Key Points
PTPRK binds to STAT3 and directly dephosphorylates phospho-STAT3 at Tyr705.
Loss of PTPRK, located in the deleted 6q region, leads to STAT3 activation and contributes to nasal-type NK/T-cell lymphoma pathogenesis.
Abstract
Nasal-type natural killer/T-cell lymphoma (NKTCL) is an aggressive disease characterized by frequent deletions on 6q, and constitutive activation of signal transducer and activator of transcription 3 (STAT3). Phosphorylation at Tyr705 activates STAT3, inducing dimerization, nuclear translocation, and DNA binding. In this study, we investigated whether receptor-type tyrosine-protein phosphatase κ (PTPRK), the only protein tyrosine phosphatase at 6q that contains a STAT3-specifying motif, negatively regulates STAT3 activation in NKTCL. PTPRK was highly expressed in normal NK cells but was underexpressed in 4 of 5 (80%) NKTCL cell lines and 15 of 27 (55.6%) primary tumors. Significantly, PTPRK protein expression was inversely correlated with nuclear phospho-STAT3Tyr705 expression in NKTCL cell lines (P = .025) and tumors (P = .040). PTPRK restoration decreased nuclear phospho-STAT3Tyr705 levels, whereas knockdown of PTPRK increased such levels in NKTCL cells. Phosphatase substrate-trapping mutant assays demonstrated the binding of PTPRK to STAT3, and phosphatase assays showed that PTPRK directly dephosphorylated phospho-STAT3Tyr705. Restoration of PTPRK inhibited tumor cell growth and reduced the migration and invasion ability of NKTCL cells. Monoallelic deletion and promoter hypermethylation caused underexpression of PTPRK messenger RNA in NKTCL, and methylation of the PTPRK promoter significantly correlated with inferior overall survival (P = .049) in NKTCL patients treated with the steroid-dexamethasone, methotrexate, ifosfamide, l-asparaginase, and etoposide regimen. Altogether, our findings show that PTPRK underexpression leads to STAT3 activation and contributes to NKTCL pathogenesis.
Introduction
Extranodal nasal-type natural killer/T-cell lymphoma (NKTCL) primarily develops in the nasal cavity but can also occur in extranasal sites, either as a primary extranasal or disseminated disease.1,2 NKTCL histology is characterized by the angiocentric and angioinvasive infiltration of lymphoma cells and prominent necrosis.1 NKTCLs can be widely disseminated in the leukemic phase1,2 and are invariably associated with Epstein-Barr virus (EBV) infection in lymphoma cells.1 Epidemiologically, NKTCL is prevalent in Asia and South America but is rare in the United States and Europe.1
NKTCL is an aggressive disease1 characterized by frequent deletions on chromosome 6q.3-14 Through the use of array-based comparative genomic hybridization and gene expression profiling, oncogenic pathways operating in NKTCL have been delineated by various groups,14-19 and the PRDM1, ATG5, AIM1, HACE1, and FOXO3 genes have been identified as tumor-suppressor gene candidates in the most frequently deleted region in NKTCL, 6q21, by functional analyses.14-17
Recent studies have demonstrated that constitutive activation of signal transducer and activator of transcription 3 (STAT3), a known pro-oncogenic transcription factor, is common in NKTCL and is an early feature in NKTCL pathogenesis.15,20 STAT3 activation is accompanied by tyrosine phosphorylation at Tyr705, which induces dimerization and nuclear translocation, where STAT3 molecules activate the transcription of a wide array of genes that play critical roles in development, immunity, and proliferation.21 Recently, it was reported that STAT3 activation in NKTCL most often results from constitutive janus kinase 3 (JAK3) phosphorylation at Tyr980, and in a proportion of NKTCL primary tumors, constitutive JAK3 activation is associated with acquired mutations in the JAK3 pseudokinase domain.22,23 Protein phosphorylation is a reversible process, and the degree of phosphorylation of a target protein reflects a net balance between the competing actions of protein kinases and phosphatases. Because recent large-scale genetic analyses of various human tumors have highlighted the relevance of protein tyrosine phosphatases (PTPases) as putative tumor suppressors24,25 , in this study, we explored whether the loss of expression of the receptor-type tyrosine-protein phosphatase κ (PTPRK), the only PTPase among the 3 PTPases located in the commonly deleted 6q region (see supplemental Table 1 on the Blood Web site) that contains a STAT3-specifying YXXQ motif26 at the first catalytic domain, leads to STAT3 activation and plays a role in NKTCL oncogenesis. The PTPRK gene, located at 6q22.2-q22.3,27 belongs to the receptor PTPase (R-PTPase)-type R2B subfamily and is characterized by an extracellular adhesion molecule-like domain, a single transmembrane helix, and a cytoplasmic tyrosine phosphatase domain.28-32
In this study, we provide a novel molecular mechanism leading to STAT3 deregulation in NKTCL. We show that PTPRK binds to STAT3 and directly dephosphorylates phospho-STAT3 at Tyr705 and regulates the levels of phospho-STAT3Tyr705 in NKTCL. PTPRK, located in the commonly deleted 6q region, is frequently underexpressed in NKTCL due to monoallelic loss and promoter hypermethylation, and PTPRK loss leads to STAT3 activation and contributes to NKTCL pathogenesis.
Materials and methods
NKTCL biopsy specimens
Archival paraffin and frozen tumor blocks from 27 Chinese patients diagnosed with NKTCL from 1996 to 2010 were retrieved for this study from the Department of Pathology at Queen Mary Hospital, Hong Kong. Twenty-two of the primary NKTCL cases included in this study have been previously described.33-35 Clinical features were available for all 27 NKTCL patients. Treatment outcome data were available for the 17 NKTCL patients treated with the steroid-dexamethasone, methotrexate, ifosfamide, l-asparaginase, and etoposide (SMILE) regimen,33-35 and the remaining 10 patients were treated with other multiagent chemotherapies, with or without radiotherapy. The SMILE protocol for the treatment of NKTCL patients was approved by the University of Hong Kong/Hospital Authority Hong Kong West Cluster Institutional Review Board.33-35 Research was conducted in accordance with the Declaration of Helsinki.
Viral transductions of NKTCL cell lines
The Moloney murine leukemia virus-based retroviral vector pBMN-GFP (Orbigen Inc., San Diego, CA), containing a downstream internal ribosomal entry site for green fluorescent protein (GFP) expression, and a RetroNectin-coated plate system were used for retroviral gene transduction. Full-length PTPRK complementary DNA (cDNA) (4341 bp) with a Myc-DDK tag sequence at the carboxyl-terminus was subcloned into the retroviral vector (DDK sequence: DYKDDDDK) (supplemental Figure 1Ai). NKYS and KHYG cells were successfully transduced with a retroviral vector containing PTPRK cDNA with transduction efficiencies of ∼12% and 7%, respectively (supplemental Figure 1Aii). The in vitro functional assays were performed on GFP-sorted cells following validation of PTPRK expression in PTPRK cDNA transduced NKYS cells (supplemental Figure 2).
PTPRK knockdown was accomplished using a CMV-puro (pLKO.1puro-CMV-TagRFP) lentiviral plasmid (Sigma-Aldrich, St. Louis, MO) for PTPRK short hairpin RNA (shRNA) transduction in SNK6 cells (supplemental Figure 1Bi), and the transduction efficiency was ∼20% (supplemental Figure 1Bii). In vitro functional assays were performed on phycoerythrin-sorted SNK6 cells, following validation that PTPRK expression was partially knocked down to ∼ 30% of the original level in sorted SNK6 cells using 2 independent shRNAs (supplemental Figure 3).
PTPRK promoter methylation analysis
The methylation status of the PTPRK promoter was analyzed via methylation-specific polymerase chain reaction (PCR) (or MSP) and further confirmed with bisulfite genomic sequencing (BGS). The cytosine guanine dinucleotide (CpG) sites in the PTPRK promoter (−300 bp to +300 bp) and the position of the MSP and BGS primers (supplemental Table 2) used in this study are presented in supplemental Figure 4.
Detailed descriptions of the various methods employed in this study are provided in the supplemental “Materials and methods.”
Results
PTPRK messenger RNA (mRNA) and PTPRK protein are frequently underexpressed in NKTCL
Semiquantitative reverse transcription-polymerase chain reaction (RT-PCR) analysis showed that PTPRK mRNA was not expressed in 4 of 5 (80%) NKTCL cell lines but was highly expressed in 2 preparations of normal NK cells and an NKTCL cell line (SNK6) (Figure 1A). Significantly lower levels of PTPRK mRNA expression were detected in 15 of 27 (55.6%) NKTCL primary tumors, which is less than ∼ 30% of the levels in normal NK cells (Figure 1B). Immunohistochemical staining showed that PTPRK protein expression in tumor cells (Figure 2A) correlated with its mRNA expression in the 27 NKTCL primary tumors (P < .034; supplemental Table 3).
PTPRK mRNA is frequently underexpressed in NKTCLs. Gel images show the PTPRK mRNA expression levels as detected by semiquantitative RT-PCR in 2 samples of normal NK cells, 5 NKTCL cell lines (A), and 27 NKTCL primary tumors (B). The mRNA expression in each sample was normalized to β-actin expression. PTPRK mRNA was highly expressed in normal NK cells, but frequently not detected in NKTCL cell lines and significantly underexpressed in more than half of the NKTCL primary tumors.
PTPRK mRNA is frequently underexpressed in NKTCLs. Gel images show the PTPRK mRNA expression levels as detected by semiquantitative RT-PCR in 2 samples of normal NK cells, 5 NKTCL cell lines (A), and 27 NKTCL primary tumors (B). The mRNA expression in each sample was normalized to β-actin expression. PTPRK mRNA was highly expressed in normal NK cells, but frequently not detected in NKTCL cell lines and significantly underexpressed in more than half of the NKTCL primary tumors.
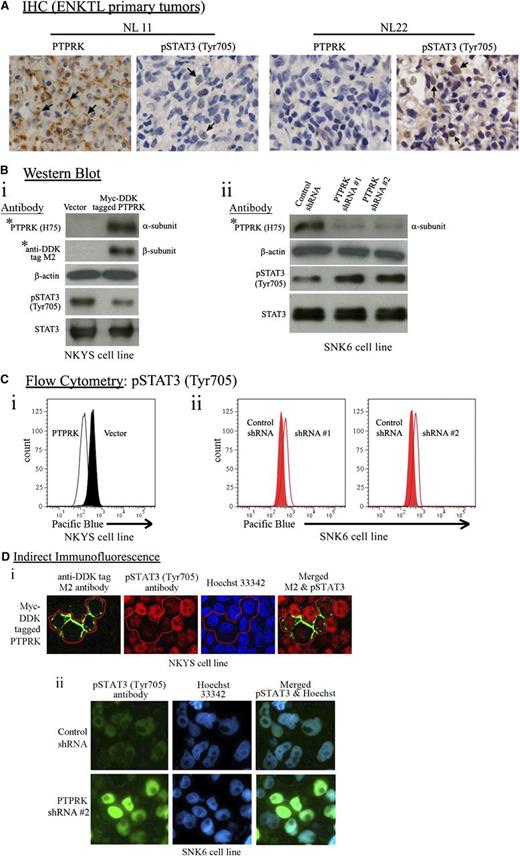
PTPRK protein expression inversely correlates with STAT3 activation. (A) Immunohistochemical staining of paraffin sections of representative NKTCL primary tumors with high (NL11) and low (NL22) PTPRK expression using a rabbit polyclonal PTPRK (H75) antibody raised against amino acids 27-101 mapping within an N-terminal extracellular domain of human PTPRK (brown cellular membranous staining between the cell-cell contact and faint cytoplasmic staining is indicated by arrows), and a phospho-STAT3Tyr705 antibody (brown nuclear staining is indicated by arrows). The immunostaining demonstrated that PTPRK protein expression inversely correlated with phospho-STAT3Tyr705 levels in NKTCL primary tumors. Original magnification ×400. (B) Western blot analyses of PTPRK and phospho-STAT3Tyr705 expression in PTPRK nonexpressing NKYS cells with reconstitution of PTPRK expression (i) and in PTPRK-expressing SNK6 cells with partial knockdown of PTPRK expression (ii), using the indicated antibodies. Restoration of PTPRK expression significantly decreased phospho-STAT3Tyr705 levels in NKYS cells, whereas partial knockdown of PTPRK significantly increased phospho-STAT3Tyr705 levels in SNK6 cells. (Top of 2Bi) The blot was probed first with a PTPRK antibody and was then stripped and reprobed with the anti-DDK tag M2 antibody to detect the PTPRK-tag fusion protein. β-Actin was used as an internal loading control. The levels of STAT3/phospho-STAT3Tyr705 proteins were analyzed using STAT3 and phospho-STAT3Tyr705 antibodies. The entire western blot images showing precursor and mature subunits of PTPRK are presented in supplemental Figures 2A,3. (C) Flow cytometry analyses on (i) PTPRK-transduced (white) vs mock vector-transduced (black) NKYS cells, and (ii) PTPRK-specific shRNA-treated (white) vs control shRNA-treated (red) SNK6 cells using a phospho-STAT3Tyr705 antibody. (D) (i) PTPRK nonexpressing NKYS cells were infected with a retrovirus expressing PTPRK with a myc-DDK tag at the 3′ end. Double indirect immunofluorescence was performed to detect ectopic PTPRK (detected using an anti-DDK tag M2 antibody, and green fluorescence cellular membranous and cytoplasmic staining) and phospho-STAT3Tyr705 proteins (red fluorescence nuclear staining). Hoechst 33342 was used to stain the nuclei (blue fluorescence nuclear staining). The images were captured using a Carl Zeiss LSM 510 confocal microscope (original magnification ×600). (ii) PTPRK-expressing SNK6 cells were infected with lentivirus expressing PTPRK-targeting shRNA. Indirect immunofluorescence was performed to detect phospho-STAT3Tyr705 protein (green fluorescence nuclear staining). Hoechst 33342 was used to stain the nuclei. The image was captured using a Leica Q550CW microscope (original magnification ×400).
PTPRK protein expression inversely correlates with STAT3 activation. (A) Immunohistochemical staining of paraffin sections of representative NKTCL primary tumors with high (NL11) and low (NL22) PTPRK expression using a rabbit polyclonal PTPRK (H75) antibody raised against amino acids 27-101 mapping within an N-terminal extracellular domain of human PTPRK (brown cellular membranous staining between the cell-cell contact and faint cytoplasmic staining is indicated by arrows), and a phospho-STAT3Tyr705 antibody (brown nuclear staining is indicated by arrows). The immunostaining demonstrated that PTPRK protein expression inversely correlated with phospho-STAT3Tyr705 levels in NKTCL primary tumors. Original magnification ×400. (B) Western blot analyses of PTPRK and phospho-STAT3Tyr705 expression in PTPRK nonexpressing NKYS cells with reconstitution of PTPRK expression (i) and in PTPRK-expressing SNK6 cells with partial knockdown of PTPRK expression (ii), using the indicated antibodies. Restoration of PTPRK expression significantly decreased phospho-STAT3Tyr705 levels in NKYS cells, whereas partial knockdown of PTPRK significantly increased phospho-STAT3Tyr705 levels in SNK6 cells. (Top of 2Bi) The blot was probed first with a PTPRK antibody and was then stripped and reprobed with the anti-DDK tag M2 antibody to detect the PTPRK-tag fusion protein. β-Actin was used as an internal loading control. The levels of STAT3/phospho-STAT3Tyr705 proteins were analyzed using STAT3 and phospho-STAT3Tyr705 antibodies. The entire western blot images showing precursor and mature subunits of PTPRK are presented in supplemental Figures 2A,3. (C) Flow cytometry analyses on (i) PTPRK-transduced (white) vs mock vector-transduced (black) NKYS cells, and (ii) PTPRK-specific shRNA-treated (white) vs control shRNA-treated (red) SNK6 cells using a phospho-STAT3Tyr705 antibody. (D) (i) PTPRK nonexpressing NKYS cells were infected with a retrovirus expressing PTPRK with a myc-DDK tag at the 3′ end. Double indirect immunofluorescence was performed to detect ectopic PTPRK (detected using an anti-DDK tag M2 antibody, and green fluorescence cellular membranous and cytoplasmic staining) and phospho-STAT3Tyr705 proteins (red fluorescence nuclear staining). Hoechst 33342 was used to stain the nuclei (blue fluorescence nuclear staining). The images were captured using a Carl Zeiss LSM 510 confocal microscope (original magnification ×600). (ii) PTPRK-expressing SNK6 cells were infected with lentivirus expressing PTPRK-targeting shRNA. Indirect immunofluorescence was performed to detect phospho-STAT3Tyr705 protein (green fluorescence nuclear staining). Hoechst 33342 was used to stain the nuclei. The image was captured using a Leica Q550CW microscope (original magnification ×400).
PTPRK expression inversely correlates with nuclear phospho-STAT3Tyr705 in NKTCL
The immunohistochemical staining of phospho-STAT3Tyr705 (brown nuclear staining) and PTPRK (brown cellular membranous staining between the cell-cell contact and faint cytoplasmic staining) in 5 NKTCL cell lines and 27 tumors showed high levels of phospho-STAT3Tyr705 in the 4 PTPRK nonexpressing cell lines, whereas a very low level of phospho-STAT3Tyr705 was detected in PTPRK-expressing SNK6 cells (supplemental Table 4). There was also a reciprocal correlation between nuclear phospho-STAT3Tyr705 and PTPRK levels in tumor cells of the 27 NKTCL primary tumors examined (P < .040) (Figure 2A; supplemental Table 4).
Restoration of PTPRK significantly decreases nuclear phospho-STAT3Tyr705 levels, whereas knockdown of PTPRK significantly increases nuclear phospho-STAT3Tyr705 levels
Western blot and flow cytometry analysis using an anti–phospho-STAT3Tyr705 antibody showed decreased phospho-STAT3Tyr705 levels in PTPRK-transduced NKYS cells (Figure 2Bi-Ci), whereas partial knockdown of PTPRK (∼70%) significantly increased the levels of phospho-STAT3Tyr705 in SNK6 cells transduced with 2 independent PTPRK-specific shRNAs (Figure 2Bii-Cii). Each of the shRNAs (#1 and #2) independently provided efficient knockdown of PTPRK expression.
Indirect immunofluorescence experiments demonstrated lower levels of nuclear phospho-STAT3Tyr705 in NKYS cells with reconstituted PTPRK expression compared with the surrounding nontransduced cells (Figure 2Di), whereas SNK6 cells transduced with PTPRK-specific shRNA #2 showed substantially higher nuclear phospho-STAT3Tyr705 levels than the surrounding nontransduced cells (Figure 2Dii).
STAT3 is a substrate of PTPRK
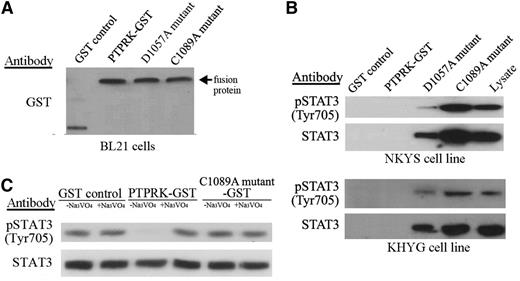
To determine whether phospho-STAT3Tyr705 dephosphorylation was directly mediated by PTPRK, a substrate-trapping assay was performed. Three glutathione S-transferase (GST) fusion proteins (wild-type [WT], mutant 1, and mutant 2) containing the first phosphatase domain of PTPRK were constructed. The mutant 1 and mutant 2 proteins contained mutations predicted to result in substrate trapping (D1057A in mutant 1 and C1089A in mutant 2). The fusion proteins were expressed in Escherichia coli, and equal amounts of each protein were attached to beads (Figure 3A). The beads were then incubated with cell lysates from NKYS and KHYG cells. The phosphorylated form of STAT3 was specifically bound by the 2 substrate-trapping mutants (the C1089A mutant coprecipitated more STAT3 and phospho-STAT3Tyr705 than did the D1057A mutant), and did not bind to the WT or control GST proteins (Figure 3B).
PTPRK binds to STAT3 and directly dephosphorylates phospho-STAT3 at Tyr705. (A) Immunoblot showing GST alone and GST fusion proteins (WT/mutant types) eluted from the indicated beads before incubation with cell lysates. An antibody against GST was used. The arrow indicates the fusion proteins. (B) Phosphatase substrate-trapping mutant assays. Cell lysates of NKYS (top) and KHYG (bottom) cells were incubated with beads bound to the indicated GST-fusion proteins. Proteins bound to the beads were resolved on a 7.5% sodium dodecyl sulfate/polyacrylamide gel electrophoresis gel, and western blots were performed with the indicated antibodies. (C) In vitro phosphatase assay. The immunoprecipitated STAT3/phospho-STAT3Tyr705 protein was incubated in vitro with purified WT and mutant GST-PTPRK in the absence or presence of a phosphatase inhibitor (Na3VO4). The levels of STAT3/phospho-STAT3Tyr705 proteins were analyzed by western blot using STAT3 and phospho-STAT3Tyr705 antibodies. The purified GST protein alone without the phosphatase domain of PTPRK was used as the control. The incubation of WT protein but not mutant 2 or the control, in the absence of Na3VO4 but not in the presence of Na3VO4, resulted in a significant decrease in phospho-STAT3Tyr705 levels. This result indicated that phospho-STAT3Tyr705 was a direct substrate of PTPRK.
PTPRK binds to STAT3 and directly dephosphorylates phospho-STAT3 at Tyr705. (A) Immunoblot showing GST alone and GST fusion proteins (WT/mutant types) eluted from the indicated beads before incubation with cell lysates. An antibody against GST was used. The arrow indicates the fusion proteins. (B) Phosphatase substrate-trapping mutant assays. Cell lysates of NKYS (top) and KHYG (bottom) cells were incubated with beads bound to the indicated GST-fusion proteins. Proteins bound to the beads were resolved on a 7.5% sodium dodecyl sulfate/polyacrylamide gel electrophoresis gel, and western blots were performed with the indicated antibodies. (C) In vitro phosphatase assay. The immunoprecipitated STAT3/phospho-STAT3Tyr705 protein was incubated in vitro with purified WT and mutant GST-PTPRK in the absence or presence of a phosphatase inhibitor (Na3VO4). The levels of STAT3/phospho-STAT3Tyr705 proteins were analyzed by western blot using STAT3 and phospho-STAT3Tyr705 antibodies. The purified GST protein alone without the phosphatase domain of PTPRK was used as the control. The incubation of WT protein but not mutant 2 or the control, in the absence of Na3VO4 but not in the presence of Na3VO4, resulted in a significant decrease in phospho-STAT3Tyr705 levels. This result indicated that phospho-STAT3Tyr705 was a direct substrate of PTPRK.
PTPRK directly dephosphorylates phospho-STAT3Tyr705
Both the phosphorylated and nonphosphorylated forms of STAT3 were immunoprecipitated from HEK293T cells and incubated as phosphatase substrates with equal amounts of WT or mutant 2 PTPRK protein, or with GST alone, with or without the phosphatase inhibitor Na3VO4. STAT3 was dephosphorylated by WT phosphatase but not the phosphatase-dead mutant 2 or GST alone (Figure 3C). Phosphatase activity was also inhibited by Na3VO4, which indicated that this process was phosphatase dependent (Figure 3C). These results indicate that phospho-STAT3Tyr705 is a direct substrate of PTPRK.
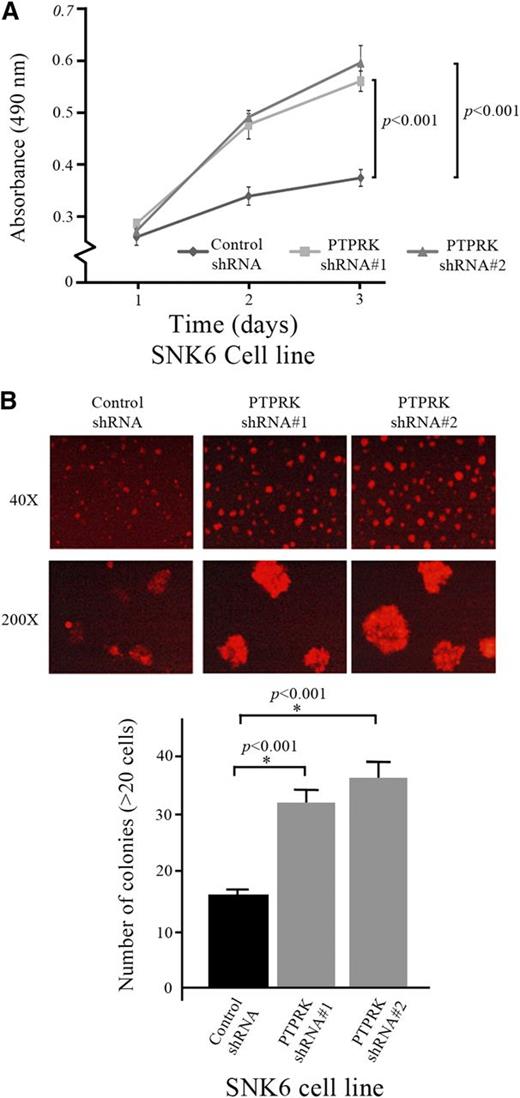
PTPRK restoration suppresses tumor cell proliferation
GFP-sorted PTPRK-transduced NKYS and KHYG cells were grown in culture medium and the restoration of PTPRK expression significantly suppressed the cellular proliferation rate (2-way analysis of variance; P < .05 for both cell lines) (Figure 4A). Moreover, when PTPRK-transduced cells were grown in methylcellulose, NKTCL cells re-expressing PTPRK formed smaller colonies and fewer colonies containing >20 cells compared with NKTCL cells transduced with the mock vector (χ2, P < .001 for NKYS, P < .005 for KHYG) (Figure 4B). These results suggested that the clonogenicity of the NKTCL cells was significantly decreased after the restoration of PTPRK expression, and these data were consistent with the tumor-suppressive properties of PTPRK. Moreover, PTPRK re-expression significantly increased the population of cells undergoing late apoptosis, as shown by Annexin-V and 7-amino-actinomycin D (7-AAD) staining, with an average increase of 20% to 40% compared with cells transduced with the mock vector (χ2, P < .001) (Figure 4C). Annexin-V staining precedes the loss of membrane integrity that later accompanies cell death, while 7-AAD can be used as a viability probe for nonviable cell exclusion methods.
Tumor suppressive properties of PTPRK. (A) Restoration of PTPRK expression suppressed the cellular proliferation rate. The graph shows the proliferation rates of GFP-sorted PTPRK-transduced, and mock vector-transduced NKYS and KHYG cells. The cell proliferation rate was determined according to the degree of absorbance in the MTS assay. The absorbance of triplicate wells was plotted against the number of days after seeding. (B) The restoration of PTPRK expression suppressed anchorage-independent growth. (Left) Anchorage-independent colony formation in GFP-sorted PTPRK-transduced, and mock vector-transduced cells in 1.2% methylcellulose. (Right) Bar charts showing the number of colonies formed in 5 randomly selected microscopic fields after 5 days. (C) The restoration of PTPRK expression increased the percentages of NKTCL cells undergoing apoptosis. (Left) Representative graphical results of the flow cytometry analyses of GFP-sorted PTPRK-transduced, and mock vector-transduced NKTCL cells stained with Cy5-conjugated Annexin-V and 7-AAD. (Right) The quantitative differences in the percentages of PTPRK-transduced and mock vector-transduced NKYS cells are shown. For (A-C), each assay was repeated 3 times. The bars indicate the mean ± SE. (D) Reconstituted PTPRK expression increased the proportion of NKYS cells undergoing G0/G1 cell-cycle arrest. (Left) Representative graphical results of GFP-sorted PTPRK-transduced, and mock vector-transduced NKYS cells in different phases of the cell cycle, as determined by flow cytometry after propidium iodide staining. The peaks corresponding to the different phases of the cell cycle are indicated on the DNA histograms. The percentages of cells in the G0/G1, S, and G2/M phases are indicated next to each graph. (Right) The quantitative differences in the percentages of PTPRK-transduced and mock vector-transduced NKYS cells in different phases of the cell cycle. (E) The restoration of PTPRK expression triggered caspase-mediated apoptosis of NKYS cells. Western blots show cleaved caspase-9, caspase-3, and poly[ADP-ribose] polymerase (PARP) in GFP-sorted PTPRK-transduced, and mock vector-transduced NKYS cells. β-Actin was used as a loading control.
Tumor suppressive properties of PTPRK. (A) Restoration of PTPRK expression suppressed the cellular proliferation rate. The graph shows the proliferation rates of GFP-sorted PTPRK-transduced, and mock vector-transduced NKYS and KHYG cells. The cell proliferation rate was determined according to the degree of absorbance in the MTS assay. The absorbance of triplicate wells was plotted against the number of days after seeding. (B) The restoration of PTPRK expression suppressed anchorage-independent growth. (Left) Anchorage-independent colony formation in GFP-sorted PTPRK-transduced, and mock vector-transduced cells in 1.2% methylcellulose. (Right) Bar charts showing the number of colonies formed in 5 randomly selected microscopic fields after 5 days. (C) The restoration of PTPRK expression increased the percentages of NKTCL cells undergoing apoptosis. (Left) Representative graphical results of the flow cytometry analyses of GFP-sorted PTPRK-transduced, and mock vector-transduced NKTCL cells stained with Cy5-conjugated Annexin-V and 7-AAD. (Right) The quantitative differences in the percentages of PTPRK-transduced and mock vector-transduced NKYS cells are shown. For (A-C), each assay was repeated 3 times. The bars indicate the mean ± SE. (D) Reconstituted PTPRK expression increased the proportion of NKYS cells undergoing G0/G1 cell-cycle arrest. (Left) Representative graphical results of GFP-sorted PTPRK-transduced, and mock vector-transduced NKYS cells in different phases of the cell cycle, as determined by flow cytometry after propidium iodide staining. The peaks corresponding to the different phases of the cell cycle are indicated on the DNA histograms. The percentages of cells in the G0/G1, S, and G2/M phases are indicated next to each graph. (Right) The quantitative differences in the percentages of PTPRK-transduced and mock vector-transduced NKYS cells in different phases of the cell cycle. (E) The restoration of PTPRK expression triggered caspase-mediated apoptosis of NKYS cells. Western blots show cleaved caspase-9, caspase-3, and poly[ADP-ribose] polymerase (PARP) in GFP-sorted PTPRK-transduced, and mock vector-transduced NKYS cells. β-Actin was used as a loading control.
PTPRK restoration triggers caspase-mediated apoptosis
We next investigated the effects of PTPRK expression on cell death and cell-cycle progression. The restoration of PTPRK expression increased the proportion of GFP-sorted PTPRK-transduced NKYS cells blocked in the G0/G1 phase (Figure 4D). Moreover, western blot analysis revealed cleavage of several members of the caspase pathway (caspase-3, caspase-9, and poly[ADP-ribose] polymerase) in PTPRK-positive NKYS cells (Figure 4E). These findings suggested that the restoration of PTPRK expression triggered caspase-mediated apoptosis in NKYS cells.
PTPRK knockdown abolishes its oncosuppressive function
Partial PTPRK knockdown (∼70%) led to a significant increase in the proliferation rate of RFP-sorted, lentiviral shRNA-transduced SNK6 cells (2-way analysis of variance, P < .001 for both shRNA sequences effective against PTPRK) (Figure 5A) and the in vitro clonogenicity of SNK6 cells (χ2, P < .001 for both shRNA sequences) (Figure 5B).
Partial PTPRK knockdown inhibits its tumor suppressive properties. (A) Knockdown of PTPRK expression significantly increased the proliferation rate of SNK6 cells. The graph shows the proliferation rates of PTPRK-specific shRNA#1- and shRNA#2-treated, and scrambled shRNA-treated SNK6 cells. The cell proliferation rate was determined as the degree of absorbance in the MTS assay. The average absorbance of triplicate wells was plotted against the number of days after seeding. (B) PTPRK expression knockdown significantly increased the anchorage-independent growth of SNK6 cells. (Top) The anchorage-independent colony formation assay was performed on PTPRK-specific shRNA#1- and shRNA#2-treated, and control shRNA-treated SNK6 cells in 0.4% methylcellulose. (Bottom) Bar chart showing the number of colonies formed in 5 randomly selected microscopic fields after 1 week. The experiments were performed in triplicate and repeated 3 times. The graph indicates the mean ± SD.
Partial PTPRK knockdown inhibits its tumor suppressive properties. (A) Knockdown of PTPRK expression significantly increased the proliferation rate of SNK6 cells. The graph shows the proliferation rates of PTPRK-specific shRNA#1- and shRNA#2-treated, and scrambled shRNA-treated SNK6 cells. The cell proliferation rate was determined as the degree of absorbance in the MTS assay. The average absorbance of triplicate wells was plotted against the number of days after seeding. (B) PTPRK expression knockdown significantly increased the anchorage-independent growth of SNK6 cells. (Top) The anchorage-independent colony formation assay was performed on PTPRK-specific shRNA#1- and shRNA#2-treated, and control shRNA-treated SNK6 cells in 0.4% methylcellulose. (Bottom) Bar chart showing the number of colonies formed in 5 randomly selected microscopic fields after 1 week. The experiments were performed in triplicate and repeated 3 times. The graph indicates the mean ± SD.
PTPRK restoration reduces tumor cell migration and invasion ability
After GFP sorting, PTPRK-transduced and mock vector-transduced NKYS cells were pelleted and resuspended in fresh culture medium; PTPRK-transduced NKYS cells rapidly formed clumps of 5 to 10 cells, whereas NKYS cells transduced with the mock vector remained primarily in a single-cell suspension (Figure 6Ai) after 6 hours. These results suggested that PTPRK restoration enhanced cell-cell contact in NKYS cells.
PTPRK inhibits the cell migration and invasion ability of NKTCL cells. (A) Images show cell-cell interactions of NKTCL cells after resuspension in fresh medium for 6 hours. (i) Clumps of 5 to 10 cells were formed by GFP-sorted PTPRK-transduced NKYS cells in culture (diffused green fluorescence over the entire cell). Mock vector-transduced NKYS cells were used as the control. (ii) Clumps of 5 to 10 cells were only formed by RFP-sorted control shRNA-transduced SNK6 cells in culture (diffused red fluorescence over the entire cell), but not cells transduced with PTPRK-specific shRNA#1 and shRNA#2. Original magnification ×400. (B) Bar charts show the migration and invasion rate of NKTCL cells according to the number of sorted cells that passed through transwell inserts containing 8-µm pore polyethylene terephthalate membranes and the number of cells that passed through inserts covered with diluted Matrigel basement membrane, respectively. The cells that migrated to the lower chambers were counted in 5 randomly selected microscopic fields (200×). Each assay was repeated 3 times. The bars indicate the mean ± SE. Re-expression of PTPRK significantly inhibited the cell migratory and invasion abilities of non–PTPRK-expressing NKYS cells (i), whereas knockdown of PTPRK enhanced SNK6 cell migration and invasion (ii).
PTPRK inhibits the cell migration and invasion ability of NKTCL cells. (A) Images show cell-cell interactions of NKTCL cells after resuspension in fresh medium for 6 hours. (i) Clumps of 5 to 10 cells were formed by GFP-sorted PTPRK-transduced NKYS cells in culture (diffused green fluorescence over the entire cell). Mock vector-transduced NKYS cells were used as the control. (ii) Clumps of 5 to 10 cells were only formed by RFP-sorted control shRNA-transduced SNK6 cells in culture (diffused red fluorescence over the entire cell), but not cells transduced with PTPRK-specific shRNA#1 and shRNA#2. Original magnification ×400. (B) Bar charts show the migration and invasion rate of NKTCL cells according to the number of sorted cells that passed through transwell inserts containing 8-µm pore polyethylene terephthalate membranes and the number of cells that passed through inserts covered with diluted Matrigel basement membrane, respectively. The cells that migrated to the lower chambers were counted in 5 randomly selected microscopic fields (200×). Each assay was repeated 3 times. The bars indicate the mean ± SE. Re-expression of PTPRK significantly inhibited the cell migratory and invasion abilities of non–PTPRK-expressing NKYS cells (i), whereas knockdown of PTPRK enhanced SNK6 cell migration and invasion (ii).
PTPRK knockdown was accomplished by lentiviral shRNA in SNK6 cells, and following RFP-sorting, lentiviral shRNA-transduced SNK6 cells were centrifuged and resuspended in fresh culture medium; the cells transduced with negative control shRNA formed clumps of >20 cells, whereas SNK6 cells transduced with shRNA targeting PTPRK remained primarily in a single-cell suspension (Figure 6Aii) after 6 hours. These results indicated a loss of homophilic interactions triggered by the receptor PTPRK after PTPRK knockdown in SNK6 cells.
Matrigel cell migration and invasion assays demonstrated that PTPRK restoration significantly reduced the rate of migration and invasion of GFP-sorted PTPRK-transduced NKYS cells (χ2, P < .001 for migration, P < .005 for invasion) (Figure 6Bii), whereas the partial knockdown of PTPRK in RFP-sorted, lentiviral shRNA-transduced SNK6 cells increased the migration and invasion rate (χ2, P < .001 for both the migration and invasion assays) (Figure 6Bii).
Demethylation with 5-aza-2’-deoxycytidine (5-aza-dC) restores PTPRK mRNA expression in non–PTPRK-expressing NKTCL cell lines
To explore whether PTPRK is epigenetically silenced in NKTCL, we first treated 5 NKTCL cell lines with pharmacologic concentrations of the DNA demethylating agent 5-aza-dC for 3 or 6 days. Semiquantitative RT-PCR analysis revealed that PTPRK mRNA expression was restored in all 4 non–PTPRK-expressing cell lines following 5-aza-dC treatment (Figure 7A). Consistent with the RT-PCR results, immunofluorescence of a non–PTPRK-expressing NKYS cell line also revealed the re-expression of PTPRK at the protein level, following 5-aza-dC treatment (Figure 7B). This 5-aza-dC–induced re-expression indicated that PTPRK is downregulated by promoter hypermethylation in NKTCL.
Underexpression of PTPRK mRNA is due to promoter hypermethylation and monoallelic deletions in NKTCL. (A) Gel images showing the PTPRK mRNA expression levels detected by semiquantitative RT-PCR in 5 NKTCL cell lines after treatment with 5-aza-dC for 3 or 6 days. Pharmacologic demethylation with 5-aza-dC restored PTPRK mRNA expression in all 4 non–PTPRK-expressing NKTCL cell lines. The mRNA expression in each sample was normalized to β-actin expression. (B) Immunofluorescence image of a non–PTPRK-expressing NKYS cell line showing the re-expression of PTPRK protein after treatment with the 5-aza-dC demethylating agent; an antibody to the N-terminus of PTPRK was used (arrows indicate green fluorescent cellular membranous and cytoplasmic staining). Original magnification ×1000. (C) (Top) Gel image revealing the methylation status of the PTPRK promoter, as determined by MSP in 2 preparations of normal NK cells and 5 NKTCL cell lines. The PTPRK promoter was unmethylated in PTPRK-expressing normal NK cells and NK malignant cell lines, whereas PTPRK was methylated in non–PTPRK-expressing NKTCL cell lines. (Bottom) Detailed methylation analysis of the CpG sites in the PTPRK promoter was performed via BGS in normal NK cells, untreated NKTCL cells, and 5-aza-dC–treated malignant cells. BGS confirmed the PTPRK promoter methylation status determined by MSP. The amount that each circle is filled represents the percentage of methylated cytosines detected via BGS from 8 to 10 sequenced colonies. (D) The PTPRK promoter was frequently methylated in NKTCL primary tumors. (Top) The composite gel images show the PTPRK promoter methylation status as determined by MSP in 27 NKTCL primary tumors. BGS confirmed the PTPRK promoter methylation status determined by MSP (bottom). The CpG sites of each representative case are presented in the row as individual circles. (E) Gel image of PTPRK gene allelic loss analysis in 6 representative cases of primary NKTCL tumors as determined by semiquantitative multiplex PCR (30 cycles) using the β-globin gene as normal allele control. Samples with a PTPRK/β-globin allelic ratio of < 0.75, as determined by densitometry, were considered to have a monoallelic PTPRK gene deletion (case #14, case #15, and case #16). (F) Methylation of the PTPRK promoter significantly correlated with inferior OS in NKTCL patients treated with the SMILE protocol. Kaplan-Meier curves indicating DFS (left panel) and OS (right panel) distributions according to methylation status of the PTPRK promoter in NKTCL patients. Both Kaplan-Meier curves were compared using the log-rank, Breslow, and Tarone-Ware tests, and the P values are presented alongside the survival plots. The results revealed that methylation of the PTPRK promoter was significantly correlated with inferior DFS and OS in NKTCL patients treated with the SMILE regimen. The treatment outcome data were available for the 17 NKTCL patients treated with the SMILE regimen who were included in this study. M, methylated; MW, molecular weight markers; U, unmethylated; UMD, universal methylated DNA; UUD, universal unmethylated DNA.
Underexpression of PTPRK mRNA is due to promoter hypermethylation and monoallelic deletions in NKTCL. (A) Gel images showing the PTPRK mRNA expression levels detected by semiquantitative RT-PCR in 5 NKTCL cell lines after treatment with 5-aza-dC for 3 or 6 days. Pharmacologic demethylation with 5-aza-dC restored PTPRK mRNA expression in all 4 non–PTPRK-expressing NKTCL cell lines. The mRNA expression in each sample was normalized to β-actin expression. (B) Immunofluorescence image of a non–PTPRK-expressing NKYS cell line showing the re-expression of PTPRK protein after treatment with the 5-aza-dC demethylating agent; an antibody to the N-terminus of PTPRK was used (arrows indicate green fluorescent cellular membranous and cytoplasmic staining). Original magnification ×1000. (C) (Top) Gel image revealing the methylation status of the PTPRK promoter, as determined by MSP in 2 preparations of normal NK cells and 5 NKTCL cell lines. The PTPRK promoter was unmethylated in PTPRK-expressing normal NK cells and NK malignant cell lines, whereas PTPRK was methylated in non–PTPRK-expressing NKTCL cell lines. (Bottom) Detailed methylation analysis of the CpG sites in the PTPRK promoter was performed via BGS in normal NK cells, untreated NKTCL cells, and 5-aza-dC–treated malignant cells. BGS confirmed the PTPRK promoter methylation status determined by MSP. The amount that each circle is filled represents the percentage of methylated cytosines detected via BGS from 8 to 10 sequenced colonies. (D) The PTPRK promoter was frequently methylated in NKTCL primary tumors. (Top) The composite gel images show the PTPRK promoter methylation status as determined by MSP in 27 NKTCL primary tumors. BGS confirmed the PTPRK promoter methylation status determined by MSP (bottom). The CpG sites of each representative case are presented in the row as individual circles. (E) Gel image of PTPRK gene allelic loss analysis in 6 representative cases of primary NKTCL tumors as determined by semiquantitative multiplex PCR (30 cycles) using the β-globin gene as normal allele control. Samples with a PTPRK/β-globin allelic ratio of < 0.75, as determined by densitometry, were considered to have a monoallelic PTPRK gene deletion (case #14, case #15, and case #16). (F) Methylation of the PTPRK promoter significantly correlated with inferior OS in NKTCL patients treated with the SMILE protocol. Kaplan-Meier curves indicating DFS (left panel) and OS (right panel) distributions according to methylation status of the PTPRK promoter in NKTCL patients. Both Kaplan-Meier curves were compared using the log-rank, Breslow, and Tarone-Ware tests, and the P values are presented alongside the survival plots. The results revealed that methylation of the PTPRK promoter was significantly correlated with inferior DFS and OS in NKTCL patients treated with the SMILE regimen. The treatment outcome data were available for the 17 NKTCL patients treated with the SMILE regimen who were included in this study. M, methylated; MW, molecular weight markers; U, unmethylated; UMD, universal methylated DNA; UUD, universal unmethylated DNA.
PTPRK mRNA is frequently downregulated by promoter methylation in NKTCL
The PTPRK promoter was unmethylated in normal NK cells and the PTPRK-expressing SNK6 cell line, whereas the PTPRK promoter was methylated in 4 non–PTPRK-expressing NKTCL cell lines (Figure 7C, top panel). In contrast to the normal NK cells and the PTPRK-expressing SNK6 cell line, the 4 non–PTPRK-expressing cell lines exhibited almost complete methylation of the entire CpG island region of PTPRK (Figure 7C, bottom panel). The CpG sites containing Sp1 (2 CpG sites; #8 and #9), heat shock factor (1 CpG site; #10), and CREB (3 CpG sites; #32, #33, and #34) were nearly completely methylated in the 4 cell lines, whereas these same sites were unmethylated or only partially methylated in normal samples and the SNK6 cell line. Moreover, reduced levels or complete loss of PTPRK mRNA expression significantly correlated with PTPRK promoter hypermethylation in the NKTCL cell lines (P = .025) (supplemental Table 5). Furthermore, treatment of the NKTCL cell lines with 5-aza-dC caused demethylation of the CpG sites in the PTPRK promoter and activated PTPRK mRNA expression (Figure 7C, bottom panel).
Next, MSP and BGS assays were performed on 27 NKTCL primary tumors and a methylated PTPRK promoter was detected in 16 of 27 tumors (59%) (Figure 7D; supplemental Table 6A). An unmethylated band was present in nearly all the tumors and was most likely due to the presence of tumor-infiltrating nontumor cells. Reduced levels or complete loss of PTPRK mRNA expression was also significantly correlated with PTPRK promoter hypermethylation in NKTCL primary tumors (P = .001) (supplemental Table 6B).
PTPRK gene monoallelic deletions are detected in a fraction of NKTCL primary tumors
PTPRK allelic deletion analysis was performed on genomic DNA from NKTCL primary tumors by semiquantitative multiplex PCR using PTPRK and β-globin (as the normal allele control) gene-specific primers (Figure 7E). Overall, monoallelic PTPRK gene deletions were detected in 8 of 27 (30%) tumors, which included 2 of 11 (18%) tumors with unmethylated PTPRK promoters and 6 of 16 (38%) tumors with methylated PTPRK promoters (supplemental Table 6A). Monoallelic PTPRK gene deletions were also detected in 3 of 5 NKTCL cell lines studied, which is consistent with the deletion of 6q23.2 as previously reported in these cell lines.17
Mutational inactivation of PTPRK is uncommon in NKTCL
The mutational analysis of PTPRK via bidirectional DNA sequencing of the entire coding sequence of PTPRK transcripts only detected 1 silent mutation (coding sequence 583, GTT>GTC) in the SNK6 cell line. No inactivating deletions, insertions, or mutations were detected in the PTPRK mRNA obtained from NKTCL primary tumors.
Underexpression of PTPRK protein in tumor cells correlates with advanced-stage disease in NKTCL patients
A retrospective correlation of tumor cell PTPRK protein levels with the clinical features of 27 NKTCL patients showed that low PTPRK levels in tumor cells were significantly correlated with a higher number of extranodal sites (1 vs >1, Pearson’s χ2-test, P = .013), (supplemental Table 7) and were associated with a high international prognostic index (IPI) (0-1 vs 2, 3, or 4-5, Pearson’s χ2-test, P = .040) (supplemental Table 7).
Tumor cell PTPRK promoter hypermethylation correlates with an unfavorable prognosis in NKTCL patients
Next, a retrospective correlation of the methylation status of tumor cell PTPRK promoter DNA was performed against the clinical features of the 27 NKTCL patients (Table 1). The univariate statistical correlation revealed that PTPRK promoter hypermethylation in tumors was highly significantly correlated with advanced-stage disease (P = .006) and with greater extranodal involvement in patients (P = .006). Moreover, patients harboring tumors with methylated PTPRK promoters exhibited a significantly higher IPI (P < .001).
Correlation of tumor cell methylation status of the PTPRK promoter with clinical features in 27 NKTCL patients
| . | PTPRK promoter . | . | |
|---|---|---|---|
| Methylated . | Unmethylated . | Pearson’s χ2-test (P) . | |
| Gender | |||
| Male | 8 | 8 | .238 |
| Female | 8 | 3 | |
| Age | |||
| ≤60 y | 11 | 8 | .824 |
| >60 y | 5 | 3 | |
| Stage | |||
| 1, 2 | 6 | 10 | .006 |
| 3, 4 | 10 | 1 | |
| Elevated serum LDH levels | |||
| No | 6 | 8 | .072 |
| Yes | 10 | 3 | |
| ECOG score | |||
| 0, 1 | 15 | 11 | .398 |
| 2, 3, or 4 | 1 | 0 | |
| Number of extranodal sites of disease | |||
| 1 | 6 | 10 | .006 |
| >1 | 10 | 1 | |
| IPI | |||
| 0-1 | 2 | 10 | <.001 |
| 2, 3, or 4-5 | 14 | 1 | |
| . | PTPRK promoter . | . | |
|---|---|---|---|
| Methylated . | Unmethylated . | Pearson’s χ2-test (P) . | |
| Gender | |||
| Male | 8 | 8 | .238 |
| Female | 8 | 3 | |
| Age | |||
| ≤60 y | 11 | 8 | .824 |
| >60 y | 5 | 3 | |
| Stage | |||
| 1, 2 | 6 | 10 | .006 |
| 3, 4 | 10 | 1 | |
| Elevated serum LDH levels | |||
| No | 6 | 8 | .072 |
| Yes | 10 | 3 | |
| ECOG score | |||
| 0, 1 | 15 | 11 | .398 |
| 2, 3, or 4 | 1 | 0 | |
| Number of extranodal sites of disease | |||
| 1 | 6 | 10 | .006 |
| >1 | 10 | 1 | |
| IPI | |||
| 0-1 | 2 | 10 | <.001 |
| 2, 3, or 4-5 | 14 | 1 | |
ECOG, Eastern Cooperative Oncology Group; LDH, lactate dehydrogenase.
Significantly, in 17 NKTCL patients treated with the SMILE protocol, an univariate Kaplan-Meier analysis revealed that patients with a tumor containing methylated PTPRK promoter exhibited a trend toward inferior disease-free survival (DFS) (log-rank P = .052, Breslow P = .024, Tarone-Ware P = .034; Figure 7F) and a significant correlation with overall survival (OS) (log-rank P = .049, Breslow P = .040, Tarone-Ware P = .044; Figure 7F).
Discussion
One of the main characteristics of NKTCL is the constitutive activation of STAT3.15,19,20 Tyrosine phosphorylation at Tyr705 is required for STAT3 to bind to specific DNA target sites, and it was recently reported that STAT3 activation results from constitutive JAK3 phosphorylation at Tyr980 in NKTCL.22,23 JAK3 was found to be constitutively phosphorylated in 87% of NKTCL primary tumors, and in 21% of these cases, this was the result of activating mutations of JAK3.22,23 It is likely that multiple mechanisms contribute to this constitutive STAT3 activation in NKTCL, as the net level of tyrosine phosphorylation of STAT3 at Tyr705 reflects a balance between the competing activities of protein kinases and phosphatases. In the present study, we describe a novel molecular mechanism leading to the observed STAT3 deregulation in NKTCL.
Importantly, we found a strong in vivo correlation between low PTPRK expression and high nuclear phospho-STAT3Tyr705 levels in NKTCL primary tumors. Moreover, our findings from phosphatase substrate-trapping mutant assays, direct phosphatase assays, and in vitro functional studies show that PTPRK binds to STAT3 and directly dephosphorylates phospho-STAT3 at Tyr705, and regulates the levels of phospho-STAT3Tyr705 in NKTCL. The loss of PTPRK expression (a negative regulator of tyrosine kinases), due to monoallelic deletions and transcriptional silencing through aberrant promoter hypermethylation, likely contributes to lymphomagenesis by activating the STAT3 oncoprotein in NKTCL cells. Previously, phospho-STAT3Tyr705 was identified as a direct substrate of 2 other R-PTPases, protein tyrosine phosphatase, receptor type T36 and protein tyrosine phosphatase, receptor type D.37 Indeed, increased phospho-STAT3TYR705 transcriptional activities due to the suppression/mutation of R-PTPases have been documented in several cancers.24,25 Together with our finding showing that phospho-STAT3Tyr705 is a true substrate of PTPRK, PTPases could be considered wardens of STAT3 signaling.38
The loss of R-PTPases was previously implicated in tumor development and progression.39,40 In our study, the restoration of PTPRK in NKTCL cells in vitro suppressed tumor cell growth and triggered caspase-mediated apoptosis, whereas the partial knockdown of PTPRK increased tumor cell growth in NKTCL cells. Furthermore, because the nonenzymatic N-terminal extracellular domain of R-PTPases provides a link between cell-cell contact and cellular signaling events involving tyrosine phosphorylation,31,32 the loss of PTPRK expression in NKTCL cells also resulted in tumor growth and migration. In NKTCL patients, we found a clinical correlation between low tumor cell PTPRK levels and high IPI, suggesting that low PTPRK expression may lead to the dissemination of lymphoma cells to extranasal sites and a possible contribution of PTPRK loss to the NKTCL aggressiveness that is associated with PTPRK non-enzymatic cell-cell contact functions. Similarly, knockdown of PTPRK was recently shown to result in increased proliferation, adhesion, invasion, and migration of breast cancer cells in vitro.41 A decrease in cell proliferation and colony formation rate was also reported in Raji (Burkitt lymphoma B-cell line) and Jurkat (T-cell leukemia cell line) cells transduced with PTPRK.42 Further evidence for the role of PTPRK in lymphomagenesis comes from previous studies of primary central nervous system lymphomas (CNSLs) with a 140-kb deletion at 6q22-q23 (which contains the PTPRK gene), as well as the observation that a loss of PTPRK expression is common in CNSLs.43 Moreover, there is a tendency for a shorter survival time in CNSL patients with tumors that lack PTPRK expression compared with patients whose tumors maintain PTPRK expression.43 Recently, decreased levels of PTPRK transcript have also been associated with a poor prognosis in breast cancer.41
PTPRK loss has also been described in other lymphoid and nonlymphoid tumors. For example, PTPRK was reported to be downregulated by EBV nuclear antigen 1 in EBV-associated Hodgkin lymphoma,44 and PTPRK loss secondary to inactivating mutations was found to contribute to glioma pathogenesis.45 However, our results showed that PTPRK inactivation by somatic mutations is not a major mechanism in NKTCL. Recently, aberrant hypermethylation of the PTPRK gene promoter was described in 47% of primary acute lymphoblastic leukemia cases,42 and our results suggest that transcriptional silencing through aberrant promoter hypermethylation exacerbates the consequences of allelic loss of the PTPRK gene in NKTCL.
A significant impact of PTPRK promoter hypermethylation on OS was described in acute lymphoblastic leukemia patients treated with the hyper-CVAD protocol.42 Traditional chemotherapeutic regimens typically result in overall response rates of only 40% to 50% for newly diagnosed NKTCL patients and are typically ineffective for relapsed cases.33 A novel protocol, SMILE, was recently developed and has resulted in overall response rates of ∼ 80%.33-35 Because prognostic indicators change when novel treatment strategies are used, our results showed that methylated PTPRK significantly correlated with advanced-stage disease, as well as unfavorable prognosis in NKTCL patients treated with the SMILE regimen. However, the clinical impact of these findings should be considered as very preliminary, as the analysis included only a small sample of 17 NKTCL patients treated with SMILE; these results therefore require future validation in a separate, larger cohort of NKTCL patients treated with SMILE to further verify the prognostic value of our findings.
In addition to the direct dephosphorylation of STAT3 at Tyr705, PTPRK may target STAT3 activation at several levels to achieve tumor suppression in NKTCL. For instance, based on our preliminary in vitro PTPRK restoration and knockdown experiments (supplemental Figure 5), we speculate that PTPRK may also dephosphorylate phospho-JAK3 at Tyr980 and that loss of PTPRK expression in NKTCL potentially provides an additional mechanism leading to constitutive JAK3 phosphorylation at Tyr980 in NKTCL, particularly in cases without JAK3 activating mutations.22,23
PTPRK has also been linked to the downregulation of other signaling pathways. It was recently reported that Raji cells transduced with PTPRK showed decreased levels of phospho-Erk1/2 (T202/Y204), phospho-Akt (S473), phospho-STAT3 (S727), and phospho-STAT5 (Y694),42 whereas increased levels of tyrosine-phosphorylated JNK were reported in PTPRK-knockdown prostate cancer cells.46 Although these studies implied that PTPRK regulates the phosphorylation of these proteins, the authors did not investigate which one of these dephosphorylation of proteins was directly mediated by PTPRK. Further studies are required to identify and characterize other substrates whose dephosphorylation is directly mediated by PTPRK, rather than through the phosphatases downstream of PTPRK, to assess the potential involvement of additional signaling pathways that may act in conjunction with STAT3 in the molecular pathogenesis of NKTCL. Indeed, a recent study identified PTPRK as a novel binding partner of CD133, which has the ability to directly dephosphorylate tyrosine phosphorylation of CD133 in colon cancer cells.47
NKTCL is an aggressive disease and the treatment outcome is suboptimal, especially for advanced stage and relapsed/refractory cases.33 As results from this study demonstrate that methylation of the PTPRK promoter can be reversed by demethylating agent 5-aza-dC, PTPRK-mediated cell signaling may be potentially targeted with epigenetic therapy in NKTCL to develop more effective therapeutic strategies. The re-expression of PTPRK and its correlations with phospho-STAT3Tyr705 levels as a biomarker could be assessed and correlated with treatment outcome in NKTCL patients treated with DNA hypomethylating agents. Hypomethylating agents are currently used in the treatment of patients with advanced myelodysplastic syndromes.48
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Junjiro Tsuchiyama for providing the NKYS cell line49 for this study. The authors also thank Dr Wing-Yan Au, Dr Chor-Sang Chim, Dr Anskar Y.H. Leung, and other members of the Division of Hematology, Department of Medicine, Queen Mary Hospital, Hong Kong, who participated in diagnosing and managing the NKTCL patients included in this study, and Silas S. Brown for his careful reading of the manuscript.
This study was supported by grants from the Research Grants Council of Hong Kong (GRF; HKU/775607M) and the University of Hong Kong (SFPBR; 200511159119).
Authorship
Contribution: Y.-W.C., T.G., and L.S. designed and performed the experiments and analyzed data; K.-Y.W., M.L.Y.W., Y.-P.C., and J.C.O.T. performed the experiments; E.T. and Y.-L.K. selected the NKTCL cases and provided data on the patients’ clinical features and treatment outcomes; F.L. selected the tumor blocks; W.P.L. and G.-D.L. selected the tumor blocks and contributed vital reagents; W.W.L.C. and R.K.H.A.-Y. analyzed the immunohistochemistry slides; N.S. contributed vital reagents; Q.T. offered valuable advice throughout the project; Y.-W.C. and T.G. wrote the manuscript; G.S. conceived the idea, designed and supervised the study, and finalized the manuscript; and all authors commented on and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gopesh Srivastava, Department of Pathology, The University of Hong Kong, Queen Mary Hospital Compound, 102 Pokfulam Rd, Hong Kong; e-mail: gopesh@pathology.hku.hk.
References
Author notes
Y.-W.C. and T.G. contributed equally to this study.


![Figure 4. Tumor suppressive properties of PTPRK. (A) Restoration of PTPRK expression suppressed the cellular proliferation rate. The graph shows the proliferation rates of GFP-sorted PTPRK-transduced, and mock vector-transduced NKYS and KHYG cells. The cell proliferation rate was determined according to the degree of absorbance in the MTS assay. The absorbance of triplicate wells was plotted against the number of days after seeding. (B) The restoration of PTPRK expression suppressed anchorage-independent growth. (Left) Anchorage-independent colony formation in GFP-sorted PTPRK-transduced, and mock vector-transduced cells in 1.2% methylcellulose. (Right) Bar charts showing the number of colonies formed in 5 randomly selected microscopic fields after 5 days. (C) The restoration of PTPRK expression increased the percentages of NKTCL cells undergoing apoptosis. (Left) Representative graphical results of the flow cytometry analyses of GFP-sorted PTPRK-transduced, and mock vector-transduced NKTCL cells stained with Cy5-conjugated Annexin-V and 7-AAD. (Right) The quantitative differences in the percentages of PTPRK-transduced and mock vector-transduced NKYS cells are shown. For (A-C), each assay was repeated 3 times. The bars indicate the mean ± SE. (D) Reconstituted PTPRK expression increased the proportion of NKYS cells undergoing G0/G1 cell-cycle arrest. (Left) Representative graphical results of GFP-sorted PTPRK-transduced, and mock vector-transduced NKYS cells in different phases of the cell cycle, as determined by flow cytometry after propidium iodide staining. The peaks corresponding to the different phases of the cell cycle are indicated on the DNA histograms. The percentages of cells in the G0/G1, S, and G2/M phases are indicated next to each graph. (Right) The quantitative differences in the percentages of PTPRK-transduced and mock vector-transduced NKYS cells in different phases of the cell cycle. (E) The restoration of PTPRK expression triggered caspase-mediated apoptosis of NKYS cells. Western blots show cleaved caspase-9, caspase-3, and poly[ADP-ribose] polymerase (PARP) in GFP-sorted PTPRK-transduced, and mock vector-transduced NKYS cells. β-Actin was used as a loading control.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/10/10.1182_blood-2014-07-588970/4/m_1589f4.jpeg?Expires=1769101661&Signature=NrDIEmIgx7YuMSzx5cg63~WdnJhVg797OdPpS-9jbIJZCN9kdsgDqu4IqrA7p-NB73FrX4nYSWx~zohkgmC1fZ8tcLYHWmZYoaQER6NfVvP2853Pqitj85XkuJrtbc7nlewATwyJg~TR9kqHOmUCCJQfAh2v0mQsYMyPIbG8p9sybf3sZNY8ktR7DsXJmulVvM3zd1fCTqdWVTxCWSmyPCAG~S~StBhIOKeRw7I1K27x5-~BdmcE9Fk4yzbb8CqBjKH1nZmz3kkK48LXqLIx0PRkgH2kNMT8Vwhp8l5vIcXWfwbumlqsSYRHfuUIkunAs2CTfFMFtCOt2j~mvZVaCA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal