Key Points
Cell autonomous BCR interactions and interactions with low-affinity autoantigens drive leukemia development in an in vivo model of CLL.
BCR signals induced by binding to external antigen can increase the aggressiveness of CLL.
Abstract
Chronic lymphocytic leukemia (CLL) is a common B-cell malignancy characterized by a highly variable course and outcome. The disease is believed to be driven by B-cell receptor (BCR) signals generated by external antigens and/or cell-autonomous BCR interactions, but direct in vivo evidence for this is still lacking. To further define the role of the BCR pathway in the development and progression of CLL, we evaluated the capacity of different types of antigen/BCR interactions to induce leukemia in the Eμ-TCL1 transgenic mouse model. We show that cell autonomous signaling capacity is a uniform characteristic of the leukemia-derived BCRs and represents a prerequisite for CLL development. Low-affinity BCR interactions with autoantigens generated during apoptosis are also positively selected, suggesting that they contribute to the pathogenesis of the disease. In contrast, high-affinity BCR interactions are not selected, regardless of antigen form or presentation. We also show that the capacity of the leukemic cells to respond to cognate antigen correlates inversely with time to leukemia development, suggesting that signals induced by external antigen increase the aggressiveness of the disease. Collectively, these findings provide in vivo evidence that the BCR pathway drives the development and can influence the clinical course of CLL.
Introduction
Chronic lymphocytic leukemia (CLL) is a common lymphoid malignancy characterized by the expansion and progressive accumulation of mature CD5+ B lymphocytes. The disease has a highly variable clinical course, ranging from rapid progression with fatal outcome to relatively indolent behavior with normal life expectancy.1
The B-cell receptor (BCR) pathway is believed to play a major role in the pathogenesis of CLL.2-4 Signals propagated through the BCR have been shown to increase leukemic cell survival in vitro,5,6 and there is growing evidence that such signals are continuously delivered to the leukemic cells in vivo. This evidence particularly refers to data obtained from gene expression profiling (GEP) studies, which have shown that freshly isolated CLL cells express high levels of genes that can be induced in normal B cells by BCR engagement.7 Such BCR target genes are especially enriched in CLL cells isolated from lymph nodes, which is an important site of antigen encounter.8 In addition, several molecules involved in BCR signal transduction, such as the kinases LYN, spleen tyrosine kinase (SYK), phosphatidylinositol 3-kinase, and protein kinase C, are constitutively active in freshly isolated CLL cells, further suggesting that the BCR pathway is aberrantly or excessively activated in CLL and may represent an important driving force behind the relentless accumulation of the malignant cells.9-12 In support of the latter possibility are data from recent clinical trials with drugs that inhibit BCR signal transduction, which have demonstrated significant activity in patients with CLL.13-15
In addition to its potential role in the development and maintenance of the disease, the BCR pathway is also believed to influence disease progression. This view is primarily supported by the significant association between the clinical course of CLL and 2 BCR related features, which are the mutational status of the immunoglobulin heavy chain variable region (IGHV) genes and expression of the BCR-associated protein tyrosine kinase ZAP-70.7,16-18 Specifically, patients with aggressive CLL typically express unmutated IGHV genes and high levels of ZAP-70, whereas the contrary is usually the case in patients with indolent disease. The mutational status of the IGHV genes reflects features of the antigen/BCR interaction, such as antigen affinity and structure, whereas expression of ZAP-70 has been associated with a greater capacity of the leukemic cells to transduce BCR signals.19 Taken together, these data suggest that the variability in the clinical course of CLL may be due to different types of antigens reacting with the leukemic cells or a different capacity of the leukemic cells to propagate the antigenic stimuli.
The antigens that potentially drive CLL in vivo have still not been identified, but recent studies have provided substantial information regarding the reactivity of the leukemic cell BCRs. In CLL with unmutated IGHV genes (U-CLL), the leukemic cells typically express polyreactive BCRs that bind with low-affinity to various autoantigens, such as nonmuscle myosin heavy chain IIA, vimentin, dsDNA, Sm, or oxidized lipoproteins, which interestingly are all neo-autoantigens generated during apoptosis or oxidation.20-25 In addition, binding of U-CLL immunoglobulins to certain microbial antigens, such as pneumococcal polysaccharides or the pUL32 protein of cytomegalovirus, has been reported.21,26
The antigen specificity of the leukemic BCRs encoded by mutated IGHV genes (M-CLL) is largely unknown, but a role for high-affinity autoantigens has been postulated based on the considerable similarities between M-CLL cells and anergic B cells from the IgHEL/sHEL transgenic mouse model of anergy.27-30 In the latter model, B cells expressing a somatically hypermutated high-affinity BCR reactive with the antigen hen egg lysozyme (HEL) are rendered anergic by continuous exposure to HEL as a soluble autoantigen.31
In addition to being capable of interacting with external antigens, it was recently shown that CLL BCRs can also interact between themselves, thus generating a signal in the absence of any external ligand.32 This cell autonomous signaling capacity is supposed to be enabled by inter- or intramolecular interactions between the leukemic CDR3 regions and intrinsic motifs located in the FR2 and FR3 regions of the immunoglobulin molecule itself.32,33
To further define the role of the various BCR signals in the pathogenesis of CLL, we investigated the capacity of different antigen/BCR interactions to induce leukemia in the Eμ-TCL1 transgenic mouse model of CLL. This model is characterized by the spontaneous development of IGHV-unmutated leukemias that recapitulate many of the main features of human CLL, including the dependence on BCR signals for growth and survival.34-39 We show in this model that cell autonomous BCR interactions and interactions with low-affinity autoantigens are positively selected during leukemia development, suggesting that these 2 types of interactions cooperate in the pathogenesis of CLL.
Materials and methods
Transgenic mice
The transgenic mouse strains Eμ-TCL1, IgPtC (6-1), IgSm (2-12H), IgHEL (MD4), sHEL (ML5), and mHEL-KK were described previously.34,40-43 All strains had been extensively backcrossed on the C57BL/6 background and were housed under conventional barrier protection. Eμ-TCL1/IgHEL transgenic mice were immunized monthly with HEL/CpG particles starting from 4 to 6 weeks and until 10 months of age; the particles were prepared as described elsewhere.44 Leukemia development was monitored by performing monthly white blood cell counts and by flow cytometry analysis with antibodies against IgMa, IgMb, IgD, CD19, B220, and CD5 using a FACSCalibur instrument (BD Biosciences). Overt leukemia was defined as >10 × 106 CD5+ B cells/mL peripheral blood. Mice were killed when they developed symptoms of advanced leukemia, such as lethargy, impaired mobility, shallow or labored breathing, and massive splenomegaly with lymphocytosis due to clonal CD5+ B cells. All animal procedures were performed in accordance with Italian national (Italian legislative decree 116/92 and European directive 8/609) and institutional guidelines.
CLL samples
Blood samples were collected from patients fulfilling standard diagnostic criteria for CLL. Informed consent was obtained from all patients according to the Declaration of Helsinki, and approval for the study was obtained from the Ethical Committee at the Catholic University Hospital “A. Gemelli” (protocol P/734/CE/2012). CLL cells were purified and stimulated as described elsewhere.5
Antigen binding analysis
Leukemic cells were stained with fluorescein-labeled phosphatidylcholine (PtC) liposomes (FormuMax Scientific, Inc.) or biotinylated Sm (AROTEC) or HEL (Sigma-Aldrich) antigens, followed by staining with streptavidin Allophycocyanin (BD Biosciences). Biotinylation was done with the FluoReporter Mini-biotin-XX protein labeling kit (Invitrogen). Cells were costained with anti-CD19, anti-CD45R/B220, or anti-CD5 (BD Biosciences).
Analysis of cell autonomous BCR signaling activity, IgD expression, IGHV/IGLV sequencing, phospho-flow, immunoblotting, immunofluorescence, and statistical analysis are described in supplemental Data available on the Blood Web site.
Results
Establishment of cohorts of Eμ-TCL1 transgenic mice with transgenic BCRs
To determine the type of antigen/BCR interaction that can drive CLL in vivo, we established 3 cohorts of Eμ-TCL1 transgenic mice that expressed transgenic heavy chains (HCs) derived from 3 different antibodies (supplemental Figure 1).
The first 2 cohorts expressed unmutated transgenic HCs encoded by the IGHV1-14/IGHD2-3/IGHJ4 or IGHV12-3/IGHD2-3/IGHJ1 combination, which when paired with appropriate endogenous light chains (LCs), generate low-affinity autoantibodies against the antigens Sm and PtC, respectively.40,41,45 Sm and PtC are both intracellular autoantigens that are translocated or exposed on apoptotic or senescent cells,46,47 thereby mimicking typical U-CLL antigens. Human and murine CLL immunoglobulins that react with Sm or PtC have been previously reported,23,35 and we also detected binding of fluorescently labeled Sm and fluorescently labeled PtC to 3 and 2 of 40 investigated primary human CLL samples, respectively (supplemental Figure 2).
The third cohort of Eμ-TCL1 transgenic mice expressed both the HC and LC from an IGHV-mutated high-affinity anti-HEL BCR (IgHEL) and was established to mimic the high-affinity antigen/BCR interactions that have been implicated in the pathogenesis of M-CLL. These mice were further crossed with transgenic mice expressing HEL as a soluble autoantigen (Eμ-TCL1/IgHEL/sHEL triple transgenic mice) to evaluate the capacity of anergy-inducing autoantigens to drive leukemia in vivo.42 In addition, to test whether leukemia could be induced by chronic interactions with high-affinity foreign antigens or intracellular autoantigens, we generated 2 additional subcohorts of Eμ-TCL1/IgHEL transgenic mice that were either repetitively immunized with HEL-coated particles conjugated with an immunostimulatory CpG oligonucleotide (HEL/CpG-ODN particles)44 or were crossed with transgenic mice that express HEL as an intracellular autoantigen translocated on apoptotic cells (Eμ-TCL1/IgHEL/mHEL-KK triple transgenic mice).43
All transgenic HCs were of the IgMa allotype, allowing easy distinction from endogenous IgMb-positive HCs by flow cytometry. The anti-PtC and anti-Sm transgenes generate only IgM HCs, whereas both IgM and IgD HCs can be generated by the anti-HEL transgene.
Eμ-TCL1/IgPtC transgenic mice develop leukemias that react with the PtC antigen
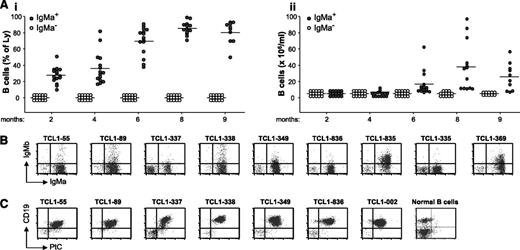
In young Eμ-TCL1/IgPtC transgenic mice (6-7 weeks old), ∼90% of peripheral blood B cells expressed the transgenic IgMa+ HCs, and more than half of them displayed binding to PtC (supplemental Figure 3). Starting from ∼4 months of age, the percentage and absolute number of these cells began to rise, and by 8 months of age, most of these animals developed overt CD5+ B-cell leukemia (Figure 1A). In all cases, the leukemic cells expressed the transgenic HC, but coexpression of endogenous HCs (IgMb+) was occasionally observed (Figure 1B). The LCs expressed by the leukemic cells were always encoded by the IGKV4-91 gene (Table 1). Previous studies have shown that this LC always gives rise to anti-PtC antibodies when paired with the transgenic IGHV12-3/IGHD2-3/IGHJ1 HC.40 Reactivity of the leukemic BCRs with PtC was further confirmed by performing flow cytometry analysis with fluorescent PtC liposomes (Figure 1C).
Leukemia development in Eμ-TCL1/IgPtC transgenic mice. (A) The percentage and absolute number of B cells in the peripheral blood of Eμ-TCL1/IgPtC transgenic mice (n = 15) are shown in panels Ai and Aii, respectively. B cells that express only an endogenous HC (IgMa−) are indicated by empty circles, whereas B cells that express only the transgenic HC (IgMa+) or coexpress the transgenic and an endogenous HC are indicated by filled circles. Each circle represents the value for an individual mouse at the indicated time point; horizontal lines represent mean values. Two mice died of leukemia between 6 and 8 months and 3 between 8 and 9 months of age. (B) Flow cytometry analysis of surface IgMa and IgMb expression in representative leukemias. Analysis was performed on the lymphocyte gate. (C) Flow cytometry analysis of leukemic cells stained with PtC-liposomes and anti-CD19. B cells obtained from the spleen of a normal mouse were used as a negative control.
Leukemia development in Eμ-TCL1/IgPtC transgenic mice. (A) The percentage and absolute number of B cells in the peripheral blood of Eμ-TCL1/IgPtC transgenic mice (n = 15) are shown in panels Ai and Aii, respectively. B cells that express only an endogenous HC (IgMa−) are indicated by empty circles, whereas B cells that express only the transgenic HC (IgMa+) or coexpress the transgenic and an endogenous HC are indicated by filled circles. Each circle represents the value for an individual mouse at the indicated time point; horizontal lines represent mean values. Two mice died of leukemia between 6 and 8 months and 3 between 8 and 9 months of age. (B) Flow cytometry analysis of surface IgMa and IgMb expression in representative leukemias. Analysis was performed on the lymphocyte gate. (C) Flow cytometry analysis of leukemic cells stained with PtC-liposomes and anti-CD19. B cells obtained from the spleen of a normal mouse were used as a negative control.
Dominant clonally rearranged IGHV and IGLV gene sequences identified in leukemias derived from the various Eμ-TCL1 cohorts
| Mouse . | Cohort . | HC . | IGHV (% hom.) . | HCDR3 . | LC . | IGLV (% hom.) . | LCDR3 . |
|---|---|---|---|---|---|---|---|
| TCL1-55 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPF-TF |
| TCL1-89 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPY-TF |
| TCL1-335 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPL-TF |
| TCL1-337 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPL-TF |
| Endogenous | IGHV1-69 (97.6%) | CAR-WGRWLDY-W | |||||
| TCL1-338 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPL-TF |
| TCL1-349 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPL-TF |
| TCL1-369 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPY-TF |
| TCL1-835 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPL-TF |
| TCL1-836 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPL-TF |
| TCL1-120 | Eμ-TCL1/IgSm | Endogenous | IGHV5-4 (99.6%) | CAR-DSPLGLYYFDY-W | Endogenous | ND. | ND |
| TCL1-121 | Eμ-TCL1/IgSm | Transgenic | IGHV1-14 (99.3%) | CAR-NGWLPPMDY-W | Endogenous | IGKV14-126 (100%) | CLQ-HGESPY-TF |
| Endogenous | IGHV2-2 (100%) | CAR-ANWDAWFAY-W | |||||
| Endogenous | IGHV11-2 (100%) | CMR-YGYDWYFDV-W | |||||
| TCL1-165 | Eμ-TCL1/IgSm | Transgenic | IGHV1-14 (99.3%) | CAR-NGWLPPMDY-W | Endogenous | IGKV4-57 (100%) | CQQ-RSSYPP-TF |
| Endogenous | IGHV5-9 (100%) | CAR-HYDFDY-W | |||||
| TCL1-166 | Eμ-TCL1/IgSm | Transgenic | IGHV1-14 (99.3%) | CAR-NGWLPPMDY-W | Endogenous | IGKV3-1 (100%) | CQQ-SRKVW-TF |
| TCL1-417 | Eμ-TCL1/IgSm | Transgenic | IGHV1-14 (99.3%) | CAR-NGWLPPMDY-W | Endogenous | IGKV5-43 (100%) | CQQ-SNSWPQL-TF |
| Endogenous | IGHV2-2 (100%) | CAR-SYDGYYGYAMDY-W | |||||
| TCL1-481 | Eμ-TCL1/IgSm | Transgenic | IGHV1-14 (99.3%) | CAR-NGWLPPMDY-W | Endogenous | IGKV5-43 (100%) | CQQ-SNSWPQL-TF |
| Endogenous | IGHV2-2 (100%) | CAR-SYDGYYGYAMDY-W | |||||
| TCL1-685 | Eμ-TCL1/IgSm | Transgenic | IGHV1-14 (99.3%) | CAR-NGWLPPMDY-W | Endogenous | IGKV4-68 (100%) | CQQ-WSSNPMY-TF |
| TCL1-699 | Eμ-TCL1/IgSm | Transgenic | IGHV1-14 (99.3%) | CAR-NGWLPPMDY-W | Endogenous | IGKV4-72 (100%) | CQQ-WSSNPW-TF |
| TCL1-48 | TCL1/IgHEL | Endogenous | IGHV12-3 (100%) | CAG-DRTGYWYFDV-W | Endogenous | IGKV1-135 (100%) | CWQ-GTHFPR-TF |
| Endogenous | IGKV3-7 (99.7%) | CQH-SWEIPL-TF | |||||
| TCL1-217 | TCL1/IgHEL | Endogenous | IGHV6-3 (100%) | CTE-GGNYFDY-W | Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF |
| Endogenous | IGHV11-1 (100%) | CAR-YYYYGSSYAMDY-W | |||||
| Endogenous | IGHV1-58 (100%) | CAR-GGGTAXFAY-W | |||||
| TCL1-324 | TCL1/IgHEL | Endogenous | IGHV5-9-1 (100%) | CTR-DWDYFDY-W | Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF |
| TCL1-603 | TCL1/IgHEL | Endogenous | IGHV5-9-1 (100%) | CTR-DWDYFDY-W | Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF |
| Endogenous | IGHV1S56 (98.3%) | CAR-GGMVYYYAMDY-W | |||||
| Endogenous | IGHV5-17 (100%) | CAS-TARATNYYAMDY-W | |||||
| TCL1-683 | TCL1/IgHEL | Endogenous | IGHV5-9-1 (100%) | CTR-DWDYFDY-W | Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF |
| Endogenous | IGHV3-8 (100%) | CAR-YYYGSMDY-W | |||||
| TCL1-941 | TCL1/IgHEL+HEL/CpG | Endogenous | IGHV1S22 (100%) | CTR-LDY-W | Transgenic | IGKV5-43 (99.3%) | CWQ-GTHFPR-TF |
| Endogenous | IGHV1S56 (98.3%) | CAR-GGMVYYYAMDY-W | |||||
| TCL1-356 | TCL1/IgHEL/sHEL | Endogenous | IGHV5-9-1 (100%) | CTR-DWDYFDY-W | Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF |
| TCL1-362 | TCL1/IgHEL/sHEL | Endogenous | IGHV5-9-1 (100%) | CTR-DWDYFDY-W | Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF |
| TCL1-739 | TCL1/IgHEL/sHEL | Endogenous | IGHV12-3 (100%) | CAG-DSDGYYWFAY-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIP-TF |
| Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF | |||||
| TCL1-947 | TCL1/IgHEL/sHEL | Endogenous | IGHV5-9-1 (100%) | CTR-DWDYFDY-W | Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF |
| Endogenous | IGHV1S56 (98.3%) | CAR-GGMVYYYAMDY-W | |||||
| Endogenous | IGHV1S22 (100%) | CTR-LDY-W | |||||
| Endogenous | IGHV5-17 (100%) | CAS-TARATNYYAMDY-W | |||||
| TCL1-749 | TCL1/IgHEL/mHEL-KK | Endogenous | IGHV12-3 (100%) | CAG-DSSSYWYFDV-W | Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF |
| Endogenous | IGHV1S81 (100%) | CAR-EDY-W | |||||
| Endogenous | IGHV1S56 (98.3%) | CAR-GGMVYYYAMDY-W |
| Mouse . | Cohort . | HC . | IGHV (% hom.) . | HCDR3 . | LC . | IGLV (% hom.) . | LCDR3 . |
|---|---|---|---|---|---|---|---|
| TCL1-55 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPF-TF |
| TCL1-89 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPY-TF |
| TCL1-335 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPL-TF |
| TCL1-337 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPL-TF |
| Endogenous | IGHV1-69 (97.6%) | CAR-WGRWLDY-W | |||||
| TCL1-338 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPL-TF |
| TCL1-349 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPL-TF |
| TCL1-369 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPY-TF |
| TCL1-835 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPL-TF |
| TCL1-836 | Eμ-TCL1/IgPtC | Transgenic | IGHV12-3 (100%) | CAG-DYDGYWYFDV-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIPL-TF |
| TCL1-120 | Eμ-TCL1/IgSm | Endogenous | IGHV5-4 (99.6%) | CAR-DSPLGLYYFDY-W | Endogenous | ND. | ND |
| TCL1-121 | Eμ-TCL1/IgSm | Transgenic | IGHV1-14 (99.3%) | CAR-NGWLPPMDY-W | Endogenous | IGKV14-126 (100%) | CLQ-HGESPY-TF |
| Endogenous | IGHV2-2 (100%) | CAR-ANWDAWFAY-W | |||||
| Endogenous | IGHV11-2 (100%) | CMR-YGYDWYFDV-W | |||||
| TCL1-165 | Eμ-TCL1/IgSm | Transgenic | IGHV1-14 (99.3%) | CAR-NGWLPPMDY-W | Endogenous | IGKV4-57 (100%) | CQQ-RSSYPP-TF |
| Endogenous | IGHV5-9 (100%) | CAR-HYDFDY-W | |||||
| TCL1-166 | Eμ-TCL1/IgSm | Transgenic | IGHV1-14 (99.3%) | CAR-NGWLPPMDY-W | Endogenous | IGKV3-1 (100%) | CQQ-SRKVW-TF |
| TCL1-417 | Eμ-TCL1/IgSm | Transgenic | IGHV1-14 (99.3%) | CAR-NGWLPPMDY-W | Endogenous | IGKV5-43 (100%) | CQQ-SNSWPQL-TF |
| Endogenous | IGHV2-2 (100%) | CAR-SYDGYYGYAMDY-W | |||||
| TCL1-481 | Eμ-TCL1/IgSm | Transgenic | IGHV1-14 (99.3%) | CAR-NGWLPPMDY-W | Endogenous | IGKV5-43 (100%) | CQQ-SNSWPQL-TF |
| Endogenous | IGHV2-2 (100%) | CAR-SYDGYYGYAMDY-W | |||||
| TCL1-685 | Eμ-TCL1/IgSm | Transgenic | IGHV1-14 (99.3%) | CAR-NGWLPPMDY-W | Endogenous | IGKV4-68 (100%) | CQQ-WSSNPMY-TF |
| TCL1-699 | Eμ-TCL1/IgSm | Transgenic | IGHV1-14 (99.3%) | CAR-NGWLPPMDY-W | Endogenous | IGKV4-72 (100%) | CQQ-WSSNPW-TF |
| TCL1-48 | TCL1/IgHEL | Endogenous | IGHV12-3 (100%) | CAG-DRTGYWYFDV-W | Endogenous | IGKV1-135 (100%) | CWQ-GTHFPR-TF |
| Endogenous | IGKV3-7 (99.7%) | CQH-SWEIPL-TF | |||||
| TCL1-217 | TCL1/IgHEL | Endogenous | IGHV6-3 (100%) | CTE-GGNYFDY-W | Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF |
| Endogenous | IGHV11-1 (100%) | CAR-YYYYGSSYAMDY-W | |||||
| Endogenous | IGHV1-58 (100%) | CAR-GGGTAXFAY-W | |||||
| TCL1-324 | TCL1/IgHEL | Endogenous | IGHV5-9-1 (100%) | CTR-DWDYFDY-W | Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF |
| TCL1-603 | TCL1/IgHEL | Endogenous | IGHV5-9-1 (100%) | CTR-DWDYFDY-W | Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF |
| Endogenous | IGHV1S56 (98.3%) | CAR-GGMVYYYAMDY-W | |||||
| Endogenous | IGHV5-17 (100%) | CAS-TARATNYYAMDY-W | |||||
| TCL1-683 | TCL1/IgHEL | Endogenous | IGHV5-9-1 (100%) | CTR-DWDYFDY-W | Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF |
| Endogenous | IGHV3-8 (100%) | CAR-YYYGSMDY-W | |||||
| TCL1-941 | TCL1/IgHEL+HEL/CpG | Endogenous | IGHV1S22 (100%) | CTR-LDY-W | Transgenic | IGKV5-43 (99.3%) | CWQ-GTHFPR-TF |
| Endogenous | IGHV1S56 (98.3%) | CAR-GGMVYYYAMDY-W | |||||
| TCL1-356 | TCL1/IgHEL/sHEL | Endogenous | IGHV5-9-1 (100%) | CTR-DWDYFDY-W | Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF |
| TCL1-362 | TCL1/IgHEL/sHEL | Endogenous | IGHV5-9-1 (100%) | CTR-DWDYFDY-W | Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF |
| TCL1-739 | TCL1/IgHEL/sHEL | Endogenous | IGHV12-3 (100%) | CAG-DSDGYYWFAY-W | Endogenous | IGKV4-91 (100%) | CQQ-GSSIP-TF |
| Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF | |||||
| TCL1-947 | TCL1/IgHEL/sHEL | Endogenous | IGHV5-9-1 (100%) | CTR-DWDYFDY-W | Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF |
| Endogenous | IGHV1S56 (98.3%) | CAR-GGMVYYYAMDY-W | |||||
| Endogenous | IGHV1S22 (100%) | CTR-LDY-W | |||||
| Endogenous | IGHV5-17 (100%) | CAS-TARATNYYAMDY-W | |||||
| TCL1-749 | TCL1/IgHEL/mHEL-KK | Endogenous | IGHV12-3 (100%) | CAG-DSSSYWYFDV-W | Transgenic | IGKV5-43 (99.3%) | CQQ-SNSWPY-TF |
| Endogenous | IGHV1S81 (100%) | CAR-EDY-W | |||||
| Endogenous | IGHV1S56 (98.3%) | CAR-GGMVYYYAMDY-W |
Eμ-TCL1/IgSm transgenic mice develop leukemias that react with the Sm antigen
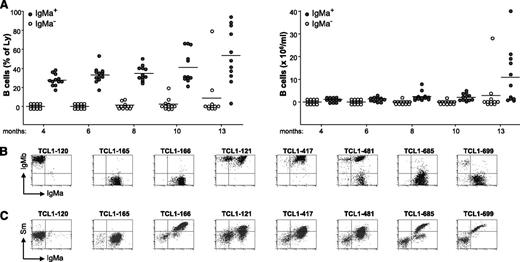
Similar to the previous cohort, 90% of peripheral blood B cells in young Eμ-TCL1/IgSm transgenic mice expressed the transgenic IgMa+ HC, and approximately half of them displayed binding to Sm (supplemental Figure 4). Starting from ∼6 months of age, the percentage and absolute number of these cells began to rise, and by 14 months of age, 8 animals had developed overt CD5+ B-cell leukemia (Figure 2A). With 1 exception (TCL1-120), all of these leukemias expressed the transgenic IgMa+ HC, either alone (n = 4) or together with endogenous IgMb+ HCs (n = 3; Figure 2B). The endogenous LCs of these leukemias were encoded by diverse IGLV genes, although in 4 cases, identical or highly similar IGLV sequences were observed (Table 1; leukemias TCL1-417 and TCL1-481 and leukemias TCL1-685 and TCL1-699). The reactivity of the leukemic BCRs was investigated by performing flow cytometry analysis with biotin-labeled Sm. As shown in Figure 2C, 6 of the 7 IgMa+ leukemias displayed binding to Sm. Altogether, the data obtained from the analysis of Eμ-TCL1/IgPtC and Eμ-TCL1/IgSm transgenic mice showed that low-affinity BCRs reactive with apoptosis-associated autoantigens are positively selected during leukemia development in Eμ-TCL1 transgenic mice.
Leukemia development in Eμ-TCL1/IgSm transgenic mice. (A) Plots depict the percentage and absolute number of IgMa+ (filled circles) and IgMa− (empty circles) B cells in the peripheral blood of Eμ-TCL1/IgSm transgenic mice (n = 12) at the indicated time points. One mouse died of leukemia at 11 months of age. (B) Flow cytometry analysis of surface IgMa and IgMb expression in representative leukemias. Analysis was performed on CD19+ cells. (C) Flow cytometry analysis of leukemic cells stained with biotinylated Sm and anti-IgMa. Analysis was performed on the lymphocyte gate.
Leukemia development in Eμ-TCL1/IgSm transgenic mice. (A) Plots depict the percentage and absolute number of IgMa+ (filled circles) and IgMa− (empty circles) B cells in the peripheral blood of Eμ-TCL1/IgSm transgenic mice (n = 12) at the indicated time points. One mouse died of leukemia at 11 months of age. (B) Flow cytometry analysis of surface IgMa and IgMb expression in representative leukemias. Analysis was performed on CD19+ cells. (C) Flow cytometry analysis of leukemic cells stained with biotinylated Sm and anti-IgMa. Analysis was performed on the lymphocyte gate.
Eμ-TCL1/IgHEL transgenic mice develop leukemias that do not react with the HEL antigen
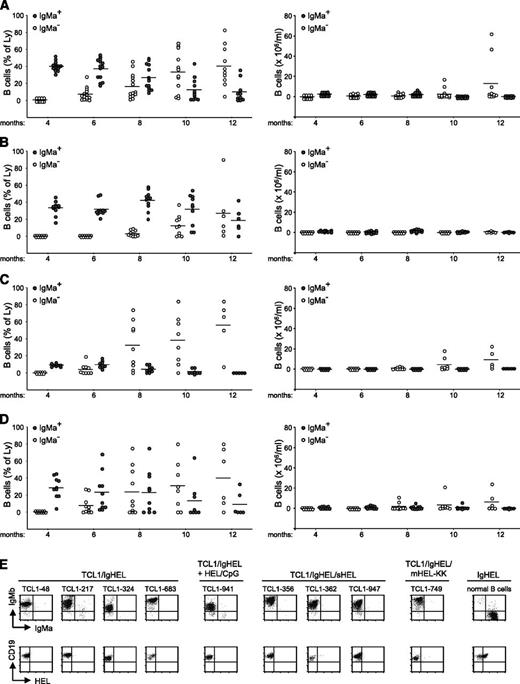
We next investigated leukemia development in Eμ-TCL1 transgenic mice that expressed the mutated high-affinity transgenic anti-HEL BCR. In the absence of HEL, Eμ-TCL1/IgHEL transgenic mice showed a time-dependent reduction in the percentage of B cells expressing the transgenic IgHEL HC with concomitant expansion of CD5+ B cells expressing endogenous HCs (Figure 3A). By 12 months of age, 4 of the 14 animals from this group had developed overt leukemia, which in all cases was IgMa−/IgMb+. A similar behavior was observed in Eμ-TCL1/ IgHEL transgenic mice repetitively immunized with HEL/CpG-ODN particles (Figure 3B). In these animals, the reduction in the percentage of IgMa+ B cells and the expansion of IgMb+ B cells was delayed with respect to the previous cohort, but on extended follow-up, most of these animals also developed overt IgMa−/IgMb+/CD5+ leukemia (data not shown). Eμ-TCL1/IgHEL/sHEL and Eμ-TCL1/IgHEL/mHEL-KK transgenic mice (Figure 3C-D, respectively) also developed IgMa−/IgMb+/CD5+ leukemias, which occurred at a similar rate as Eμ-TCL1/IgHEL double transgenic mice that were not exposed to the HEL antigen. Flow cytometry analysis of representative leukemias from each of the 4 Eμ-TCL1/IgHEL cohorts showed that none of them bind the HEL antigen, in contrast to most normal B cells from the same animals (Figure 3E).
Leukemia development in Eμ-TCL1/IgHEL transgenic mice. Plots depict the percentage and absolute number of IgMa+ (filled circles) and IgMa− (empty circles) B cells in the peripheral blood of (A) Eμ-TCL1/IgHEL transgenic mice that were not exposed to the HEL antigen (n = 14), (B) Eμ-TCL1/IgHEL transgenic mice repetitively immunized with HEL/CpG particles (n = 11), (C) Eμ-TCL1/IgHEL transgenic mice in which HEL was expressed as a soluble autoantigen (n = 9), and (D) Eμ-TCL1/IgHEL transgenic mice in which HEL was expressed as an intracellular autoantigen translocated on apoptotic cells (n = 10). (E) Flow cytometry analysis of representative leukemias stained with anti-IgMa, anti-IgMb, anti-CD19, and biotinylated HEL. Normal splenic B cells from an IgHEL transgenic mouse were used as a positive control for HEL binding.
Leukemia development in Eμ-TCL1/IgHEL transgenic mice. Plots depict the percentage and absolute number of IgMa+ (filled circles) and IgMa− (empty circles) B cells in the peripheral blood of (A) Eμ-TCL1/IgHEL transgenic mice that were not exposed to the HEL antigen (n = 14), (B) Eμ-TCL1/IgHEL transgenic mice repetitively immunized with HEL/CpG particles (n = 11), (C) Eμ-TCL1/IgHEL transgenic mice in which HEL was expressed as a soluble autoantigen (n = 9), and (D) Eμ-TCL1/IgHEL transgenic mice in which HEL was expressed as an intracellular autoantigen translocated on apoptotic cells (n = 10). (E) Flow cytometry analysis of representative leukemias stained with anti-IgMa, anti-IgMb, anti-CD19, and biotinylated HEL. Normal splenic B cells from an IgHEL transgenic mouse were used as a positive control for HEL binding.
Consistent with the flow cytometry data, transgenic HC transcripts were not detected by IGHV sequence analysis in any of the 11 investigated leukemias derived from the 4 Eμ-TCL1/IgHEL transgenic subcohorts. Rather, these leukemias were found to express single or multiple endogenous HCs that were encoded by a highly biased repertoire of unmutated IGHV genes (Table 1). Specifically, 6 of these leukemias expressed HCs encoded by the endogenous IGHV5-9-1 gene associated with the CDR3 motif DWDYFDY, 4 leukemias expressed HCs encoded by the endogenous IGHV1S56 gene associated with the CDR3 motif GGMVYYYAMDY, and 2 leukemias expressed HCs encoded by the endogenous IGHV5-17 gene associated with the CDR3 motif TARATNYYAMDY. Interestingly, in all cases, the endogenous HCs were paired with the transgenic IGKV5-43 LC. These data suggest that the leukemic BCRs in Eμ-TCL1/IgHEL transgenic mice are also selected because of particular binding properties, although they do not bind the HEL antigen.
BCRs expressed by the leukemic cells possess autonomous signaling activity
To establish the contribution of the cell autonomous BCR signal in leukemia development, we next investigated the autonomous signaling capacity of leukemic BCRs derived from the different transgenic cohorts. These included representative anti-PtC and anti-Sm BCRs, as well as BCRs encoded by endogenous IgMb+ HCs. In addition, we tested the autonomous signaling activity of the anti-HEL BCR, which was expressed by normal B cells but not by any of the leukemias that developed in Eμ-TCL1/IgHEL transgenic mice.
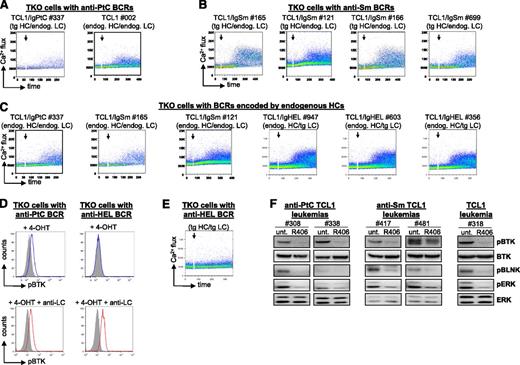
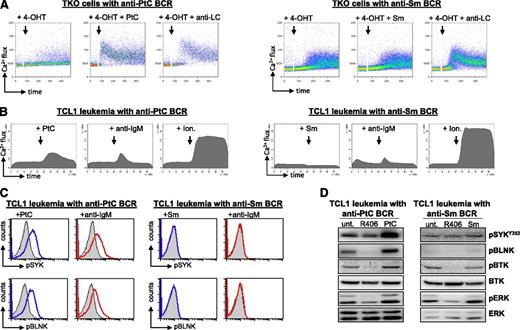
To analyze the autonomous signaling activity of these BCRs, the corresponding HCs and LCs were introduced in the BCR-negative murine B-cell line TKO, which expresses an inactive B-cell linker (BLNK) adaptor protein that becomes functional in the presence of 4-hydroxytamoxifen (4-OHT). Addition of 4-OHT to TKO cells with an autonomously active BCR results in activation of BLNK and further propagation of the BCR signal to Bruton agammaglobulinemia tyrosine kinase (BTK) and phospholipase Cγ2, which then induce an increase in intracellular Ca2+ levels.32 As shown in Figure 4A-C, activation of BLNK resulted in an increase in intracellular Ca2+ levels in all TKO lines that were transduced with a leukemia-derived BCR, suggesting that they all possess autonomous signaling activity. The capacity of the leukemic BCRs to generate an autonomous signal was validated by analyzing BTK phosphorylation, which was also induced on addition of 4-OHT (Figure 4D). In contrast, no increase in BTK phosphorylation or intracellular Ca2+ flux was observed in TKO cells that were transduced with the anti-HEL BCR, suggesting that this BCR does not possess autonomous signaling activity (Figure 4D-E).
Analysis of autonomous BCR signaling capacity. (A-C,E) Flow cytometry analysis of Ca2+ flux after activation of the ERT2-BLNK fusion protein by 4-OHT in TKO cells expressing representative leukemic BCRs encoded by transgenic or endogenous HCs. Addition of 4-OHT is marked by an arrow. (D) Analysis of BTK phosphorylation in TKO cells expressing an autonomously active anti-PtC BCR. TKO cells expressing the anti-HEL BCR were used as a negative control. (F) Analysis of pBTK, pBLNK, and phospho-ERK (pERK) levels in primary Eμ-TCL1 leukemias expressing a transgenic anti-PtC BCR, a transgenic anti-Sm BCR, or a nontransgenic BCR. Western blot analysis was performed on cellular extracts obtained from untreated cells (unt.) or cells that were treated for 2 hours with 1 μM SYK inhibitor R406.
Analysis of autonomous BCR signaling capacity. (A-C,E) Flow cytometry analysis of Ca2+ flux after activation of the ERT2-BLNK fusion protein by 4-OHT in TKO cells expressing representative leukemic BCRs encoded by transgenic or endogenous HCs. Addition of 4-OHT is marked by an arrow. (D) Analysis of BTK phosphorylation in TKO cells expressing an autonomously active anti-PtC BCR. TKO cells expressing the anti-HEL BCR were used as a negative control. (F) Analysis of pBTK, pBLNK, and phospho-ERK (pERK) levels in primary Eμ-TCL1 leukemias expressing a transgenic anti-PtC BCR, a transgenic anti-Sm BCR, or a nontransgenic BCR. Western blot analysis was performed on cellular extracts obtained from untreated cells (unt.) or cells that were treated for 2 hours with 1 μM SYK inhibitor R406.
The autonomous signaling activity of the leukemia-derived BCRs was further validated by experiments with primary leukemic B cells and by investigating additional readouts of the BCR response, such as phosphorylated BLNK and phosphorylated extracellular signal-regulated kinase (ERK). As shown in Figure 4F, inhibition of BCR signaling with an inhibitor of the BCR-proximal kinase SYK resulted in a substantial reduction in the levels of phosphorylated BTK, BLNK, and ERK in unstimulated leukemic cells.
Leukemic anti-PtC and anti-Sm BCRs display different responsiveness to external antigen
The in vivo experiments demonstrated that cell autonomous BCR interactions and BCR interactions with low-affinity external autoantigens are both positively selected during leukemia development. Because the cell autonomous interaction by itself was capable of inducing a BCR signal, we next investigated what are the consequences of stimulation with external cognate antigen. As shown in Figure 5A, stimulation of TKO cells expressing a leukemic anti-PtC BCR with the PtC antigen resulted in increased Ca2+ mobilization. The increase in signal intensity was similar to the increase induced by stimulation with anti-LC antibodies, which were used as a positive control. In contrast, stimulation with Sm did not induce any change in Ca2+ flux in TKO cells expressing a leukemic anti-Sm BCR, and only a minor increase was observed following stimulation with anti-LC antibodies.
Signaling responses induced by anti-PtC and anti-Sm BCRs on stimulation with external antigen. (A) Flow cytometry analysis of Ca2+ flux after stimulation of TKO cells expressing a leukemia-derived anti-PtC or a leukemia-derived anti-Sm BCR. Stimulations were done with the corresponding cognate antigen or with an anti-LC antibody. (B) Flow cytometry analysis of Ca2+ flux in primary leukemic cells expressing a transgenic anti-PtC or a transgenic anti-Sm BCR. The cells were stimulated with antigen, anti-IgM, or Ionomycin (Ion.) as indicated in the figure. Ionomycin was used as a positive control. (C) Phospho-flow analysis of Eμ-TCL1-derived leukemias expressing an anti-PtC or an anti-Sm BCR. Cells were stimulated for 10 minutes at 37°C with their cognate antigen or with anti-IgM. Blue lines indicate the phospho-SYK (pSYK) and phospho-BLNK (pBLNK) signal in cells stimulated with cognate antigen, red lines in cells stimulated with anti-IgM, and filled histograms in unstimulated cells. These results are representative of 7 experiments performed with leukemias expressing an anti-PtC and 5 experiments performed with leukemias expressing an anti-Sm BCR. The remaining experiments are shown in supplemental Figure 4. (D) Immunoblotting analysis of cellular extracts from primary leukemic cells expressing an anti-PtC or an anti-Sm BCR. Cells were stimulated for 10 minutes with cognate antigen or were incubated for 2 hours with 1 μM R406 prior to lysis. The levels of pSYK are not reduced by R406 treatment because the investigated Y352 residue is phosphorylated by SRC-family kinases.
Signaling responses induced by anti-PtC and anti-Sm BCRs on stimulation with external antigen. (A) Flow cytometry analysis of Ca2+ flux after stimulation of TKO cells expressing a leukemia-derived anti-PtC or a leukemia-derived anti-Sm BCR. Stimulations were done with the corresponding cognate antigen or with an anti-LC antibody. (B) Flow cytometry analysis of Ca2+ flux in primary leukemic cells expressing a transgenic anti-PtC or a transgenic anti-Sm BCR. The cells were stimulated with antigen, anti-IgM, or Ionomycin (Ion.) as indicated in the figure. Ionomycin was used as a positive control. (C) Phospho-flow analysis of Eμ-TCL1-derived leukemias expressing an anti-PtC or an anti-Sm BCR. Cells were stimulated for 10 minutes at 37°C with their cognate antigen or with anti-IgM. Blue lines indicate the phospho-SYK (pSYK) and phospho-BLNK (pBLNK) signal in cells stimulated with cognate antigen, red lines in cells stimulated with anti-IgM, and filled histograms in unstimulated cells. These results are representative of 7 experiments performed with leukemias expressing an anti-PtC and 5 experiments performed with leukemias expressing an anti-Sm BCR. The remaining experiments are shown in supplemental Figure 4. (D) Immunoblotting analysis of cellular extracts from primary leukemic cells expressing an anti-PtC or an anti-Sm BCR. Cells were stimulated for 10 minutes with cognate antigen or were incubated for 2 hours with 1 μM R406 prior to lysis. The levels of pSYK are not reduced by R406 treatment because the investigated Y352 residue is phosphorylated by SRC-family kinases.
To further explore the possibility that anti-PtC and anti-Sm BCRs differ in their ability to generate signals on binding to their cognate antigens, we performed Ca2+ flux and phospho-flow analysis using primary leukemic cells. Consistent with the previous findings, all anti-PtC leukemias showed a substantial increase in Ca2+ flux, phospho-SYK, and phospho-BLNK following stimulation with PtC or anti-IgM, whereas anti-Sm leukemias displayed only a marginal increase or did not respond to either stimulus (Figure 5B; supplemental Figure 5). Similar results were obtained by immunoblotting analysis, which showed increased phosphorylation of SYK, BLNK, BTK, and ERK in anti-PtC but not in anti-Sm expressing leukemic cells on binding to cognate antigen (Figure 5D).
Sm antigen is internalized by the leukemic cells
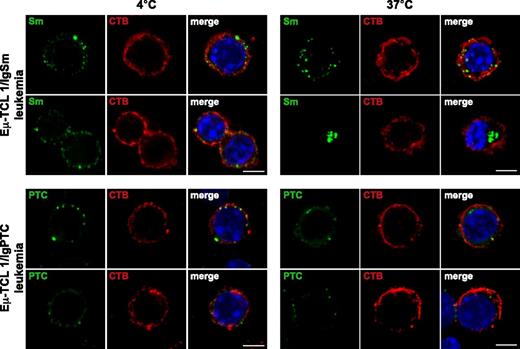
Although Sm binding did not activate BCR signaling, it remained possible that this interaction contributes to the growth of the leukemic cells by providing costimulatory signals, such as peptides for T-cell presentation or Toll-like receptor ligands. To further evaluate this possibility, we investigated internalization of Sm and PtC by primary leukemic cells that expressed the corresponding BCR. As shown in Figure 6, both antigens were internalized following incubation at 37°C.
Internalization of Sm and PtC by primary leukemic cells. Eμ-TCL1/IgSm leukemia cells were stained at 4°C with biotin-labeled Sm, washed, and then stained with streptavidin-FITC. Eμ-TCL1/IgPtC leukemia cells were stained at 4°C with fluorescein-labeled PtC-liposomes. Cells were then incubated for 20 minutes at 4°C or 37°C, fixed, and costained with cholera toxin B (CTB)-Alexa 633 to identify the cellular membranes and Hoechst 33332 to mark the nuclei. Images were acquired with a laser scanning confocal microscope (TCS SP5; Leica Microsystems, Mannheim, Germany) using a ×40 (NA = 1.25) oil immersion lens with optical pinhole at 1 AU. Confocal Z-stacks collected at a magnification of ×40 with a 0.29-μm interval and a 5-μm total optical depth are displayed. Scale bar indicates 5 μm. Images at 4°C show Sm/CTB and PtC/CTB colocalization on the cellular membrane. In images acquired from cells incubated at 37°C, Sm and PtC can be seen dissociated from CTB and located below the cellular membrane. IgHEL-expressing normal B cells incubated either with the Sm antigen or PtC were used as a negative control in these experiments (data not shown).
Internalization of Sm and PtC by primary leukemic cells. Eμ-TCL1/IgSm leukemia cells were stained at 4°C with biotin-labeled Sm, washed, and then stained with streptavidin-FITC. Eμ-TCL1/IgPtC leukemia cells were stained at 4°C with fluorescein-labeled PtC-liposomes. Cells were then incubated for 20 minutes at 4°C or 37°C, fixed, and costained with cholera toxin B (CTB)-Alexa 633 to identify the cellular membranes and Hoechst 33332 to mark the nuclei. Images were acquired with a laser scanning confocal microscope (TCS SP5; Leica Microsystems, Mannheim, Germany) using a ×40 (NA = 1.25) oil immersion lens with optical pinhole at 1 AU. Confocal Z-stacks collected at a magnification of ×40 with a 0.29-μm interval and a 5-μm total optical depth are displayed. Scale bar indicates 5 μm. Images at 4°C show Sm/CTB and PtC/CTB colocalization on the cellular membrane. In images acquired from cells incubated at 37°C, Sm and PtC can be seen dissociated from CTB and located below the cellular membrane. IgHEL-expressing normal B cells incubated either with the Sm antigen or PtC were used as a negative control in these experiments (data not shown).
Response to external antigen correlates with leukemia development and progression
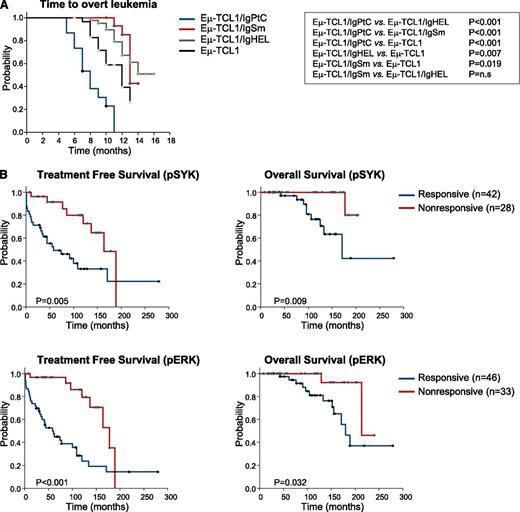
To investigate whether signals generated by external antigens can influence the aggressiveness of the disease, we compared the rate of leukemia development in Eμ-TCL1 transgenic (tg) mice with a transgenic BCR (Eμ-TCL1/IgPtC, Eμ-TCL1/IgSm, and Eμ-TCL1/IgHEL) and Eμ-TCL1 tg mice with a normal BCR repertoire. As shown in Figure 7A and supplemental Figure 6, time to overt leukemia development was significantly shorter, and the percentage and absolute number of leukemic CD5+ B cells was significantly higher in Eμ-TCL1/IgPtC tg mice compared with Eμ-TCL1/IgSm, Eμ-TCL1/IgHEL, or Eμ-TCL1 tg mice with a normal BCR repertoire. In contrast, time to overt leukemia development was significantly prolonged, and the percentage and absolute number of leukemic CD5+ B cells was significantly lower in Eμ-TCL1/IgSm and Eμ-TCL1/IgHEL tg mice compared with Eμ-TCL1 tg mice with a normal BCR repertoire. These results suggest that BCRs responsive to external antigens accelerate leukemia development, whereas BCRs that do not respond or do not bind to external antigen have an opposite effect.
BCR responsiveness and leukemia behavior in murine and human CLL. (A) Kaplan-Meier plots showing time to development of overt leukemia in Eμ-TCL1, Eμ-TCL1/IgPtC, Eμ-TCL1/IgSm, and Eμ-TCL1/IgHEL transgenic mice. (B) Treatment-free and overall survival in CLL patients stratified according to response to anti-IgM stimulation. Cases that showed a >20% increase in pSYK or pERK levels after 10-minute stimulation with anti-IgM were classified as responders. Data are presented separately for the pSYK and pERK analysis because 14% of the cases (8/59) that were analyzed for both readouts showed discordant results.
BCR responsiveness and leukemia behavior in murine and human CLL. (A) Kaplan-Meier plots showing time to development of overt leukemia in Eμ-TCL1, Eμ-TCL1/IgPtC, Eμ-TCL1/IgSm, and Eμ-TCL1/IgHEL transgenic mice. (B) Treatment-free and overall survival in CLL patients stratified according to response to anti-IgM stimulation. Cases that showed a >20% increase in pSYK or pERK levels after 10-minute stimulation with anti-IgM were classified as responders. Data are presented separately for the pSYK and pERK analysis because 14% of the cases (8/59) that were analyzed for both readouts showed discordant results.
To further address the role of BCR responsiveness in modulating the aggressiveness of the disease, we investigated treatment-free and overall survival in a large series of CLL patients that had previously been analyzed in our laboratory for the capacity to respond to BCR stimulation (Figure 7B). Sixty percent (42/70) of the patients were classified as responders based on an increase in pSYK and 58% (46/79) based on an increase in pERK levels. A significant difference was observed between the 2 groups both in terms of treatment-free and overall survival. Collectively, these data suggest that the capacity to respond to external antigen influences leukemia behavior in both murine and human CLL.
Discussion
Chronic antigen stimulation plays an important role in the pathogenesis of several indolent B-cell malignancies that arise in the setting of chronic bacterial or viral infections, such as gastric mucosa-associated lymphoid tissue (MALT) lymphomas associated with Helicobacter pylori infection or splenic marginal zone and lymphoplasmacytoid lymphomas associated with hepatitis C virus infection.48,49 In these malignancies, the pathogenic role of antigen is further corroborated by clinical studies, which have shown that elimination of the infectious agent at early stages of the disease can lead to tumor regression.50,51 In CLL, a large amount of circumstantial evidence supports a role for an aberrant antigen-driven immune response in the pathogenesis of the disease, but the nature of the antigens that drive this response has still not been established.
We now show that 2 types of BCR interactions are positively selected during leukemia development in a well-established in vivo murine model of the disease. These include the recently discovered cell autonomous BCR interaction, which is a typical feature of both U-CLL and M-CLL BCRs and interactions with low-affinity autoantigens, which are considered primarily a feature of U-CLL BCRs. Both interactions involve binding to endogenous antigenic determinants, suggesting that CLL, and in particular U-CLL, develops as a consequence of an aberrant autoantigen-driven response.
We also show that high-affinity BCR interactions are not positively selected during leukemia development in this model, regardless whether the cognate antigen is provided as a foreign antigen, as a soluble autoantigen or as an intracellular autoantigen exposed on apoptotic cells. The inability of such interactions to induce leukemia in vivo suggests that they do not play a role in the pathogenesis of M-CLL. It should be noted, however, that the high-affinity anti-HEL BCR that was investigated in our study did not possess cell autonomous activity, so we cannot exclude the possibility that high-affinity antigen/BCR interactions could cooperate with cell autonomous BCR interactions in driving M-CLL. In line with this possibility, high-affinity interactions between 3 stereotyped M-CLL BCR subsets and the fungal antigen β-(1,6)-glucan or the Fc portion of human IgG were recently reported.52-54 Follow-up studies in the Eμ-TCL1 tg model using high-affinity BCRs with autonomous signaling capacity should be undertaken to further resolve this issue.
The relative contribution of the cell autonomous and cell external BCR interactions in the pathogenesis of CLL is still unclear, but the fact that both interactions were positively selected suggests independent roles. The role of the cell autonomous BCR interaction could be to prolong the survival of the leukemic cells, as suggested by in vitro experiments demonstrating increased spontaneous apoptosis of both human and murine CLL cells treated with kinase inhibitors or siRNA molecules that target BCR pathway components.10,36,39 The role of the low-affinity interaction with external autoantigen could be to increase the intensity of the BCR signal above the threshold required for cellular activation or to facilitate the recruitment of costimulatory signals by internalizing ligands for T-helper cells or Toll-like receptors. In support of the latter possibility, confocal microscopy experiments showed that even leukemic cells that do not generate a discernable BCR signal are capable of efficiently internalizing antigen. A similar observation was recently made by Bergh et al, who showed that human CLL cells belonging to the large stereotyped U-CLL BCR subset 1 efficiently bind and internalize their cognate antigen oxidized low density lipoprotein without activating downstream BCR signaling pathways.55 Additional support for the possibility that recruitment of costimulatory signals is an important function of CLL BCRs and a possible reason for their positive selection comes from recent studies demonstrating that autologous T-helper cells are required to drive the proliferation of CLL cells in a murine xenograft model in vivo and that such autologous T-helper cells can be efficiently activated by CLL cells in an antigen-specific manner.56,57 Interestingly, the study of Os et al57 also showed that T-helper cells specific for epitopes within the CLL BCR are present in patients with CLL, suggesting that in some cases the cell autonomous BCR interaction by itself may provide all the signals required for leukemic cell proliferation.
Another interesting finding of this study was the observation that the ability of the leukemic BCRs to respond to external low-affinity antigen correlated inversely with the rate of leukemia development. Specifically, Eμ-TCL1/IgPtC tg mice developed leukemia earlier than Eμ-TCL1 tg mice with a normal BCR repertoire, whereas a delay in leukemia development was observed in Eμ-TCL1/IgSm and Eμ-TCL1/IgHEL tg mice. Whereas the delay in leukemia development in Eμ-TCL1/IgHEL tg mice could be explained by the lack of autonomous signaling capacity of the transgenic IgHEL HC and the consequent smaller starting population of B cells amenable to transformation, this was not the case of Eμ-TCL1/IgSm transgenic mice, which until 4 months of age had a similar percentage of IgMa+ B cells as Eμ-TCL1/IgPtC tg mice. A similar correlation between the capacity to respond to autoantigen stimulation and increased disease aggressiveness was also observed in a recent study that investigated the evolution of PtC-reactive Eμ-TCL1 leukemic cells after serial transfers in SCID mice.58 Altogether, these data suggest that signals induced by external antigen can accelerate the course of the disease and may also underlie the variability in the clinical course of human CLL. Consistent with the latter possibility, an association between the capacity to respond to BCR engagement and shorter treatment-free survival was also reported in 2 recently published series of patients with CLL59,60 and was confirmed in a larger series of patients investigated in our study.
In conclusion, the data presented in this study provide further evidence that the BCR pathway is a major driving force in CLL. They show that signals generated by cell autonomous BCR interactions are essential for leukemia development, but interactions with external low-affinity autoantigens can modulate the course of the disease and may provide important costimulatory signals. Further studies are warranted to determine whether the continuous BCR signals generated by the cell autonomous interaction and the intermittent BCR signals generated by interactions with external antigens are equally targeted by drugs that inhibit BCR signal transduction, as such studies may lead to the development of novel rational combinations for CLL treatment.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Italian Association for Cancer Research (project AIRC IG_12939), the Leukemia & Lymphoma Society (project R6170-10), the Nelia et Amadeo Barletta Foundation, and the Deutsche Krebshilfe (project 108935).
Authorship
Contribution: S.I., E.H., S.B., M.D.-v.M., S.G., H.J., and D.G.E. designed the study; S.I., E.H., S.B., M.D.-v.M., A.R., S.G., M.S., D.B., and G.B. performed the experiments; F.A., S.S., and L.L. provided patient specimens and clinical data; J.E.-D., V.N., H.W., R.J.C., S.H.C., and C.M.C. provided vital reagents and conceptual advice; and S.I., E.H., S.B., M.D.-v.M., A.R., S.G., D.B., F.B., H.J., and D.G.E. analyzed data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.S. is Molecular Biomedicine, Faculty of Engineering and Natural Sciences, International University Sarajevo, Bosnia and Herzegovina.
Correspondence: Dimitar G. Efremov, ICGEB, Molecular Hematology Unit, CNR Campus “Adriano Buzzati-Traverso,” Via E. Ramarini 32, I-00016 Monterotondo Scalo (Rome), Italy; e-mail: efremov@icgeb.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal