Key Points
SIRT1 is highly expressed in subsets of patients with acute myeloid leukemia harboring activating mutations in signaling pathways and is regulated at the protein levels.
Targeting SIRT1 sensitizes leukemic blast to tyrosine kinase inhibitor treatment or chemotherapy via restoration of p53 activity.
Abstract
SIRT1 is an important regulator of cellular stress response and genomic integrity. Its role in tumorigenesis is controversial. Whereas sirtuin 1 (SIRT1) can act as a tumor suppressor in some solid tumors, increased expression has been demonstrated in many cancers, including hematologic malignancies. In chronic myeloid leukemia, SIRT1 promoted leukemia development, and targeting SIRT1 sensitized chronic myeloid leukemia progenitors to tyrosine kinase inhibitor treatment. In this study, we investigated the role of SIRT1 in acute myeloid leukemia (AML). We show that SIRT1 protein, but not RNA levels, is overexpressed in AML samples harboring activating mutations in signaling pathways. In FMS-like tyrosine kinase 3-internal tandem duplication (FLT3-ITD)+-cells protein, expression of SIRT1 is regulated by FLT3 kinase activity. In addition, SIRT1 function is modulated via the ATM-DBC1-SIRT1 axis in a FLT3-ITD-dependent manner. In murine leukemia models driven by MLL-AF9 or AML1-ETO coexpressing FLT3-ITD, SIRT1 acts as a safeguard to counteract oncogene-induced stress, and leukemic blasts become dependent on SIRT1 activity. Pharmacologic targeting or RNAi-mediated knockdown of SIRT1 inhibited cell growth and sensitized AML cells to tyrosine kinase inhibitor treatment and chemotherapy. This effect was a result of the restoration of p53 activity. Our data suggest that targeting SIRT1 represents an attractive therapeutic strategy to overcome primary resistance in defined subsets of patients with AML.
Introduction
Sirtuins, also referred to as class III histone deacetylases, are NAD+-dependent protein deacetylases. The founding member, silent information regulator 2 (Sir2), was originally identified in Saccharomyces cerevisiae and has been linked to longevity.1 Mammalian sirtuin 1 (SIRT1) plays an important role in the regulation of gene expression and the maintenance of genomic integrity. For example, SIRT1 deacetylates histones H4K16Ac and H3K9Ac and promotes H3K9 trimethylation, resulting in the formation of heterochromatin and gene silencing under conditions of oxidative stress.2,3 Further, SIRT1 contributes to the regulation of DNA damage response (DDR) and repair. Upon an ataxia teleangiectasia mutated (ATM)-dependent recruitment to double-strand breaks, SIRT1 induces epigenetic changes, participates in chromatin remodeling, and directly modulates several nonhistone proteins involved in DDR.4-7 In addition, SIRT1 plays an important role in integrating and coordinating cellular stress response. Sirt1 deacetylates p53 at lysine 382 and reduces its transcriptional activity, followed by the loss of p53-dependent apoptosis in response to cell damage.8,9 Further, SIRT1 prevents stress-mediated induction of apoptosis via deacetylation of FOXO3 or KU70.10,11 Altogether, these data indicate that SIRT1 promotes cell survival under conditions of stress via inhibition of apoptosis and maintenance of genomic integrity. Therefore, SIRT1 is thought to act as a tumor suppressor protein in normal cells. Indeed, depletion of Sirt1 in murine embryonic stem cells resulted in an increase in spontaneous chromosomal abnormalities caused by compromised DDR, and haploinsufficiency of Sirt1 facilitated tumorigenesis in p53+/− mice.4,12 Vice versa, conditional overexpression of Sirt1 suppressed intestinal tumor formation in Ag-presenting cell (APC)−/+ mice and reduced the frequency of fatal tumor development in p53+/− mice on γ irradiation.4,13
Contradictorily, there is rising evidence that SIRT1 can also contribute to tumorigenesis. For example, SIRT1 is overexpressed in various cancer types,14 and negative regulators of SIRT1, such as hypermethylated in cancer 1 (HIC1), miRNA-34Z and deleted in breast cancer 1 (DBC1), are often inactivated or lost.15-18 DBC1 directly binds to the catalytic domain of SIRT1 and inhibits p53 deacetylation, followed by apoptosis.17,18 On genotoxic stress, DBC1 is activated via ATM- or ATM- and Rad3-related protein (ATR)-mediated phosphorylation at threonine 454.19,20 In hematologic malignancies, SIRT1 has been found to be overexpressed in T-cell acute lymphoblastic leukemia,21 chronic lymphocytic leukemia,22 diffuse large B-cell lymphoma,23 and chronic myeloid leukemia (CML).24 Pharmacologic inhibition or short hairpin RNA (shRNA)-mediated knockdown of SIRT1 induced apoptosis in T-cell acute lymphoblastic leukemia and CML cells.21,25 Depletion of SIRT1 suppressed BCR-ABL-mediated transformation in bone marrow (BM) transplantation assays and increased the sensitivity to tyrosine kinase inhibitors (TKIs) in vitro and in vivo. Li et al demonstrated that overexpression of SIRT1 in primary human CD34+-CML cells conferred survival and resistance to TKI therapy in a p53-dependent manner, and targeting of SIRT1 eradicated leukemic stem cells.24 These data indicate that overexpression of SIRT1 may be important to overcome BCR-ABL-mediated oncogenic stress during the process of malignant transformation and that leukemic stem cells become addicted to SIRT1 function. The role of SIRT1 in acute myeloid leukemia (AML) is less clear. SIRT1 has been shown to be consistently overexpressed in primary AML samples26 ; however, the functional consequence as well as the association with oncogenic kinases or RAS-guanosine triphosphatases has not been investigated. In this study, we analyzed expression and regulation of SIRT1 in several distinct AML models and investigated the functional role of SIRT1 in the context of defined genetic backgrounds.
Materials and methods
Cell lines and reagents
Cell lines were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen or American Type Culture Collection. Molm-14 cells were kindly provided by the Scott A. Armstrong laboratory, Memorial Sloan Kettering Center, NY. Cells were cultured in fully supplemented RPMI1640 media (Gibco) and kept at 37°C in a humidified atmosphere of 5% CO2. The interleukin 3-dependent murine cell lines 32Dcl3 and BA/F3 were cultured in 10% WEHI-conditioned, fully supplemented RPMI1640 medium. BA/F3 cells expressing different oncogenes were kindly provided by the D. Gary Gilliland laboratory (previous location, Howard Hughes Medical Institute, Boston, MA). FMS-like tyrosine kinase 3-internal tandem duplication (FLT3-ITD)-expressing 32Dcl3 and M-07e cells were generated as previously described.27 PKC412 (midostaurin) was provided by Novartis Pharmaceuticals. Tenovin-6 (TV-6; Cayman Biopharmaceuticals), EX527 (Seleckchem), and protein kinase B (AKT) Inhibitor VIII and PD98059 (Calbiochem) were purchased.
Patient samples
Heparin-treated BM and peripheral blood samples were obtained from patients with AML treated at the University Medical Center of Mainz, Germany. Informed consent was obtained in accordance with the Declaration of Helsinki, and laboratory experiments were performed with approval from the local ethics committee institutional review boards. Mononuclear cells (MNCs) enriched in AML blasts were isolated by means of Ficoll-Hypaque (Seromed). For cell death assays, MNCs were maintained in Iscove modified Dulbecco medium (Sigma-Aldrich) supplemented with interleukin 3 (10 ng/mL), stem cell factor (50 ng/mL), interleukin 6 (50 ng/mL), and FLT3-L (50 ng/mL). Normal CD34pos hematopoietic progenitor cells were isolated from cryopreserved leukapheresis products. CD34pos cells were enriched by positive selection, using immunomagnetic microbeads (Miltenyi Biotec), according to the manufacturer´s protocol.
Animals
Four- to 6-week-old C57BL/6J mice were obtained from the local animal facility. Flt3ITD/ITD knock-in mice were generated as previously described.28 Conditional AML1-ETO-knock-in mice (kindly provided by James R. Downing, St. Jude Children's Research Hospital, Memphis, TN.) were crossed to C57BL/6J Mx1-Cre mice and injected with polyinosinic-polycytidylic acid to induce AML1-ETO expression. Genotyping and verification of Cre-mediated recombination were performed as described.29 All animal studies were conducted in compliance with institutional guidelines and were approved by regulatory authorities.
Retroviral transduction and BM transplantation
Generation of retroviral supernatants, transduction procedures, and BM transplantations were performed as previously described.30
Gene knockdown experiments
shRNAs targeting SIRT1 and STAT5 were cloned into Tet-pLKO.1-puro vectors (kindly provided by Dimitri Wiederschain, Novartis Institutes for BioMedical Research, Cambridge, MA). Tet-on-inducible pTRIPZ (clone V3THS_314004 targeting KRAS, RHS4743 expressing scrambled-shRNA) and pLKO.1 vectors targeting FLT3 (clones TRCN.772 and 773) and AKT1 (clones TRCN.39794 and 39797) were purchased from Thermo Scientific. Lentiviral particles were produced by cotransfection of 293FT cells with psPAX2, pMD2.G, and indicated lentiviral expression vectors. Transfections were carried out using TransIT (Mirus), as per the manufacturer’s instructions. Transduction was carried out in the presence of 5 μg/mL polybrene. After transduction, cells were selected with 1.5 μg/mL puromycin (Sigma-Aldrich).
Xenotransplantation assays
For cell line experiments, 1×106 MV4-11 cells transduced with Tet-pLKO.1 vectors expressing a tet-on-inducible miR-shRNA directed against SIRT1 or scrambled shRNA were transplanted via tail vein injection into sublethally irradiated (1.5 Gy) NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Jackson Laboratory). Ten days after transplantation, treatment with oral doxycycline was initiated. PKC412 treatment (100 mg/kg per day) by oral gavage was initiated 15 days after transplantation. For secondary transplantations, 1×106 cells were transplanted into sublethally irradiated NSG mice.
For engraftment assay, primary MNCs derived from 2 different FLT3-ITD-positive AML patients were cultured for 48 hours in cytokine-supplemented media and treated with TV-6, PKC412, or both in combination. Via tail vein injection, 5×106 cells were transplanted into sublethally irradiated NSG mice. Six weeks after transplantation, mice were euthanized and BM and spleen cells were analyzed for human CD45 expression.
Statistics
Unless otherwise specified, data are presented as mean ± standard deviation (SD). Comparisons between 2 groups were performed using the unpaired Student t-test or Mann-Whitney test for continuous variables and using Fisher’s exact test or χ-square test for categorical variables. A P value of <.05 was considered significant. For animal studies, Kaplan-Meier survival analysis was performed and survival was calculated using the log-rank test. Statistical computations were performed using GraphPad Prism software, version 5.0.
Full methods for flow cytometry, quantitative reverse-transcription polymerase chain reaction, colony-forming unit assays, immunoblotting, coimmunoprecipitation, gene expression profiling, apoptosis, and proliferations assays are available in supplemental Methods on the Blood Web site.
Results
SIRT1 is overexpressed in AML samples harboring oncogenic tyrosine kinases or RAS mutations
To assess whether SIRT1 plays a role in AML, we first performed expression analyses at protein and RNA levels in primary AML samples. Immunoblot analysis demonstrated SIRT1 protein expression in 31 of 40 AML probes (Figure 1A). Twenty of 31 patients with detectable SIRT1 expression harbored mutations within the receptor tyrosine kinases FLT3 and KIT or the NRAS gene (Figure 1A; supplemental Table 1). As previously described, SIRT1 was highly expressed in blast crisis CML and only barely detectable in chronic-phase CML samples (Figure 1A, lanes 20 and 21).24 To precisely compare SIRT1 protein expression between FLT3-mutated and FLT3-WT AML samples, we determined SIRT1 protein expression levels relative to Actin by densitometry (Figure 1B; supplemental Table 1). We noticed significantly increased SIRT1 expression levels in FLT3-mutated compared with FLT3-wild-type (WT) samples. SIRT1 protein expression was also detected in 10 of 13 AML cell lines (Figure 1C). In all SIRT1-positive cell lines, activating mutations in FLT3, NRAS, or KRAS have been described (supplemental Table 2).31 Subsequently, we investigated SIRT1 expression in BA/F3 cells overexpressing distinct mutated tyrosine kinases or their WT counterparts. Consistent with the expression data in established leukemic cells, we observed a substantial increase in SIRT1 expression in cells transduced with TEL-PDGFRβ, c-KitD816V, JAK2V617F, or FLT3-ITD compared with parental BA/F3 cells or JAK2-WT- and FLT3-WT-expressing cells (Figure 1D). Similar findings were observed in M-07e and 32D cells expressing mutant FLT3 (supplemental Figure 1A-B). To investigate whether SIRT1 expression is regulated at the transcriptional level, we performed RQ-PCR analysis. There was no difference in SIRT1 messenger RNA (mRNA) levels between samples with high, medium, and low/no SIRT1 protein expression (Figure 1E). Samples with very high SIRT1 protein levels had rather low RNA expression levels. Most of these samples harbored mutations in FLT3, KIT, or RAS (Figure 1F). In contrast, in primary AML samples or cell lines without activating mutations, SIRT1 protein expression correlated well with mRNA levels. Further, BA/F3-FLT3-ITD cells showed decreased Sirt1 mRNA expression levels compared with BA/F3-WT cells (supplemental Figure 1C). These data were also confirmed by the analysis of SIRT1 mRNA expression in 436 AML samples with normal karyotypes. Patients harboring FLT3-ITD mutations had significantly lower SIRT1 mRNA expression levels compared with FLT3-WT patients (Figure 1G).
SIRT1 protein levels are highly expressed in AML samples harboring activated signaling pathways. (A) Protein expression levels of SIRT1, DBC1, and Actin in cell lysates of primary AML or CML samples, normal BM cells (lane 41), and CD34pos cells (lanes 42 and 43). (B) SIRT1 protein expression was quantified by densitometric analysis and normalized to Actin expression. Shown are normalized SIRT1 expression levels of FLT3-ITD (n = 16), FLT3-TKD (n = 3), and FLT3-WT (n = 18) AML samples. ***P < .001; ns, not significant; unpaired Student t test. (C-D) Protein expression levels of SIRT1, DBC1, and Actin in human leukemic cell lines (C) and in BA/F3 cells (D) transduced with different oncogenes, as indicated. (E) SIRT1 mRNA expression was determined by real-time quantitative polymerase chain reaction (RQ-PCR) analysis and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. SIRT1 protein expression was quantified by densitometric analyses and normalized to Actin expression. Shown are mRNA levels relative to SIRT1 protein expression. (F) Shown is the correlation of SIRT1 mRNA and SIRT1 protein expression in FLT3-ITD and RAS-mutated cells (left; ns, not significant) and FLT3-WT and RAS-WT cells (right; P = .033). Black boxes indicate primary patient samples, white boxes indicate cell lines. (G) Log2 transformed mean centered SIRT1 gene expression values (IMAGE clone IMAGE:796114) are plotted for FLT3-ITD (n = 108) vs no ITD AML cases (n = 307).
SIRT1 protein levels are highly expressed in AML samples harboring activated signaling pathways. (A) Protein expression levels of SIRT1, DBC1, and Actin in cell lysates of primary AML or CML samples, normal BM cells (lane 41), and CD34pos cells (lanes 42 and 43). (B) SIRT1 protein expression was quantified by densitometric analysis and normalized to Actin expression. Shown are normalized SIRT1 expression levels of FLT3-ITD (n = 16), FLT3-TKD (n = 3), and FLT3-WT (n = 18) AML samples. ***P < .001; ns, not significant; unpaired Student t test. (C-D) Protein expression levels of SIRT1, DBC1, and Actin in human leukemic cell lines (C) and in BA/F3 cells (D) transduced with different oncogenes, as indicated. (E) SIRT1 mRNA expression was determined by real-time quantitative polymerase chain reaction (RQ-PCR) analysis and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. SIRT1 protein expression was quantified by densitometric analyses and normalized to Actin expression. Shown are mRNA levels relative to SIRT1 protein expression. (F) Shown is the correlation of SIRT1 mRNA and SIRT1 protein expression in FLT3-ITD and RAS-mutated cells (left; ns, not significant) and FLT3-WT and RAS-WT cells (right; P = .033). Black boxes indicate primary patient samples, white boxes indicate cell lines. (G) Log2 transformed mean centered SIRT1 gene expression values (IMAGE clone IMAGE:796114) are plotted for FLT3-ITD (n = 108) vs no ITD AML cases (n = 307).
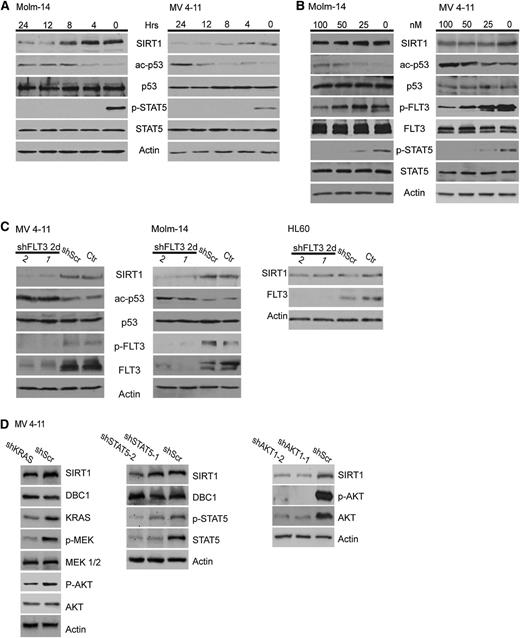
SIRT1 expression is regulated by aberrant tyrosine kinase activity
The observed association with oncogenic signaling pathways prompted us to investigate whether SIRT1 expression is regulated by tyrosine kinase activity. Therefore, we treated the FLT3-ITD-positive cell lines Molm-14 and MV4-11 with the FLT3-inhibitor PKC412. We observed a time- and dose-dependent decrease of SIRT1 protein expression (Figure 2A-B) accompanied by an increase in p53 acetylation, a well-known target protein of SIRT1. Similar results were obtained with PKC412-treated M-07e-FLT3-ITD and BA/F3-FLT3-ITD cells (supplemental Figure 2A-B). In contrast, SIRT1 protein levels remained unchanged in BA/F3 cells expressing the PKC412-resistant FLT3-ITD_N676K mutant (supplemental Figure 1B).32 To rule out PKC412 off-target effects, we transduced MV4-11 and Molm-14 cells with lentiviral shRNAs directed against FLT3 or control shRNAs. Again, SIRT1 protein expression declined on FLT3-knockdown (Figure 2C). Concomitantly, p53 acetylation levels increased. These effects were not observed on knockdown of WT FLT3 in HL-60 cells (Figure 2C). FLT3-ITDs aberrantly activate several downstream pathways including STAT5, RAS/RAF/MAPK/ERK, and PI3K/AKT. To explore whether these pathways contribute to the regulation of SIRT1 expression, we performed knockdown and inhibitor experiments. shRNA-mediated knockdown of KRAS, STAT5, and AKT1, as well as treatment with the AKT inhibitor VIII, caused downregulation of SIRT1 protein levels, whereas treatment with the MEK inhibitor PD98059 had no effect (Figure 2D; supplemental Figure 2C-D). These data indicate that STAT5 and RAS signaling, particularly via activation of the PI3K-AKT1 axis, are involved in the regulation of SIRT1 and likely act in concert, as inhibition of a single pathway was not able to inhibit SIRT1 expression as efficiently as direct inhibition of FLT3 (Figure 2A-C). We next asked whether inhibition of FLT3 tyrosine kinase activity causes downregulation of SIRT1 mRNA levels. Molm-14 and MV4-11 cells were treated with 100 nM PKC412 for 24 hours. At this time, FLT3 and STAT5 activation was substantially suppressed (Figure 2A-B); however, we did not observe obvious differences in SIRT1 mRNA expression levels in treated samples compared with untreated controls (supplemental Figure 2E). These data support the hypothesis that SIRT1 expression is regulated at posttranscriptional levels.
SIRT1 protein expression is regulated by FLT3-ITD tyrosine kinase activity. (A-B) Molm-14 and MV4-11 cells were treated with 100 nM PKC412 for different periods (A) and different concentrations for 24 hours (B), as indicated. SIRT1 expression, p53 acetylation, and tyrosine phosphorylation of FLT3 and STAT5 were analyzed by immunoblotting. (C) Molm-14, MV4-11, and HL-60 cells were transduced with lentiviral vectors expressing nonsilencing scrambled shRNA or with 2 different FLT3 shRNA clones. Forty-eight hours after transduction, SIRT1 expression, p53 acetylation, FLT3 tyrosine phosphorylation, and expression were analyzed. (D) MV4-11 cells were transduced with lentiviral vectors expressing a doxycycline-inducible nonsilencing miR-shRNA (shScr) or a miR-shRNA targeting KRAS (sh_KRAS; left), STAT5 (sh_STAT5-1 and sh_STAT5-2; middle), or lentiviral vectors expressing 2 different AKT1 shRNA clones (right). Immunoblot analysis was performed 72 hours after doxycycline treatment or straight knockdown, respectively.
SIRT1 protein expression is regulated by FLT3-ITD tyrosine kinase activity. (A-B) Molm-14 and MV4-11 cells were treated with 100 nM PKC412 for different periods (A) and different concentrations for 24 hours (B), as indicated. SIRT1 expression, p53 acetylation, and tyrosine phosphorylation of FLT3 and STAT5 were analyzed by immunoblotting. (C) Molm-14, MV4-11, and HL-60 cells were transduced with lentiviral vectors expressing nonsilencing scrambled shRNA or with 2 different FLT3 shRNA clones. Forty-eight hours after transduction, SIRT1 expression, p53 acetylation, FLT3 tyrosine phosphorylation, and expression were analyzed. (D) MV4-11 cells were transduced with lentiviral vectors expressing a doxycycline-inducible nonsilencing miR-shRNA (shScr) or a miR-shRNA targeting KRAS (sh_KRAS; left), STAT5 (sh_STAT5-1 and sh_STAT5-2; middle), or lentiviral vectors expressing 2 different AKT1 shRNA clones (right). Immunoblot analysis was performed 72 hours after doxycycline treatment or straight knockdown, respectively.
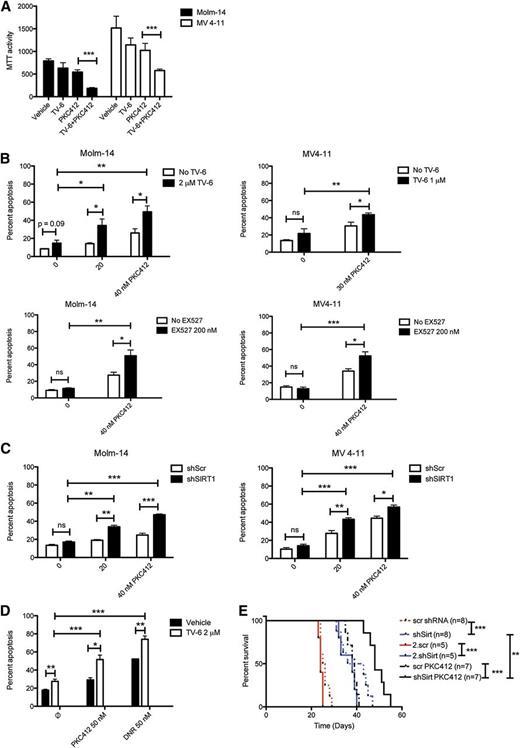
Targeting SIRT1 sensitizes AML cells to PKC412 treatment in vitro and confers increased overall survival in vivo
To evaluate the biological consequences of SIRT1 overexpression, we treated Molm-14 and MV4-11 cells with TV-6, a small-molecule inhibitor of SIRT1 and SIRT2, with and without PKC412 and monitored proliferation and apoptosis. In preliminary cell survival assays, TV-6-50% inhibitory concentration values for MV4-11 and Molm-14 cells were shown to be 2.1 and 5.0 µM, respectively (supplemental Figure 3). To identify synergistic/additive effects between TV-6 and PKC412, sublethal concentrations for both compounds were used. Treatment with TV-6 and PKC412 at low concentrations alone had only minor effects in both cell lines. However, the combination significantly inhibited cell growth (Figure 3A). Consistently, treatment with TV-6 alone slightly induced apoptosis, whereas combined treatment enhanced apoptotic cell death (Figure 3B). Similar results were obtained after treatment with the specific SIRT1 inhibitor EX527 (Figure 3B).33 To rule out off-target effects on pharmacologic SIRT1 inhibition, we transduced Molm-14 and MV4-11 cells with lentivirus expressing a doxycycline-inducible shRNA directed against SIRT1 or scrambled shRNA. Immunoblot analysis confirmed effective knockdown accompanied by an increase in p53 acetylation (supplemental Figure 4). Five days after initiation of doxycycline treatment, we observed a decline in cell proliferation compared with control cells (supplemental Figure 4). In line with our inhibitor data, SIRT1 knockdown alone induced only marginal apoptosis but sensitized both cell lines to PKC412 (Figure 3C). We next wanted to confirm our results in primary AML patient samples. First, MNCs derived from FLT3-ITD-positive AML patients were transduced with lentivirus expressing either shRNA directed against FLT3 or scrambled shRNA. Immunoblot analysis confirmed efficient FLT3 knockdown (supplemental Figure 5A). Loss of FLT3 was accompanied by a decrease in SIRT1 protein levels and increased p53 acetylation. Similar effects were observed on treatment of FLT3-ITD-positive blasts with PKC412. Single-agent TV-6 treatment with 2 and 5 µM caused an increase of p53 acetylation (supplemental Figure 5B-C). To examine whether TV-6 can augment the effects of PKC412 and daunorubicin, primary blasts were treated with either agent alone or in combination. We observed a slight increase in apoptosis on treatment with TV-6 alone in all samples tested (Figure 3D; supplemental Figure 6). Again, targeting SIRT1 significantly sensitized FLT3-ITD-positive primary blasts to PKC412 treatment but also to unspecific cytotoxic drugs as daunorubicin (Figure 3D). These data indicate that SIRT1 protects leukemic blasts against cytotoxic drug effects and that targeting SIRT1 uncouples FLT3-ITD-mediated drug resistance. Further, 2 primary AML samples were evaluated for their potential to engraft NSG mice. MNCs were treated with TV-6 or PKC412 or in combination for 48 hours, and viable cells were transplanted into NSG mice. On treatment with TV-6 or PKC412, engraftment of human CD45+ cells in BM and spleen of recipient mice was impaired (supplemental Figure 7). The combination of TV-6 and PKC412 further reduced engraftment efficacy in both AML samples compared with either agent alone. Finally, we investigated the effect of SIRT1 knockdown in vivo, using xenotransplantation experiments. MV4-11 cells stably expressing doxycycline-inducible SIRT1 or scrambled shRNA were injected into NSG mice. Knockdown of SIRT1 resulted in significantly longer overall survival compared with control mice (Figure 3E). Efficient knockdown of SIRT1 in spleen-infiltrating MV4-11 cells was confirmed by immunoblot analysis (supplemental Figure 8). Combined treatment with PKC412 significantly prolonged survival of mice transplanted with MV4-11 cells expressing SIRT1-shRNA compared controls. To investigate whether the observed increase in survival is a result of diminished self-renewal or reduced cell growth, we performed secondary transplantation assays. No further increase in survival was observed in mice transplanted with shSIRT-MV4-11 cells, indicating reduced proliferation after depletion of SIRT1 in this highly aggressive xenotransplantation model.
Targeting SIRT1 sensitizes AML cell lines and primary AML patient cells to TKI treatment. (A) Cell proliferation assay of Molm-14 (black bars) and MV4-11 (white bars). Cells were treated with PKC412 (35 nM), TV-6 (2 and 1 µM, respectively), or a combination for 48 hours. Shown are mean values ± SD of 3 independent experiments. (B) Molm-14 and MV4-11 cells were treated with PKC412, TV-6 (top), or EX527 (bottom) or in combination, as indicated. The percentage of apoptotic cells was determined by Annexin V expression. Shown are mean values ± SD of 3 independent experiments. (C) Molm-14 (left) and MV4-11 (right) cells were transduced with lentiviral vectors expressing a doxycycline-inducible nonsilencing miR-shRNA (shScr) or a miR-shRNA targeting SIRT1 (shSIRT1-1 and shSIRT1-2), incubated with doxycycline for 72 hours, followed by PKC412 treatment, as indicated. The percentage of sub-G1 cells corresponding to apoptotic cells was determined by flow cytometry. Data represent mean values ± SD of 3 independent experiments. (E) Primary AML blasts were treated with PKC412, TV-6, and daunorubicin alone or in combination for 48 hours, as indicated. The percentage of apoptotic cells in blast gate was determined by Annexin V expression. Shown is the pooled analysis of apoptotic cell death in primary AML samples treated with PKC412 (n = 5) or daunorubicin (DNR; n = 3) with and without TV-6. *P < .05; **P < .01; ***P < .001; unpaired Student t test. (F) Kaplan-Meier survival curves of NSG mice transplanted with MV4-11 cells expressing either a doxycycline-inducible nonsilencing miR-shRNA or a miR-shRNA targeting SIRT1. Oral doxycycline treatment was initiated 10 days after transplantation, treatment with PKC412 (100 mg/kg per day) by oral gavage was started 15 days after transplantation. *P < .05, **P < .01, ***P < .001; log-rank test.
Targeting SIRT1 sensitizes AML cell lines and primary AML patient cells to TKI treatment. (A) Cell proliferation assay of Molm-14 (black bars) and MV4-11 (white bars). Cells were treated with PKC412 (35 nM), TV-6 (2 and 1 µM, respectively), or a combination for 48 hours. Shown are mean values ± SD of 3 independent experiments. (B) Molm-14 and MV4-11 cells were treated with PKC412, TV-6 (top), or EX527 (bottom) or in combination, as indicated. The percentage of apoptotic cells was determined by Annexin V expression. Shown are mean values ± SD of 3 independent experiments. (C) Molm-14 (left) and MV4-11 (right) cells were transduced with lentiviral vectors expressing a doxycycline-inducible nonsilencing miR-shRNA (shScr) or a miR-shRNA targeting SIRT1 (shSIRT1-1 and shSIRT1-2), incubated with doxycycline for 72 hours, followed by PKC412 treatment, as indicated. The percentage of sub-G1 cells corresponding to apoptotic cells was determined by flow cytometry. Data represent mean values ± SD of 3 independent experiments. (E) Primary AML blasts were treated with PKC412, TV-6, and daunorubicin alone or in combination for 48 hours, as indicated. The percentage of apoptotic cells in blast gate was determined by Annexin V expression. Shown is the pooled analysis of apoptotic cell death in primary AML samples treated with PKC412 (n = 5) or daunorubicin (DNR; n = 3) with and without TV-6. *P < .05; **P < .01; ***P < .001; unpaired Student t test. (F) Kaplan-Meier survival curves of NSG mice transplanted with MV4-11 cells expressing either a doxycycline-inducible nonsilencing miR-shRNA or a miR-shRNA targeting SIRT1. Oral doxycycline treatment was initiated 10 days after transplantation, treatment with PKC412 (100 mg/kg per day) by oral gavage was started 15 days after transplantation. *P < .05, **P < .01, ***P < .001; log-rank test.
Targeting SIRT1 inhibits colony growth and mitigates self-renewal in primary murine leukemia models
SIRT1 is a well-known regulator of cellular stress, including replicative stress mediated by oncogenes.14 To investigate the role of SIRT1 in leukemogenesis, we used different, genetically defined murine leukemia models. BM cells of Flt3-ITD-knock-in or WT mice were transduced with MLL-AF9 (M/A9)-expressing retroviruses (hereafter F3-M/A9 and M/A9). Alternatively, BM cells derived from AML1-ETO_Mx1-Cre mice were infected with retrovirus expressing FLT3-ITD (A/E-F3-ITD), FLT3-WT (A/E-F3-WT), or empty vector control (A/E-EV) (Figure 4A). M/A9 alone is able to induce AML, and coexpression of FLT3-ITD results in shortened disease latency.34 In contrast, A/E confers self-renewal activity but does not cause malignant transformation, whereas coexpression of FLT3-ITD has been shown to induce AML.29,35 Infected cells were transplanted into irradiated recipient mice, and BM cells were collected when mice showed the first signs of leukemia. We first investigated Sirt1 protein and mRNA expression in leukemic blasts. In M/A9 cells, we observed robust Sirt1 protein expression, with higher levels in Flt3-ITD coexpressing cells (Figure 4B). In A/E-EV cells, Sirt1 proteins were barely detectable, whereas coexpression of FLT3-WT increased and that of FLT3-ITD substantially increased Sirt1 expression. At mRNA levels, M/A had only modest effects (Figure 4C).
Targeting SIRT1 inhibits proliferation and self-renewal in murine leukemia models. (A) Illustration of experimental procedures. (B) Western blot of BM cells from leukemic FLT3wt/wt-MLL/AF9 (B6-M/A9), FLT3ITD/ITD-MLL/AF9 (F3-M/A9), AML1-ETO_Mx1-Cre-MSCV-FLT3-ITD (A/E-F3-ITD), and preleukemic AML1-ETO_Mx1-Cre-MSCV-FLT3-WT (A/E-F3-WT) or AML1-ETO_Mx1-Cre-MSCV (A/E-EV) mice probed with Sirt1, phospho-Flt3, Flt3, and Actin-specific antibodies. (C) Quantitative analysis of SIRT1 mRNA levels in leukemic BM cells of the indicated murine leukemia models. SIRT1 expression was normalized to GAPDH expression. Data shown are mean values (n = 3; M/A9 [left], A/E [right]). (D) Serial replating assays of leukemic BM cells derived from the indicated leukemia models, and 1×105 cells were plated in M3434 methylcellulose and treated with vehicle, TV-6, PKC412, or in combination as indicated. After 8 days, cells were pooled and replated at 1×105 cells in each round. Results shown are mean values ± SD of duplicate measurements of 3 independent experiments.
Targeting SIRT1 inhibits proliferation and self-renewal in murine leukemia models. (A) Illustration of experimental procedures. (B) Western blot of BM cells from leukemic FLT3wt/wt-MLL/AF9 (B6-M/A9), FLT3ITD/ITD-MLL/AF9 (F3-M/A9), AML1-ETO_Mx1-Cre-MSCV-FLT3-ITD (A/E-F3-ITD), and preleukemic AML1-ETO_Mx1-Cre-MSCV-FLT3-WT (A/E-F3-WT) or AML1-ETO_Mx1-Cre-MSCV (A/E-EV) mice probed with Sirt1, phospho-Flt3, Flt3, and Actin-specific antibodies. (C) Quantitative analysis of SIRT1 mRNA levels in leukemic BM cells of the indicated murine leukemia models. SIRT1 expression was normalized to GAPDH expression. Data shown are mean values (n = 3; M/A9 [left], A/E [right]). (D) Serial replating assays of leukemic BM cells derived from the indicated leukemia models, and 1×105 cells were plated in M3434 methylcellulose and treated with vehicle, TV-6, PKC412, or in combination as indicated. After 8 days, cells were pooled and replated at 1×105 cells in each round. Results shown are mean values ± SD of duplicate measurements of 3 independent experiments.
In contrast, Sirt1 mRNA was upregulated 8.5-fold and 25-fold in A/E-EV and A/E-F3-ITD cells, respectively (Figure 4C). We next investigated whether Sirt1 represents a therapeutic target in our different leukemia models. BM cells were plated in cytokine-supplemented methylcellulose and treated with TV-6 or PKC412 alone or in combination. In M/A9-positive cells, TV-6 treatment significantly inhibited colony growth and marginally diminished replating activity (Figure 4D, top). Treatment with PKC412 either alone or in combination with TV-6 had no effect on colony growth of Flt3-WT cells. Coexpression of Flt3-ITD sensitized M/A9 cells to PKC412 treatment, but inhibition of Flt3 alone did not affect replating capacity. In contrast, combined treatment with TV-6 and PKC412 caused a continuous and significant decline of self-renewal activity. In leukemic A/E-F3-ITD cells, treatment with TV-6 and PKC412 alone only inhibited colony growth, whereas combined treatment suppressed replating activity (Figure 4D, bottom). No effects on colony growth or self-renewal capacity were observed in A/E-F3-WT cells. In contrast to the blast-like appearance of M/A9 colonies, A/E colonies were loose and differentiated into all lineages (data not shown). Further, the total number of colonies was lower in untreated A/E-cells compared with M/A9-cells, indicating reduced proliferative capacity. On coexpression of FLT3-ITD, A/E colonies acquired the typical blast-like shape, increased in number, showed increased Sirt1 protein expression, and became sensitive to pharmacological SIRT1-inhibition. These data suggest that SIRT1 is preferentially required in fully transformed cells and that leukemic blasts become partially dependent on SIRT1 function to counteract oncogene-induced replicative stress and maintain their self-renewal capacity.
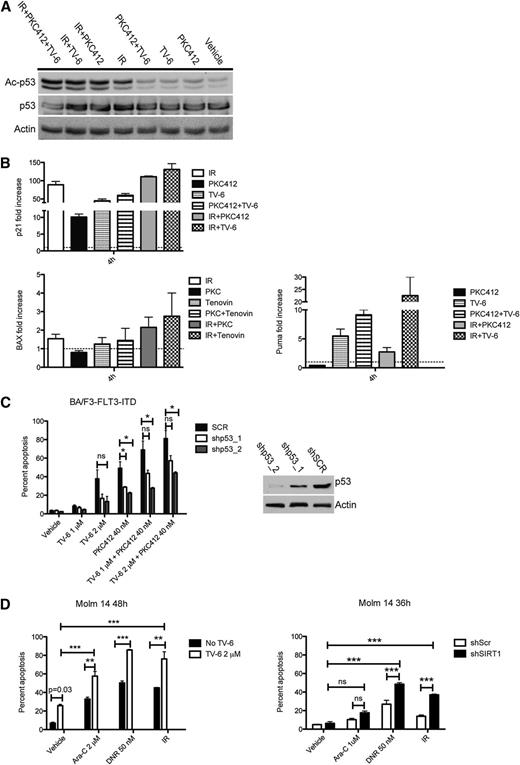
SIRT1 attenuates activation of p53 and inhibits genotoxic stress-induced apoptosis
In precancerous lesions, oncogene-mediated stress has been shown to induce double-strand breaks, followed by the activation of DDR pathways, and finally apoptosis or senescence.36 The tumor-suppressor gene TP53 is thought to play a key role in this process. As a consequence, loss of p53 function is a major cause of tumor progression in many solid tumors. In AML, p53 mutations or chromosome 17 abnormalities are rare events.37,38 However, other mechanisms such as posttranslational modifications might be involved in mitigating p53 function. Acetylation at Lys382 has been shown to be important for optimal p53 function. To investigate the relationship of SIRT1 and p53 activity, we treated p53-WT Molm-14 cells with irradiation and PKC412 or TV-6 alone or in combination. Treatment with PKC412 and TV-6 alone slightly induced de novo acetylation of p53, whereas irradiation alone induced strong p53 acetylation (Figure 5A). Consistent with p53 acetylation levels, we observed increased expression of the p53 target genes p21, BAX, and Puma (Figure 5B). p53 acetylation was further enhanced on combined treatment with PKC412 plus irradiation as well as TV-6 plus irradiation and also resulted in increased expression of p53 target genes (Figure 5A-B). Our data indicate that single-agent treatment does not induce complete p53 acetylation, and only combined treatment strategies optimize p53 activity. To accurately explore the role of p53 in SIRT1-mediated effects, we performed p53-knockdown experiments. BA/F3-FLT3-ITD cells, infected with lentivirus expressing either shRNAs directed against p53 or scrambled shRNA, were treated with TV-6 and PKC412 either alone or in combination. As shown in Figure 5C, depletion of p53 partially rescued BA/F3-FLT3-ITD cells from TV-6- or PKC412-induced apoptotic cell death, indicating that p53 represents a major downstream target of SIRT1. We next asked whether targeting SIRT1 could also enhance sensitivity to unspecific genotoxic agents similar to our observation on FLT3-TKI treatment. Therefore, Molm-14 cells were treated with irradiation, cytarabine, daunorubicin alone, or in combination with TV-6. In all combinations, TV-6 significantly increased apoptosis (Figure 5D, left). Consistent with pharmacologic SIRT1 inhibition, shRNA-induced SIRT1 knockdown also caused enhanced cell death (Figure 5D, right). Similar effects were observed in MV4-11 and 32D-FLT3-ITD cells (supplemental Figure 9).
Inhibition of SIRT1 sensitizes leukemic cells to genotoxic stress-mediated p53 activation and apoptosis. (A) Western blot of Molm-14 cells treated for 4 hours, as indicated, probed with antibodies directed against acetylated p53, p53, and Actin. (B) RQ-PCR analysis of p21 (top) and BAX and Puma (bottom) in Molm-14 cells treated as indicated. Target gene expression was normalized to GAPDH, and fold change is shown relative to untreated control (p21 and BAX) or irradiation only (Puma; because of no detectable expression in untreated cells). Data represent mean values ± SD of 3 independent experiments. (C) Analysis of apoptosis in BA/F3-FLT3-ITD cells infected with lentivirus expressing 2 different shRNA clones directed against p53 or scrambled shRNA after treatment with TV-6 and PKC412 (left). Immunoblot analysis demonstrating p53 knockdown efficacy (right). (D) Analysis of apoptosis after treatment with TV-6 (left) or SIRT1-knockdown (right) and genotoxic agents, as indicated. The percentage of sub-G1 cells corresponding to apoptotic cells was determined by flow cytometry. Data represent the mean values ± SD of 3 independent experiments. *P < .05, **P < .01, ***P < .001; unpaired Student t test.
Inhibition of SIRT1 sensitizes leukemic cells to genotoxic stress-mediated p53 activation and apoptosis. (A) Western blot of Molm-14 cells treated for 4 hours, as indicated, probed with antibodies directed against acetylated p53, p53, and Actin. (B) RQ-PCR analysis of p21 (top) and BAX and Puma (bottom) in Molm-14 cells treated as indicated. Target gene expression was normalized to GAPDH, and fold change is shown relative to untreated control (p21 and BAX) or irradiation only (Puma; because of no detectable expression in untreated cells). Data represent mean values ± SD of 3 independent experiments. (C) Analysis of apoptosis in BA/F3-FLT3-ITD cells infected with lentivirus expressing 2 different shRNA clones directed against p53 or scrambled shRNA after treatment with TV-6 and PKC412 (left). Immunoblot analysis demonstrating p53 knockdown efficacy (right). (D) Analysis of apoptosis after treatment with TV-6 (left) or SIRT1-knockdown (right) and genotoxic agents, as indicated. The percentage of sub-G1 cells corresponding to apoptotic cells was determined by flow cytometry. Data represent the mean values ± SD of 3 independent experiments. *P < .05, **P < .01, ***P < .001; unpaired Student t test.
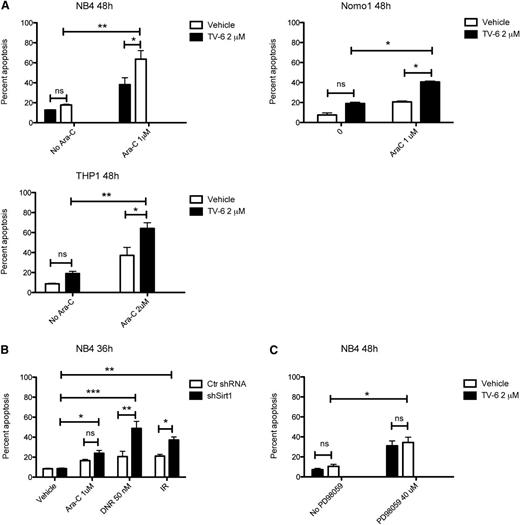
In addition to FLT3-mutant AML blasts, increased SIRT1 protein levels were also observed in KRAS-mutant cells (Figure 1), and treatment with TV-6 inhibited cell growth in most of these cell lines (supplemental Figure 3). To assess whether targeting SIRT1 also sensitizes KRAS-mutant cells to genotoxic agents, we treated NB4, Nomo1, and THP1 cells with cytarabine and TV-6, either alone or in combination. Inhibition of SIRT1 significantly increased cytarabine-induced apoptosis (Figure 6A). Again, knockdown of SIRT1 sensitized NB4 cells to cytarabine, daunorubicin, or irradiation, suggesting a specific role of SIRT1 in attenuating genotoxic stress-mediated apoptosis (Figure 6B). In line with our previous data, inhibition of MEK1 had no effect on cell survival (Figure 6C). Interestingly, all tested KRAS-mutant cell lines harbor TP53 mutations, suggesting alternative SIRT1-dependent mechanisms to counterbalance genotoxic stress. Recently, deacetylation of KU70 has been shown to mediate SIRT1 effects in TP53-mutated, BCR-ABL-positive KCL-22 cells.25
Inhibition of SIRT1 sensitizes KRAS-mutant leukemic cells to cytarabine-induced cell death. (A) NB4, Nomo1, and THP1 cells were treated with vehicle, TV-6, and cytarabine (Ara-C) alone or in combination, as indicated. (B) NB4 cells were transduced with lentiviral vectors expressing a doxycycline-inducible nonsilencing miR-shRNA (shScr) or a miR-shRNA targeting SIRT1 (shSirt1), incubated with doxycycline for 72 hours followed by treatment with cytarabine (Ara-C), daunorubicin (DNR), and irradiation (IR; 2 Gy), as indicated. (C) NB4 cells were treated with the selective MEK-inhibitor PD98059 or TV-6 alone or in combination, as indicated. The percentage of sub-G1 cells corresponding to apoptotic cells was determined by flow cytometry. Data represent mean values ± SD of 3 independent experiments. *P < .05, **P < .01, ***P < .001; unpaired Student t test.
Inhibition of SIRT1 sensitizes KRAS-mutant leukemic cells to cytarabine-induced cell death. (A) NB4, Nomo1, and THP1 cells were treated with vehicle, TV-6, and cytarabine (Ara-C) alone or in combination, as indicated. (B) NB4 cells were transduced with lentiviral vectors expressing a doxycycline-inducible nonsilencing miR-shRNA (shScr) or a miR-shRNA targeting SIRT1 (shSirt1), incubated with doxycycline for 72 hours followed by treatment with cytarabine (Ara-C), daunorubicin (DNR), and irradiation (IR; 2 Gy), as indicated. (C) NB4 cells were treated with the selective MEK-inhibitor PD98059 or TV-6 alone or in combination, as indicated. The percentage of sub-G1 cells corresponding to apoptotic cells was determined by flow cytometry. Data represent mean values ± SD of 3 independent experiments. *P < .05, **P < .01, ***P < .001; unpaired Student t test.
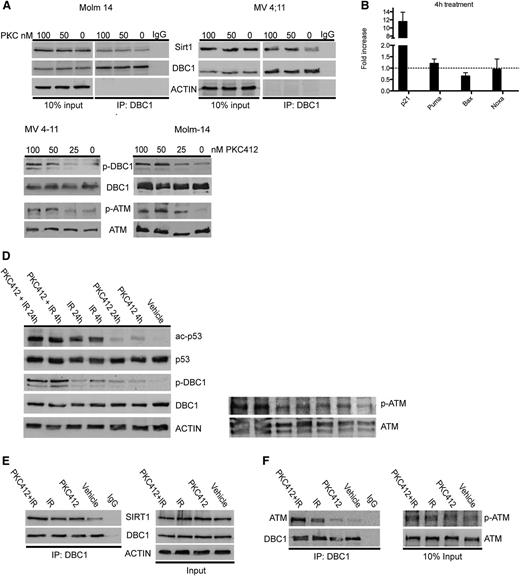
FLT3-ITD regulates p53 acetylation via the ATM-DBC1-SIRT1 axis
Treatment with PKC412 alone for 4 hours had minimal effects on p53 acetylation and no effects on SIRT1 protein expression (Figure 2A). However, combined treatment with irradiation enhanced p53 acetylation compared with each agent alone (Figure 5A). Similar effects were observed when TV-6 was combined with irradiation. These data indicate that apart from downregulation of SIRT1 protein expression, alternative mechanisms are involved in the regulation of SIRT1 function. DBC1, which inhibits the enzymatic activity of SIRT1 by binding to its catalytic domain,17,18 was highly expressed in SIRT1-positive primary AML samples and cell lines (Figure 1A-C). Therefore, we investigated whether aberrant FLT3 tyrosine kinase activity modulates DBC1 binding to SIRT1. DBC1 did not interact with SIRT1 in untreated MV4-11 and Molm-14 cells (Figure 7A). In contrast, inhibition of FLT3 tyrosine kinase activity induced binding of DBC1 to SIRT1 (Figure 7A). These data correlated well with an increase in p53 acetylation (Figures 2A and 5A) and expression of the p53 target gene p21 (Figure 7B), suggesting a DBC1-mediated inhibitory effect on SIRT1 function. In response to DNA damage, DBC1 is phosphorylated at Thr454 in an ATM/ATR-dependent manner.19,20 Treatment of MV4-11 and Molm-14 cells with PKC412 caused a dose-dependent increase of DBC1 phosphorylation at Thr454 and of ATM phosphorylation at Ser1981 (Figure 7C). Further, ATM and DBC1 activation was also modulated after treatment with unspecific cytotoxic agents. MV4-11 cells were treated with PKC412, cytarabine, or irradiation alone or in combination. Single-agent treatment induced an increase in DBC1 and ATM phosphorylation, as well as p53 acetylation. These effects were further enhanced on combination therapies (Figure 7D and supplemental Figure 10). DBC1 phosphorylation after irradiation correlated well with an increased binding to SIRT1, which was augmented on combined treatment with PKC412 (Figure 7E). Finally, we asked whether inhibition of aberrant FLT3 tyrosine kinase activity or genotoxic stress directly modulates the interaction of ATM with DBC1. MV4-11 cells were treated with PKC412 and irradiation alone or in combination, and coimmunoprecipitation analysis was performed. Single-agent treatment induced binding of ATM to DBC1, which was further enhanced on combined treatments (Figure 7F). These data indicate that aberrant FLT3 kinase activity regulates SIRT1 activity via the ATM-DBC1-SIRT1 axis and that either targeting of FLT3 or direct inhibition of SIRT1 can restore sensitivity to genotoxic agents.
Mutated FLT3 regulates the activity of SIRT1 via the ATM-DBC1 axis. (A) MV4-11 and Molm-14 cells were treated with PKC412 for 4 hours, as indicated, and coimmunoprecipitation was performed using an anti-DBC1 antibody. (B) Quantitative RQ-PCR analysis of p21, BAX, Puma, and Noxa expression on treatment of MV4-11 cells with PKC412 for 4 hours. Expression of target genes was normalized to GAPDH. Shown is the fold increase compared with untreated control of 3 independent experiments. (C-D) Western blots of MV4-11 and Molm-14 cells treated with PKC412 for 24 hours, as indicated (C), or with 100 nM PKC412, irradiation (2 Gy; IR), or in combination (D) probed with the indicated antibodies. (E-F) Coimmunoprecipitation using an anti-DBC1-antibody was performed in MV4-11 cells treated with 100 nM PKC412, irradiation (2 Gy; IR), or in combination for 4 hours. Immunoblots were probed as indicated.
Mutated FLT3 regulates the activity of SIRT1 via the ATM-DBC1 axis. (A) MV4-11 and Molm-14 cells were treated with PKC412 for 4 hours, as indicated, and coimmunoprecipitation was performed using an anti-DBC1 antibody. (B) Quantitative RQ-PCR analysis of p21, BAX, Puma, and Noxa expression on treatment of MV4-11 cells with PKC412 for 4 hours. Expression of target genes was normalized to GAPDH. Shown is the fold increase compared with untreated control of 3 independent experiments. (C-D) Western blots of MV4-11 and Molm-14 cells treated with PKC412 for 24 hours, as indicated (C), or with 100 nM PKC412, irradiation (2 Gy; IR), or in combination (D) probed with the indicated antibodies. (E-F) Coimmunoprecipitation using an anti-DBC1-antibody was performed in MV4-11 cells treated with 100 nM PKC412, irradiation (2 Gy; IR), or in combination for 4 hours. Immunoblots were probed as indicated.
Discussion
Depending on the cellular and genetic background, SIRT1 has been referred to as a tumor suppressor or oncogene. In this report, we investigated the role of SIRT1 in AML. In AML cells harboring mutated receptor tyrosine kinases or RAS-GTPases, SIRT1 protein levels were upregulated compared with control AML samples. This increased expression did not involve transcriptional activity, as RNA levels were rather decreased in several experimental models but were dependent on aberrant FLT3 tyrosine kinase activity. Inhibition of mutated FLT3 involved the FLT3-ITD downstream pathways STAT5 and PI3K-AKT1. Recently, increased SIRT1 expression was shown in the context of BCR-ABL-positive cells.24,25 In these studies, SIRT1 was regulated at transcriptional levels via the STAT5 pathway. In our study, targeting of FLT3 associated with complete inhibition of STAT5 phosphorylation had no effect on SIRT1 mRNA levels, indicating oncogene-dependent differences in SIRT1 regulation. As knockdown of STAT5 altered SIRT1 protein levels, we argue that STAT5 is involved in the posttranslational modulation of SIRT1. Indeed, STAT5 has been shown to be involved in eIF2a regulation, a major regulator of cap-dependent translation initiation.39 Alternatively, STAT5 might directly regulate protein phosphatase expression.40 At this time, 13 SIRT1 phosphorylation sites have been identified,41 most of them linked to modulate enzymatic activity42 ; their role in regulating protein stability needs to be established. Interestingly, Yuan et al showed that expression of BCR-ABL had no additional effect on cells in which SIRT1 was already expressed.25 This finding suggests the existence of alternative pathways that initiate SIRT1 expression. Our data provide evidence that in AML cells, other mechanisms can operate to induce SIRT1 expression. For example, expression of AML1-ETO caused strong upregulation of SIRT1 mRNA levels in preleukemic and leukemic cells.
Pharmacologic inhibition or knockdown of SIRT1 modestly inhibited cellular proliferation of FLT3-mutated AML cells, and despite increased p53 acetylation, only a small fraction of cells underwent apoptotic cell death. In contrast, inhibition of SIRT1 sensitized leukemic cells to TKIs, irradiation, or chemotherapy and significantly enhanced apoptosis, indicating that inhibition of SIRT1 primes leukemic cells to genotoxic stress-induced cell death via either optimized activation of p53 or alternative mechanisms. Similar effects were observed in CML blasts treated with TV-6 in combination with imatinib.24,25 Recently, SIRT2 has been shown to be upregulated in AML blasts and inhibition of SIRT2 induced apoptosis.43 Interestingly, single-agent treatment with the specific SIRT1-inhibitor EX527 had no effect on leukemic blasts (Figure 3B), but treatment with the SIRT1/2-inhibitor TV-6 slightly induced apoptosis. These data indicate redundant functions of SIRT1 and SIRT2. However, we observed no differences in apoptosis when each inhibitor was combined with genotoxic agents strongly supporting an essential role of SIRT1 in sensitizing AML blasts.
Targeting SIRT1 in primary murine BM cells transduced with M/A9 significantly decreased colony formation and progressively reduced the number of colonies with each round of replating. Coexpression of M/A9 and Flt3-ITD conferred sensitivity to PKC412, and combined treatment further suppressed colony growth and replating activity compared with each agent alone. Interestingly, TV-6 treatment had no effect on A/E-expressing cells. Although A/E-expressing cells acquired replating activity, colonies did not show the typically blast-like shape indicating a preleukemic state. On coexpression of FLT3-ITD and complete malignant transformation, A/E cells became sensitive to TV-6 or PKC412, and combined treatment attenuated self-renewal activity. These data, and in particular our observations in preleukemic and leukemic A/E cells, suggest that SIRT1 acts as safeguard during leukemogenesis to counteract oncogene-mediated replication stress.36 This function likely involves inhibition of p53 activity. The acquisition of defects in the p53 pathway has been shown to represent a prerequisite in the process of malignant transformation in many tumor models.44,45 Interestingly, even after complete transformation, leukemic blasts are partly dependent on SIRT1 function. Whereas chromosomal loss of p53 or inactivating mutations are difficult to target, inhibition of aberrant SIRT1 activity might be able to restore p53 activity.
Recently, comprehensive genomic analysis of AML patient samples revealed that about 60% of all AML patients harbor mutations within signaling transduction pathways.46 FLT3-ITD mutations are one of the most common genetic alterations in AML.47 Aberrant FLT3-ITD signaling activates multiple downstream pathways finally causing resistance to apoptosis; for example, mediated by the antiapoptotic proteins MCL1 or survivin.48,49 Interestingly, p53 mutations or loss of p17 are rare events in FLT3-mutated AML.37,38 Here we provide evidence for a direct link between FLT3-ITD signaling and disturbed p53 activity via the ATM-DBC1-SIRT1 axis. ATM has been shown to act as an upstream activator of DBC1 on genotoxic stress.19,20 Targeting FLT3 caused an increase in ATM phosphorylation, followed by recruitment and phosphorylation of DBC1, binding of DBC1 to SIRT1, and p53 acetylation. These effects were further potentiated on irradiation in combination with PKC412, indicating that treatment with FLT3-TKIs primes leukemic blasts for complete p53 activation and apoptosis. Finally, direct inhibition of SIRT1 is able to circumvent the described FLT3-ITD-mediated block in p53 activation and renders leukemic blasts sensitive to commonly used chemotherapeutic agents.
In summary, our data indicate that targeting SIRT1 represents an attractive therapeutic strategy for a subset of AML patients. Inhibition of SIRT1 reactivates the p53 pathway and, in combination therapy with specific FLT3-inhibitors or conventional chemotherapy, might eradicate leukemia propagating cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Claudia Scholl for providing reagents and Florina Sasca, Kerstin Kunz, and Birgit Enders for technical assistance.
This work was supported by the Deutsche Krebshilfe (108821) and internal funding of the Johannes Gutenberg University of Mainz and is partly based on the doctoral thesis of D.S. L.B. was supported in part by the German Research Foundation (Heisenberg-Stipendium BU 1339/3-1).
Authorship
Contribution: D.S. designed experiments, performed research, analyzed data, and wrote the manuscript; P.S.H., S.S., D.S., and L.B. performed research and analyzed data; K.K., J.S., O.K., and S.V.P. performed research; M.T. provided reagents and wrote the manuscript; and T.K. designed experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas Kindler, University Medical Center of Mainz, Langenbeckstraße 1, D-55131 Mainz, Germany; e-mail thomas.kindler@unimedizin-mainz.de.


![Figure 4. Targeting SIRT1 inhibits proliferation and self-renewal in murine leukemia models. (A) Illustration of experimental procedures. (B) Western blot of BM cells from leukemic FLT3wt/wt-MLL/AF9 (B6-M/A9), FLT3ITD/ITD-MLL/AF9 (F3-M/A9), AML1-ETO_Mx1-Cre-MSCV-FLT3-ITD (A/E-F3-ITD), and preleukemic AML1-ETO_Mx1-Cre-MSCV-FLT3-WT (A/E-F3-WT) or AML1-ETO_Mx1-Cre-MSCV (A/E-EV) mice probed with Sirt1, phospho-Flt3, Flt3, and Actin-specific antibodies. (C) Quantitative analysis of SIRT1 mRNA levels in leukemic BM cells of the indicated murine leukemia models. SIRT1 expression was normalized to GAPDH expression. Data shown are mean values (n = 3; M/A9 [left], A/E [right]). (D) Serial replating assays of leukemic BM cells derived from the indicated leukemia models, and 1×105 cells were plated in M3434 methylcellulose and treated with vehicle, TV-6, PKC412, or in combination as indicated. After 8 days, cells were pooled and replated at 1×105 cells in each round. Results shown are mean values ± SD of duplicate measurements of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/1/10.1182_blood-2013-11-538819/4/m_121f4.jpeg?Expires=1769079781&Signature=P7gI1Qa~VSHcrMqPCgt10RhQuW40TGJc6ZDlhfyq-xMUtZk9fOry3hUGNl-po3wMCklpOpA3D6Paht-zBZ8GfIGyZUnmgVIuUKHFSw17da7ggVkxuIAyOJ0aF0LX9fAaRmi9X3~x-uQFYgDKsq~9IY8s0L58MkSvuMTPbXzpXCYdEED7fM6gnQCC1IOIcOLU0BXU2~5kkmOJ-7p1tkj73NumLT0mRvBQUrP2bE2bYLuntp7~qCyEMx4n7-P7gTStkQsG4q06e6FIVRe7mIUzfHnNQkJV4XNk1oDzUHxpmUzlUfbg2wJg4I7QQsXlCIDYOhIIWmcwOBRZPXFZPkZTSQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)