZip-scramble-GFP or miRZip-anti-486-5p vectors (expressing GFP) and pLKO.1-FoxO1 shRNA and pLKO.1-scramble control vectors (expressing RFP). The CD34+RFP+GFP+ cells were sorted and cultured in GEMM medium for 3 days. The number of annexin V+ cells (F) and total cell numbers (G) were determined (n = 3). (H) CD34+ cells were transfected with siRNAs to potential miR-486-5p target genes, cultured in GEMM medium for 3 days and GPA+ cells detected by FACS. The percentage of GPA+ cells compared with control siRNA were determined (n = 3). Cumulative results represent the mean ± SEM. *P < .05, **P < .01, ***P < .001.
Potential miR-486-5p targets reported in TargetScan and miRanDa databases upregulated in anti-miR-486-5p expressing CD34+ cells
| Gene . | Description . | Log2 (FC) . | AE . |
|---|---|---|---|
| PTEN | Phosphatase and tensin homolog | 0.157 | 10.95 |
| FOXO1 | Forkhead box O1 | 0.230 | 8.68 |
| GABRE | γ-aminobutyric acid A receptor, ε | 0.887 | 7.83 |
| TWF1 | Twinfilin, actin-binding protein, homolog 1 (Drosophila) | 0.206 | 9.13 |
| AFF3 | AF4/FMR2 family, member 3 | 0.133 | 6.68 |
| ADARB1 | Adenosine deaminase, RNA-specific, B1 (RED1 homolog) | 0.104 | 7.98 |
| ANKRD12 | Ankyrin repeat domain 12 | 0.175 | 8.964 |
| TOB1 | Transducer of ERBB2, 1 | 0.175 | 8.54 |
| RFFL | RING finger and FYVE-like domain containing 1 | 0.198 | 8.79 |
| SLC4A8 | Solute carrier family 4, sodium bicarbonate | 0.097 | 7.77 |
| ARID4B | AT-rich interactive domain 4B (RBP1-like) | 0.200 | 7.57 |
| PIK3AP1 | PI3K adaptor protein 1 | 0.216 | 10.44 |
| FOXP1 | Forkhead box P1 | 0.232 | 8.72 |
| CREBL2 | Cyclic adenosine 5′-monophosphate responsive element binding protein-like 2 | 0.189 | 8.86 |
| SLC10A7 | Solute carrier family 10 (sodium/bile acid cotransporter) | 0.167 | 10.16 |
| EMP1 | Epithelial membrane protein 1 | 0.077 | 7.70 |
| WDFY3 | WD repeat and FYVE domain containing 3 | 0.289 | 7.71 |
| UBASH3B | Ubiquitin-associated and SH3 domain containing, B | 0.095 | 9.74 |
| DCBLD2 | Discoidin, CUB, and LCCL domain containing 2 | 0.254 | 10.02 |
| Gene . | Description . | Log2 (FC) . | AE . |
|---|---|---|---|
| PTEN | Phosphatase and tensin homolog | 0.157 | 10.95 |
| FOXO1 | Forkhead box O1 | 0.230 | 8.68 |
| GABRE | γ-aminobutyric acid A receptor, ε | 0.887 | 7.83 |
| TWF1 | Twinfilin, actin-binding protein, homolog 1 (Drosophila) | 0.206 | 9.13 |
| AFF3 | AF4/FMR2 family, member 3 | 0.133 | 6.68 |
| ADARB1 | Adenosine deaminase, RNA-specific, B1 (RED1 homolog) | 0.104 | 7.98 |
| ANKRD12 | Ankyrin repeat domain 12 | 0.175 | 8.964 |
| TOB1 | Transducer of ERBB2, 1 | 0.175 | 8.54 |
| RFFL | RING finger and FYVE-like domain containing 1 | 0.198 | 8.79 |
| SLC4A8 | Solute carrier family 4, sodium bicarbonate | 0.097 | 7.77 |
| ARID4B | AT-rich interactive domain 4B (RBP1-like) | 0.200 | 7.57 |
| PIK3AP1 | PI3K adaptor protein 1 | 0.216 | 10.44 |
| FOXP1 | Forkhead box P1 | 0.232 | 8.72 |
| CREBL2 | Cyclic adenosine 5′-monophosphate responsive element binding protein-like 2 | 0.189 | 8.86 |
| SLC10A7 | Solute carrier family 10 (sodium/bile acid cotransporter) | 0.167 | 10.16 |
| EMP1 | Epithelial membrane protein 1 | 0.077 | 7.70 |
| WDFY3 | WD repeat and FYVE domain containing 3 | 0.289 | 7.71 |
| UBASH3B | Ubiquitin-associated and SH3 domain containing, B | 0.095 | 9.74 |
| DCBLD2 | Discoidin, CUB, and LCCL domain containing 2 | 0.254 | 10.02 |
AE, average expression; FC, fold change.
Because PTEN and FoxO1 are important components of the phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathway and are known to regulate hematopoietic stem/progenitor cell proliferation and survival, we evaluated these targets in greater detail. Expression of anti-miR-486-5p in CB CD34+ cells significantly upregulated FoxO1 and PTEN protein levels and reduced phospho-AKT (p-AKT) levels (Figure 5D and supplemental Figure 3A). Conversely, overexpression of miR-486-5p in TF1 hematopoietic cells reduced FoxO1 and PTEN expression and increased p-AKT expression (Figure 5E and supplemental Figure 3B). To further evaluate the role of FoxO1 in miR-486-5p–mediated hematopoietic regulation, CB CD34+ cells were cotransduced with vectors expressing anti-miR-486-5p with GFP and shRNA to FoxO1 and RFP. CD34+RFP+GFP+ cells were sorted and cultured for 3 days. The apoptotic and total cells number were evaluated. Whereas significantly increased apoptosis and inhibition of growth of CD34+ cells was seen with anti-miR-486-5p, this was significantly reversed following concomitant knockdown of FoxO1, suggesting that miR-486-5p effects are mediated at least in part by FoxO1 suppression (Figure 5F-G). However, FoxO1 shRNA did not influence erythroid differentiation of CD34+ cells in GEMM culture (supplemental Figure 3C). Because shRNA-mediated inhibition of PTEN was toxic to CB CD34+ cells, it was not possible to determine whether inhibition of PTEN expression could rescue anti-miR-486-5p effects.
Because erythroid differentiation of CB CD34+ cells was not significantly altered following FoxO1 and PTEN inhibition, we screened other miR-486-5p targeted genes for effects on erythroid differentiation. CB CD34+ cells were transfected with individual pools of siRNA to 17 additional miR-486-5p target genes identified by gene expression and bioinformatics analysis as described above. Knockdown of 3 miR-486-5p target genes, RING finger and FYVE-like domain–containing E3 ubiquitin protein ligase (RFFL), epithelial membrane protein 1 (EMP1), and discoidin, CUB, and LCCL domain–containing 2 (DCBLD2) inhibition significantly increased erythroid differentiation of CB CD34+ cells (Figure 5H). Although the function of these genes in hematopoiesis is not well recognized, our results suggest that their inhibition by miR-486-5p could contribute to enhanced erythroid differentiation of CD34+ cells.
Effect of miR-486-5p on growth and imatinib sensitivity of BCR-ABL–transformed hematopoietic cells
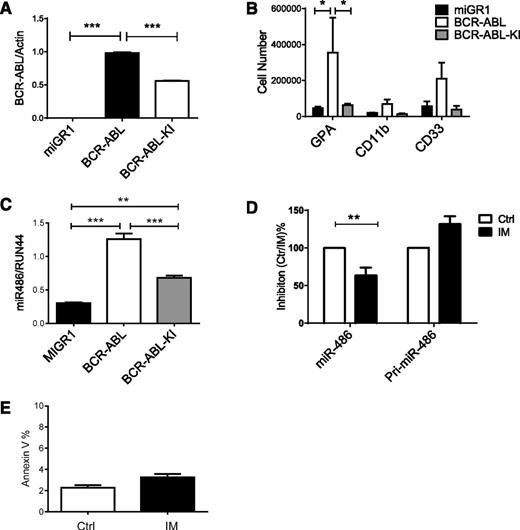
CB CD34+ cells were transduced with retrovirus vectors expressing BCR-ABL or kinase-inactive (KI) BCR-ABL (BA-KI) genes. BCR-ABL expression in transduced cells was confirmed by qPCR (Figure 6A). BCR-ABL increased erythroid differentiation of CB CD34+ cells, whereas BCR-ABL KI did not (Figure 6B). Ectopic BCR-ABL expression increased miR-486 expression in CB CD34+ cells compared with control vector–transduced cells (Figure 6C). Expression of BCR-ABL KI increased miR-486-5p expression in CB CD34+ cells, but to a significantly lesser extent than in wild-type BCR-ABL–expressing cells. Treatment of CML CD34+ cells with IM (1 µM) partially reduced expression of miR-486-5p in CML CD34+ cells (Figure 6D). These results suggest that both BCR-ABL kinase-dependent and kinase-independent mechanisms contribute to upregulated miR-486-5p expression in CML CD34+ cells. We did not observe concomitant reduction in pri-mir-486 levels, suggesting that decreased miR-486 expression is not related to reduced transcription but could be related to altered processing or degradation. The levels of apoptosis in IM-treated CD34+ cells after 24 hours were low and unlikely to account for altered miRNA expression levels (Figure 6E).
Effect of BCR-ABL on miR-486-5p expression. (A) CB CD34+ cells were transduced with MIG-R1, MIG-R1-BCR-ABL, and MIG-R1-BCR-ABL-KI retrovirus, and CD34+GFP+ cells were sorted and analyzed for BCR-ABL expression by qPCR. (B) CB CD34+ cells transduced with MIG-R1, MIG-R1-BCR-ABL, and MIG-R1-BCR-ABL-KI retrovirus were cultured in GEMM medium for 3 days, and the total number of GPA, CD11b, CD33 cells was determined by FACS analysis (n = 3). (C) The expression of miR-486-5p in CB CD34+ cells transduced with MIG-R1, MIG-R1-BCR-ABL, and MIG-R1-BCR-ABL-KI retrovirus was examined by qPCR. (D) CML CD34+ cells were treated with IM at a concentration of 1 µM for 24 hours, and miR-486-5p and pri-miR-486 expression was examined by qPCR. (E) CML CD34+ cells were treated with IM at a concentration of 1 µM for 24 hours, and annexin V+ cells were detected. Cumulative results represent the mean ± SEM. *P < .05, **P < .01, ***P < .001. Ctrl, control.
Effect of BCR-ABL on miR-486-5p expression. (A) CB CD34+ cells were transduced with MIG-R1, MIG-R1-BCR-ABL, and MIG-R1-BCR-ABL-KI retrovirus, and CD34+GFP+ cells were sorted and analyzed for BCR-ABL expression by qPCR. (B) CB CD34+ cells transduced with MIG-R1, MIG-R1-BCR-ABL, and MIG-R1-BCR-ABL-KI retrovirus were cultured in GEMM medium for 3 days, and the total number of GPA, CD11b, CD33 cells was determined by FACS analysis (n = 3). (C) The expression of miR-486-5p in CB CD34+ cells transduced with MIG-R1, MIG-R1-BCR-ABL, and MIG-R1-BCR-ABL-KI retrovirus was examined by qPCR. (D) CML CD34+ cells were treated with IM at a concentration of 1 µM for 24 hours, and miR-486-5p and pri-miR-486 expression was examined by qPCR. (E) CML CD34+ cells were treated with IM at a concentration of 1 µM for 24 hours, and annexin V+ cells were detected. Cumulative results represent the mean ± SEM. *P < .05, **P < .01, ***P < .001. Ctrl, control.
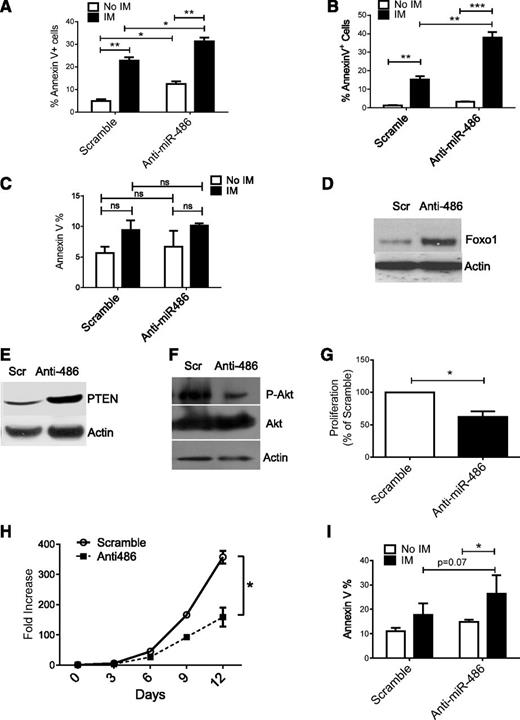
To determine the impact of miR-486-5p on growth of leukemia cells, we transduced BCR-ABL-expressing TF1 cells (TF1-BA cells) with miRZip-anti-486-5p and control vectors. Expression of anti-miR-486-5p modestly enhanced apoptosis in TF1-BA cells. IM treatment resulted in enhanced apoptosis of anti-miR-486-5p–expressing TF1-BA compared with control cells (Figure 7A). Similarly, expression of anti-miR-486-5p in BCR-ABL–transduced CD34+ cells resulted in a modest increase in apoptosis, with significantly increased apoptosis after IM (2.5 µM) treatment (Figure 7B). In contrast, anti-miR-486-5p did not significantly increase apoptosis of control MIG-R1–transduced cells (Figure 7C). Interestingly, anti-miR-486-5p expression induced apoptosis of cells expressing the imatinib-resistant BCR-ABL T315I mutant (supplemental Figure 5A). As with CB CD34+ cells, expression of anti-miR-486-5p increased FoxO1 and PTEN (Figure 7D-E and supplemental Figure 4B) and reduced p-AKT expression in CML CD34+ cells (Figure 7F and supplemental Figure 4C). Inhibition of miR-486-5p significantly reduced CML CD34+ cell division (Figure 7G and supplemental Figure 4D) and cell growth (Figure 7H). Expression of anti-miR-486-5p significantly increased apoptosis of CML CD34+ cells after IM (2.5 µM) treatment (Figure 7I). These results indicate that miR-486-5p expression contributes to survival of BCR-ABL–transformed cells after IM treatment and that inhibition of miR-486-5p enhances the sensitivity of CML progenitors to IM-mediated apoptosis.
miR-486-5p modulates survival and imatinib sensitivity of BCR-ABL–transformed cells. (A) Apoptosis of BCR-ABL–expressing TF-1 cells transduced with miRZip-anti486-5p or scramble control vectors and treated with IM (2.5 μM) for 48 hours. (B) Apoptosis of CB CD34+ cells transduced with BCR-ABL and then miRZip-anti486-5p or scramble control vectors and treated with IM (2.5 µM) for 48 hours. (C) Apoptosis of CB CD34+ cells transduced with MIG-R1 control vector followed by miRZip-anti486 or scramble control vectors and treated with IM (2.5 µM) for 48 hours. (D-F) CML CD34+ cells were transduced with miRZip-anti-486-5p and scramble vector and selected with puromycin in HGF medium for 3 days. FoxO1(D), PTEN (E), and p-AKT (F) expression was detected by western blotting. (G) CML CD34+ cells were transduced with scramble control and miRZip-Ef1-anti-486-5p vector, respectively. The transduced cells were stained with Dye670 and cultured in HGF for 3 days. The proliferation index was determined. Inhibition of proliferation as compared with scramble sequences is shown (n = 3). (H) The fold increase of cell number of scramble and miRZip-EF1-anti486-5p–transduced CML CD34+ cells in the SFEM with HGF. (I) Apoptosis of CML CD34+ cells transduced with miRZip-anti486 or scramble control vectors and treated with IM (2.5 µM) for 48 hours. Cumulative results represent the mean ± SEM. *P < .05, **P < .01, ***P < .001.
miR-486-5p modulates survival and imatinib sensitivity of BCR-ABL–transformed cells. (A) Apoptosis of BCR-ABL–expressing TF-1 cells transduced with miRZip-anti486-5p or scramble control vectors and treated with IM (2.5 μM) for 48 hours. (B) Apoptosis of CB CD34+ cells transduced with BCR-ABL and then miRZip-anti486-5p or scramble control vectors and treated with IM (2.5 µM) for 48 hours. (C) Apoptosis of CB CD34+ cells transduced with MIG-R1 control vector followed by miRZip-anti486 or scramble control vectors and treated with IM (2.5 µM) for 48 hours. (D-F) CML CD34+ cells were transduced with miRZip-anti-486-5p and scramble vector and selected with puromycin in HGF medium for 3 days. FoxO1(D), PTEN (E), and p-AKT (F) expression was detected by western blotting. (G) CML CD34+ cells were transduced with scramble control and miRZip-Ef1-anti-486-5p vector, respectively. The transduced cells were stained with Dye670 and cultured in HGF for 3 days. The proliferation index was determined. Inhibition of proliferation as compared with scramble sequences is shown (n = 3). (H) The fold increase of cell number of scramble and miRZip-EF1-anti486-5p–transduced CML CD34+ cells in the SFEM with HGF. (I) Apoptosis of CML CD34+ cells transduced with miRZip-anti486 or scramble control vectors and treated with IM (2.5 µM) for 48 hours. Cumulative results represent the mean ± SEM. *P < .05, **P < .01, ***P < .001.
Discussion
We identified miRNAs that are differentially expressed in CML and normal CD34+ cells. In addition to confirming overexpression of the cancer-associated miR-17-92 and miR-106b-25 clusters, we discovered miR-486-5p to be significantly overexpressed in CML CD34+ cells. Although miR-486-5p was first identified in human fetal liver,22 an important site for hematopoiesis during development, its role in hematopoietic regulation has not been explored. We show that miR-486-5p levels are increased after erythroid differentiation in normal CD34+ cells. Ectopic expression of miR-486-5p enhanced generation of erythroid cells from CD34+ cells, and miR-486-5p inhibition reduced proliferation, erythroid differentiation, and survival of normal CD34+ cells. Inhibition of miR-486-5p expression also significantly reduced growth and erythroid differentiation of CML CD34+ cells. These results indicate an important role for miR-486-5p in modulating normal and leukemic hematopoietic progenitor growth and erythroid differentiation. Another recent study also reported increased miR-486-3p expression during erythroid differentiation in culture.23 Importantly, the relevance of miR-486 to regulation of MEP growth has been confirmed by Shaham et al,24 who show that miR-486 is selectively expressed in myeloid leukemia of Down syndrome, also characterized by expression of erythroid markers. They show that miR-486 is important for both myeloid leukemia of Down syndrome cell survival and the unique erythroid phenotype. They further show that miR-486 expression is regulated by GATA1s binding. Interestingly, miR-486 is located within the last intron of the ankyrin gene (ANK-1), an integral red cell-membrane cytoskeletal protein.25 Previous studies have shown that GATA-1 binds and regulates the ANK-1 promoter.26 Although not directly shown, these findings suggest that miR-486 may be regulated by GATA1 binding to the ANK-1 promoter. Intronic miRNAs have also been observed to have regulatory effects on their host genes.27
Muscle-expressed miR-486-5p has been shown to regulate PI3K/AKT signaling by targeting PTEN and FoxO128 and to regulate the insulin-like growth factor 1/AKT/mTOR pathway.29 Our results indicate that miR-486-5p enhances PI3K/AKT signaling in hematopoietic cells in association with reduction in PTEN and FoxO1 levels. PTEN is a negative regulator of PI3K, and its deletion causes HSC activation and deregulation of the cell cycle.30 PTEN-mutant mice have increased myeloid and T-lymphoid cells and develop a myeloproliferative disorder.31 The FoxO family of transcription factors plays an important role in regulating cell-cycle progression and is inhibited following phosphorylation by AKT.32 Conditional deletion of FoxO1, FoxO3, and FoxO4 in adult HSCs results in myeloid lineage expansion, increased HSC cell cycling and apoptosis, and decreased long-term HSCs.33 FoxO1 plays an important role in the regulation of insulin signaling and in B-lymphoid specification.34-36 Our results suggest that miR-486-5p–mediated inhibition of FoxO1 contributes to its effects on hematopoietic progenitor proliferation and survival but do not support a role for PTEN in this process.
Enhanced AKT signaling could contribute to the effects of miR-486-5p on hematopoietic cell survival and proliferation. PI3K/AKT signaling plays an important role in erythropoietin signal transduction, and its activation by miR-486 could also contribute to the erythroid phenotype.37 Because miR-486-5p is most highly expressed in the erythrocytic cells, the effects of its inhibition may be most pronounced in that lineage. On the other hand, FoxO1 knockdown did not significantly reverse effects of miR-486-5p knockdown on erythroid differentiation, suggesting that other AKT targets may be involved or that AKT may not be the sole mediator of miR-486-5p effects on erythropoiesis and that effects may be mediated through multiple targets in a coordinated way. Using a combination of gene expression, bioinformatics, and functional screening, we identified 3 novel candidate miR-486-5p target genes (RFFL, EMP1, and DCBLD2) that may contribute to its effects on erythroid differentiation. RFFL regulates CASP8 and CASP10 levels by targeting them for proteasomal degradation, and it has antiapoptotic activity.38 Interestingly, global gene expression analysis of erythroid progenitors identified RFFL as being one of 12 novel transcripts that were continuously upregulated in maturing human primary erythroid cells. EMP1 is a widely expressed membrane protein that is associated with tumors including breast cancer and T-cell lymphoma.39,40 Finally, DCBLD2 is a membrane protein cloned from vascular cells, which is upregulated after vascular injury and considered to regulate vascular cell growth and have a variety of functions in other tissues.41 Additional studies are warranted to determine the role of these genes in erythroid development.
miR-486-5p has been reported to have both tumor-suppressive and tumor-promoting effects. miR-486-5p is downregulated in osteosarcomas, lung cancers, and hepatomas,42,43 and genomic loss is associated with progression of gastric cancers.44 On the other hand, miR-486-5p is overexpressed in invasive and metastatic pancreatic ductal adenocarcinomas45 and enhances survival of cutaneous T-cell lymphoma cells.46 Our observations that miR-486-5p is overexpressed in CML progenitors and that inhibition of miR-486-5p expression significantly reduced the growth and erythroid differentiation of BCR-ABL–transformed CB CD34+ cells and primary CML CD34+ cells support a tumor-promoting role. Both BCR-ABL kinase-dependent and kinase-independent mechanisms appear to contribute to miR-486-5p overexpression. The mechanisms underlying miR-486-5p overexpression in CML cells are unclear, although in colorectal cancer, expression has been associated with aberrant Ras activation,47 which can also occur downstream of BCR-ABL signaling. Increased miR-486-5p expression in CML compared with normal MEPs suggests that the effects of BCR-ABL on miR-486-5p expression are enhanced in the context of erythroid differentiation.
Although TKIs represent a very effective treatment of CML,48 they do not eliminate CML stem cells. Inhibition of miR-486-5p significantly enhanced IM-mediated apoptosis of CML progenitors. Enhanced targeting of CML progenitors by combined anti-miR-486-5p and IM treatment suggests that residual miR-486-5p expression may enhance sensitivity of CML progenitors to TKI-mediated apoptosis. miR-486-5p inhibition also reduced survival of cells expressing a TKI-resistant BCR-ABL mutant. Inhibition of miR-486-5p significantly increased PTEN and FoxO1 in CML CD34+ cells. Previous studies have suggested a possible role for PTEN and FoxO1 in CML pathogenesis. PTEN can function as a tumor suppressor in human Philadelphia chromosome–positive leukemia.49 Similarly, loss of FoxO1 function could contribute to oncogenic transformation in CML. Therefore, inhibition of PTEN and FoxO1 could play a role in miR-486’s effects on CML progenitors, although the role of additional miR-486-5p targets cannot be ruled out.
In conclusion, our studies identify a novel miRNA-dependent mechanism of regulation of normal hematopoiesis, perturbation of which in leukemia progenitors may contribute to altered growth and therapeutic resistance. These studies raise the possibility of targeting miRNA-regulated pathways to improve therapeutic outcomes.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge the assistance of the integrated genomics core, animal research center, and analytical cytometry core. The authors thank StemCyte for their gift of CB samples.
This work was supported by National Institutes of Health National Cancer Institute grants R01 CA95684 and P30CA033572 and Chinese ‘863’ and National Science Foundation of China grants 2012AA02A211 and 81170460.
Authorship
Contribution: L.-S.W. designed and performed experiments, analyzed and interpreted data, and wrote the manuscript; Liang L., Ling L., S.C., H.-Y.S., J.X., F.-J.X., and Y.H. performed experiments; K.-D.S. and M.L. analyzed data; G.S. and J.J.R. designed experiments and interpreted data; and R.B. designed research, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ravi Bhatia, Division of Hematology-Oncology, Department of Medicine, University of Alabama Birmingham, Birmingham, AL 35233; e-mail: rbhatia1@uab.edu.