Key Points
MLL oncoproteins downregulate RUNX1/CBFβ by the CXXC domain and flanking region as a critical step in the development of MLL-related leukemias.
RUNX1/CBFβ (core binding factor [CBF]) is a heterodimeric transcription factor complex that is frequently involved in chromosomal translocations, point mutations, or deletions in acute leukemia. The mixed lineage leukemia (MLL) gene is also frequently involved in chromosomal translocations or partial tandem duplication in acute leukemia. The MLL protein interacts with RUNX1 and prevents RUNX1 from ubiquitin-mediated degradation. RUNX1/CBFβ recruits MLL to regulate downstream target genes. However, the functional consequence of MLL fusions on RUNX1/CBFβ activity has not been fully understood. In this report, we show that MLL fusion proteins and the N-terminal MLL portion of MLL fusions downregulate RUNX1 and CBFβ protein expression via the MLL CXXC domain and flanking regions. We confirmed this finding in Mll-Af9 knock-in mice and human M4/M5 acute myeloid leukemia (AML) cell lines, with or without MLL translocations, showing that MLL translocations cause a hypomorph phenotype of RUNX1/CBFβ. Overexpression of RUNX1 inhibits the development of AML in Mll-Af9 knock-in mice; conversely, further reducing Runx1/Cbfβ levels accelerates MLL-AF9–mediated AML in bone marrow transplantation assays. These data reveal a newly defined negative regulation of RUNX1/CBFβ by MLL fusion proteins and suggest that targeting RUNX1/CBFβ levels may be a potential therapy for MLLs.
Introduction
The CBF (core binding factor) heterodimeric transcription factor complex, composed of RUNX1 and CBFβ subunits, has been shown to play critical roles in both embryogenesis and hematopoiesis.1,-3 RUNX1 binds to its DNA target sequences through an evolutionarily conserved runt domain4 and can both activate and repress the transcription of its target genes. CBFβ does not bind to DNA directly. However, CBFβ induces a conformational change in RUNX1, which increases RUNX1 DNA-binding ability.5,6 Dysregulation of RUNX1/CBFβ, through chromosome translocations, mutations, microdeletions, or transcriptional downregulation, is leukemogenic.7
The stability of RUNX1 is tightly regulated. We have demonstrated that CBFβ enhances RUNX1 stability by preventing its ubiquitin-mediated degradation in the proteasome.8 Furthermore, it has been shown that the CHIP/Stub1 E3 ubiquitin ligase can mediate RUNX1 ubiquitination and degradation independently of the molecular chaperones Hsp70/90.9 mSin3A interacts with unphosphorylated RUNX1 and protects it from proteasome-mediated degradation. Extracellular signal-regulated kinase–dependent RUNX1 phosphorylation induces the release of RUNX1 from mSin3A, resulting in degradation of RUNX1 in a time-dependent manner.10,11 RUNX1 protein levels are also regulated during the cell cycle. RUNX1 protein is increased in S and G2/M phase cells compared with G1 phase cells, and this regulation is independent of cytokine-induced RUNX1 phosphorylation.12
The mixed lineage leukemia (MLL) protein is a mammalian homolog of the trithorax proteins,13,14 which are critical for hematopoiesis and the self-renewal of adult stem cells.15 Full-length MLL is a large multidomain protein of 3969 amino acid (aa) residues including 3 AT-hook DNA-binding motifs, a cysteine-rich CXXC domain with homology to DNA methyltransferases, 4 plant homeodomain (PHD) finger motifs, a PHD-flanking bromodomain, a C-terminal transactivation domain, and a Suvar3-9, enhancer-of-zeste, trithorax (SET) domain.16,17 As with its Drosophila homolog, trx, MLL regulates target gene expression by methylating lysine 4 of histone H3 (H3K4) through the methyltransferase activity of its C-terminal SET domain; methylation of H3K4 is closely associated with transcriptional activation.18 MLL can be cleaved to an N-terminal 300-kDa fragment and a C-terminal 180-kDa fragment by taspase.19 The human MLL gene is located on chromosome 11q23 and is often involved in chromosome translocations with various partner chromosomes, generating MLL fusion proteins.20,-22 More than 70 MLL fusion proteins have been documented in leukemia patients.23,24 In almost all fusion proteins, MLL breaks within an 8.3-kb break point cluster region (BCR),25 which results in the deletion of PHD finger region but also the maintenance of the MLL CXXC domain within the fusion protein. Interestingly, similar break points are also found in MLL partial tandem duplications (MLL-PTDs), which result from partial duplication within the 5′ region of the MLL gene. These duplications consist of an in-frame repetition of MLL exons in the 5′-3′ direction and produce an elongated protein.26 The incidence of MLL-PTD was 6.4% in unselected adult and childhood acute myeloid leukemia (AML) and 5% in myelodysplastic syndromes.27,28
MLL regulates many targets involved in self-renewal, proliferation, survival, and differentiation.22,29,30 The most well-studied targets are found in the HOXA gene cluster. MLL may bind to DNA or chromatin directly or be recruited to target loci by DNA-binding transcription factors.18,31,32 Our recent study showed that MLL, RUNX1, and CBFβ interact and form a complex.33 MLL interacts with the N terminus of RUNX1 (51-106 aa), and prevents RUNX1 from ubiquitin-mediated degradation. Although CBFβ does not interact with MLL directly, it can strongly enhance the interaction between RUNX1 and MLL. RUNX1/CBFβ recruits MLL to the regulatory regions of the PU.1 gene, which is important for maintaining the H3K4 trimethylation of the PU.1 upstream regulatory region and promoter regions.34 However, the functional consequence of MLL fusions on RUNX1/CBFβ activity has not been fully understood.
In this study, we investigated the effects of MLL truncation mutants and its fusion proteins on RUNX1/CBFβ. We found that RUNX1 protein was not only downregulated by MLL fusion proteins, but also by MLL aas 1-1406, which are common to MLL fusion proteins). We confirmed this finding in Mll-Af9 knock-in mice and human M4/M5 MLL fusion–expressing AML cell lines. Using Runx1+/−Cbfβ+/− mice as a Runx1/Cbfβ hypomorph model, we found significant hematopoietic/stem progenitor cell (HSPC) expansion and higher repopulation activity. Overexpression of RUNX1 inhibits the development of AML in Mll-Af9 knock-in mice HSPCs. Conversely, reducing Runx1/Cbfb levels accelerates MLL-AF9–mediated AML in bone marrow transplantation (BMT) assays. Our data suggest that downregulation of RUNX1/CBFβ is critical for the development and maintenance of MLL translocation-related leukemia; therefore, targeting RUNX1/CBFβ levels may be a potential therapy for MLLs.
Methods
Methods and materials used in this study can be found in the supplemental data on the Blood Web site. All animal studies were conducted according to approved Institutional Animal Care and Use Committee protocol and federal regulations.
Results
MLL-BP and MLL fusion proteins decrease RUNX1 and CBFβ protein levels
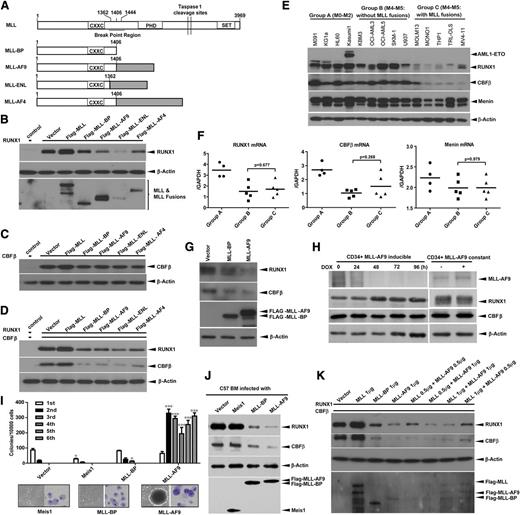
To understand the impact of MLL fusion protein expression on RUNX1 and CBFβ, either MLL, MLL-BP (1-1406), or MLL fusions were coexpressed with RUNX1, CBFβ, or both RUNX1 and CBFβ in 293T cells (Figure 1A). We found that MLL-BP and the 3 MLL fusion proteins all decreased RUNX1 levels, and MLL-eleven nineteen leukemia (ENL) caused a greater decrease in RUNX1 compared with MLL-AF9 and MLL-AF4 fusion proteins (Figure 1B and supplemental Figure 1A). CBFβ protein was mildly decreased by MLL-BP and MLL fusions when expressed alone (Figure 1C and supplemental Figure 1B); however, when CBFβ was coexpressed with RUNX1, it was significantly decreased, indicating that the full decrease in CBFβ by MLL-BP and MLL fusions depends on RUNX1 (Figure 1D and supplemental Figure 1C). We also coexpressed either GATA-1 or C/EBPα with MLL-BP. The level of each transcription factor remained unaltered by the coexpression of MLL-BP (supplemental Figure 2), which suggests that MLL-BP has specificity for RUNX1/CBFβ. To confirm this finding, we transduced retroviruses containing MLL-BP and MLL-AF9 into U937 cells and found that both of them, but not empty virus, downregulated RUNX1 and CBFβ proteins in U937 cells (Figure 1G and supplemental Figure 1D).
MLL-BP and MLL fusion proteins downregulate RUNX1 and CBFβ. (A) Schematic diagram demonstrating full-length MLL, MLL-BP (the N-terminal 1406 aa of MLL that is the common part of MLL fusions), and 3 of the most common MLL fusion proteins found in AML patients (MLL-AF9, MLL-ENL, MLL-AF4) used in this study. Immunoblot of RUNX1 (B), CBFβ (C), or RUNX1+CBFβ (D) transiently coexpressed with blank vector, full-length MLL, MLL-BP, MLL-AF9, MLL-ENL, or MLL-AF4 in 293T cells. (E) Human leukemia cell lines were separated into 3 groups: group A, M0-M2 subtypes; group B, M4-M5 subtypes without MLL translocations; and group C, M4-M5 subtypes with MLL translocations. Endogenous RUNX1, CBFβ, and Menin protein expression in these myeloid leukemia cell lines were determined by western blotting. (F) Summary of relative RUNX1, CBFβ, and Menin mRNA levels in the cell lines shown in panel E, which were measured by quantitative real-time polymerase chain reaction and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels using the Δ−CT method. (G) U937 cells were infected with recombinant amphotropic retroviruses carrying either MSCV-PGK-eGFP vector alone or with MLL-BP/MLL-AF9. eGFP+ cells were sorted and lysed for western blotting. (H) Endogenous RUNX1 and CBFβ protein expression in human umbilical cord blood CD34+ cells with Tet-off–driven MLL-AF9 expression. (I) Colony formation of hematopoietic progenitors in wild-type BM cells infected with empty, Meis1, MLL-BP, or MLL-AF9–expressing retroviruses (*P < .05; ***P < .001). The representative image of colonies and cytospin of the first round of plating from Meis1-, MLL-BP–, or MLL-AF9–infected BM cells were shown in the lower panel. (J) Endogenous RUNX1 and CBFβ protein expression in the first round of plating in panel I. (K) Immunoblot of RUNX1 and CBFβ, which were transiently coexpressed with different ratios of MLL and MLL-AF9 in 293T cells. Blank vector was added to make sure 2 μg of total plasmids were transfected in each sample. All of these experiments were repeated 3 times and representative data are shown. DOX, doxycycline; h, hours.
MLL-BP and MLL fusion proteins downregulate RUNX1 and CBFβ. (A) Schematic diagram demonstrating full-length MLL, MLL-BP (the N-terminal 1406 aa of MLL that is the common part of MLL fusions), and 3 of the most common MLL fusion proteins found in AML patients (MLL-AF9, MLL-ENL, MLL-AF4) used in this study. Immunoblot of RUNX1 (B), CBFβ (C), or RUNX1+CBFβ (D) transiently coexpressed with blank vector, full-length MLL, MLL-BP, MLL-AF9, MLL-ENL, or MLL-AF4 in 293T cells. (E) Human leukemia cell lines were separated into 3 groups: group A, M0-M2 subtypes; group B, M4-M5 subtypes without MLL translocations; and group C, M4-M5 subtypes with MLL translocations. Endogenous RUNX1, CBFβ, and Menin protein expression in these myeloid leukemia cell lines were determined by western blotting. (F) Summary of relative RUNX1, CBFβ, and Menin mRNA levels in the cell lines shown in panel E, which were measured by quantitative real-time polymerase chain reaction and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels using the Δ−CT method. (G) U937 cells were infected with recombinant amphotropic retroviruses carrying either MSCV-PGK-eGFP vector alone or with MLL-BP/MLL-AF9. eGFP+ cells were sorted and lysed for western blotting. (H) Endogenous RUNX1 and CBFβ protein expression in human umbilical cord blood CD34+ cells with Tet-off–driven MLL-AF9 expression. (I) Colony formation of hematopoietic progenitors in wild-type BM cells infected with empty, Meis1, MLL-BP, or MLL-AF9–expressing retroviruses (*P < .05; ***P < .001). The representative image of colonies and cytospin of the first round of plating from Meis1-, MLL-BP–, or MLL-AF9–infected BM cells were shown in the lower panel. (J) Endogenous RUNX1 and CBFβ protein expression in the first round of plating in panel I. (K) Immunoblot of RUNX1 and CBFβ, which were transiently coexpressed with different ratios of MLL and MLL-AF9 in 293T cells. Blank vector was added to make sure 2 μg of total plasmids were transfected in each sample. All of these experiments were repeated 3 times and representative data are shown. DOX, doxycycline; h, hours.
To validate this finding in human AML, various leukemia cell lines were collected and separated into 3 groups based on AML subtype. RUNX1 and CBFβ protein levels were higher in the cells lines that do not have MLL translocations (groups A and B) than in the cell lines with MLL translocations (group C) (Figure 1E and supplemental Figure 1E). The Kasumi-1 cell line, which contains only 1 wild-type RUNX1 allele, exhibited higher RUNX1 protein levels than the cells lines with MLL translocations and 2 RUNX1 alleles. However, expression levels of Menin, a transcription factor that interacts with the N terminus of MLL, did not differ among these groups of leukemia cell lines (Figure 1E and supplemental Figure 1E). We found no significant difference in the levels of RUNX1, CBFβ, and Menin messenger RNA (mRNA) between the groups of M4/M5 AMLs with or without MLL translocations (Figure 1F). We also analyzed RUNX1, CBFβ, and MEN1 (Menin) mRNA levels in published AML patient samples.35 Meta-analysis showed that RUNX1, CBFβ, and MEN1 mRNA levels are not decreased in M4/M5 subtypes with MLL translocations compared with the same subtype without MLL translocations, consistent with our AML cell line data (supplemental Table 2 and supplemental Figure 3A,C).
We further tested whether MLL fusion proteins could downregulate endogenous RUNX1 and CBFβ levels in human cord blood CD34+ cells using a Tet-off inducible expression system.36 Forty-eight hours after adding doxycycline, MLL-AF9 protein levels decreased, whereas RUNX1 protein levels increased. However, neither RUNX1 nor CBFβ protein levels were changed by the presence of doxycycline in CD34+-derived cell line with stable expression of MLL-AF9 (Figure 1H).
To examine the effects of MLL-BP and MLL fusions on endogenous Runx1 and Cbfβ expression in murine cells, bone marrow (BM) cells from wild-type C57BL/6 mice were transduced with retroviruses containing either Meis1, MLL-BP, or MLL-AF9 and plated for colony-forming unit (CFU) assays. BM cells transduced with MLL-BP virus had enhanced replating potential relative to those transduced with empty or Meis1 virus in the third plating, but there were no colonies in the fourth plating. Only MLL-AF9–transduced cells had replating ability, indicating a gain-of-function activity of MLL-AF9 (Figure 1I). We confirmed that Runx1 and Cbfβ were both downregulated in MLL-BP– and MLL-AF9–expressing cells, but not in Meis1-expressing cells (Figure 1J). Our data indicate that Runx1/Cbfβ downregulation is MLL-BP dependent but is independent of the MLL downstream target, Meis1.
We have shown that wild-type MLL can stabilize the RUNX1/CBFβ complex.33 In MLL fusion leukemias, only 1 allele of MLL is truncated and fused with another gene, whereas the other allele is intact. A dominant-negative function of the MLL fusions over the remaining wild-type allele has been suggested; thus, we researched whether MLL fusion protein could downregulate RUNX1/CBFβ in the presence of wild-type MLL. We cotransfected RUNX1/CBFβ with different MLLs:MLL-AF9 ratios and found that MLL-AF9 has a strong dominant-negative effect over MLL, even at high MLL:MLL-AF9 ratios (Figure 1K).
Ubiquitin-proteasome pathway blockade cannot fully rescue RUNX1 and CBFβ from MLL-BP and MLL fusion protein–mediated downregulation
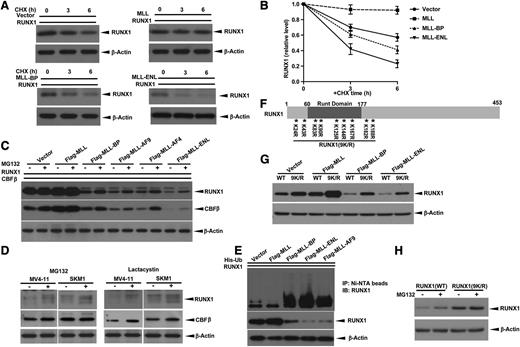
We used cycloheximide to determine whether MLL fusion proteins or MLL-BP alters the half-life of RUNX1 protein. RUNX1 had a shorter half-life in the presence of MLL-BP and an even shorter half-life in the presence of MLL-ENL, compared with the vector control (Figure 2A-B). Conversely, we noted that RUNX1 half-life was prolonged by full-length MLL (Figure 2A-B), consistent with our previous findings.33
Blocking ubiquitin-proteasome pathway does not completely rescue MLL-BP and MLL fusion–mediated RUNX1 downregulation. (A) RUNX1 expression vector was cotransfected with blank vector, full-length MLL, MLL-BP, or MLL-ENL expression vector in 293T cells. At 36 hours’ posttransfection, protein synthesis inhibitor cycloheximide (CHX) was added to a final concentration of 10 μg/mL. Cells were then harvested at indicated time points (0, 3, and 6 hours) and RUNX1 levels were determined by western blotting. β-Actin was used as a loading control. (B) Graph showing relative RUNX1 band densities normalized by β-Actin in panel A. The ratio was set to 1 at time point zero. (C) Immunoblot of transient expression of RUNX1 and CBFβ with blank vector, full-length MLL, MLL-BP, MLL-AF9, MLL-AF4, or MLL-ENL in 293T cells. Cells were treated with dimethylsulfoxide (DMSO) or 10 μmol/L MG132 for 6 hours before harvest. (D) MV4-11 and SKM1 cells were treated with DMSO (solvent, as control), 10 μmol/L MG132, or 10 μmol/L lactacystin for 6 hours. The endogenous RUNX1 and CBFβ proteins were measured by western blotting. β-Actin was used as a loading control. (E) Ubiquitination assay of RUNX1 coexpressed with MLL, MLL-BP, MLL-ENL, and MLL-AF9. (F) Schematic representation of RUNX1 (9K/R) construct used in this study. The positions of 9 lysines in RUNX1 are indicated by asterisks with numbered markers. (G) Immunoblot of wild-type (WT) RUNX1 or RUNX1 (9K/R) in which all lysines were mutated to arginines transiently coexpressed with blank vector, full-length MLL, MLL-BP, MLL-AF9, and MLL-ENL in 293T cells. (H) Immunoblot of RUNX1 in 293T cells transiently expressing WT RUNX1 or RUNX1 (9K/R) treated with DMSO or 10 μmol/L MG132 for 6 hours. All of these experiments were repeated 3 times and representative data are shown.
Blocking ubiquitin-proteasome pathway does not completely rescue MLL-BP and MLL fusion–mediated RUNX1 downregulation. (A) RUNX1 expression vector was cotransfected with blank vector, full-length MLL, MLL-BP, or MLL-ENL expression vector in 293T cells. At 36 hours’ posttransfection, protein synthesis inhibitor cycloheximide (CHX) was added to a final concentration of 10 μg/mL. Cells were then harvested at indicated time points (0, 3, and 6 hours) and RUNX1 levels were determined by western blotting. β-Actin was used as a loading control. (B) Graph showing relative RUNX1 band densities normalized by β-Actin in panel A. The ratio was set to 1 at time point zero. (C) Immunoblot of transient expression of RUNX1 and CBFβ with blank vector, full-length MLL, MLL-BP, MLL-AF9, MLL-AF4, or MLL-ENL in 293T cells. Cells were treated with dimethylsulfoxide (DMSO) or 10 μmol/L MG132 for 6 hours before harvest. (D) MV4-11 and SKM1 cells were treated with DMSO (solvent, as control), 10 μmol/L MG132, or 10 μmol/L lactacystin for 6 hours. The endogenous RUNX1 and CBFβ proteins were measured by western blotting. β-Actin was used as a loading control. (E) Ubiquitination assay of RUNX1 coexpressed with MLL, MLL-BP, MLL-ENL, and MLL-AF9. (F) Schematic representation of RUNX1 (9K/R) construct used in this study. The positions of 9 lysines in RUNX1 are indicated by asterisks with numbered markers. (G) Immunoblot of wild-type (WT) RUNX1 or RUNX1 (9K/R) in which all lysines were mutated to arginines transiently coexpressed with blank vector, full-length MLL, MLL-BP, MLL-AF9, and MLL-ENL in 293T cells. (H) Immunoblot of RUNX1 in 293T cells transiently expressing WT RUNX1 or RUNX1 (9K/R) treated with DMSO or 10 μmol/L MG132 for 6 hours. All of these experiments were repeated 3 times and representative data are shown.
Having demonstrated that RUNX1 can be degraded through the proteasome pathway,8 we tested whether MLL-BP and MLL fusions downregulate RUNX1 protein stability via the proteasome pathway. We found that MG132 can only partially rescue RUNX1/CBFβ from downregulation by MLL-BP and MLL fusion proteins (Figure 2C). We also assessed RUNX1/CBFβ protein levels in the MV4-11 (MLL-AF4+) and SKM1 (no MLL translocation) cell lines using MG132 or another proteasome inhibitor, lactacystin. Both proteasome inhibitors increased endogenous RUNX1 and CBFβ protein levels in these 2 cell lines, suggesting that proteasome inhibitors effectively stabilize RUNX1/CBFβ, regardless of the presence of MLL fusion proteins (Figure 2D). In contrast, several common protease inhibitors did not rescue RUNX1 protein degradation in the MLL fusion expressing the human leukemia cell lines MV4-11 (MLL-AF4+) and THP1 (MLL-AF9+) (supplemental Figure 4). Having shown that MLL decreases the poly-ubiquitination of RUNX1,33 we tested whether MLL-BP or MLL fusion proteins could alter RUNX1 ubiquitination; MLL-BP, MLL-AF9, and MLL-ENL all led to increased poly-ubiquitination of RUNX1 (Figure 2E and supplemental Figure 1F).
Because lysine residues are the target sites for ubiquitination, we generated a mutant RUNX1 construct in which all 9 lysine residues were mutated to arginine residues, RUNX1 (9K/R) (Figure 2F), and examined how RUNX1 (9K/R) protein stability was affected by MLL fusion proteins. As expected, RUNX1 (9K/R) was expressed at higher levels than wild-type RUNX1 (Figure 2G and data not shown). However, RUNX1 (9K/R) could still be stabilized by full-length MLL and be degraded by MLL-BP and MLL-ENL. To rule out the possibility that ubiquitin molecules were attaching to residues other than lysine on RUNX1 (for example, the N-terminal aa), we used MG132 to treat 293T expressing either wild-type RUNX1 or RUNX1 (9K/R). Wild-type RUNX1 protein levels increased when treated with MG132, whereas RUNX1 (9K/R) protein levels were minimally changed in the presence of MG132 (Figure 2H). mRNA expression levels for all samples were unchanged upon MG132 treatment (data not shown). These data provide further evidence that the ubiquitin pathway, although involved in MLL-BP and MLL fusion protein–mediated RUNX1/CBFβ downregulation to some degree, is unlikely to be the major mechanism by which MLL-BP or MLL fusion proteins downregulate RUNX1/CBFβ protein.
MLL’s CXXC domain and flanking region mediate RUNX1 and CBFβ protein downregulation
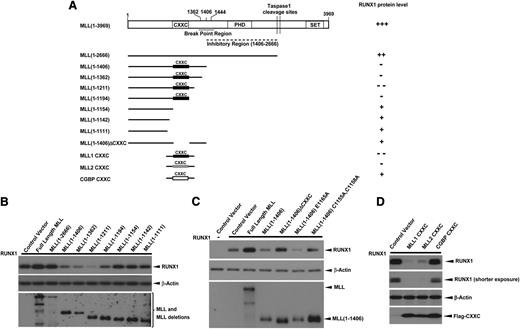
The MLL-BP is the common region for all MLL fusions and is duplicated in MLL-PTD. To address which region of MLL-BP is responsible for downregulation of RUNX1, we constructed a series of MLL deletion mutants (Figure 3A). We found that the MLL constructs—1-1406, 1-1362, 1-1211, and 1-1194—downregulated RUNX1 protein, whereas MLL deletion mutants that lacked the CXXC domain had almost no effect on RUNX1 stability (Figure 3B). This finding strongly suggests that the CXXC domain is required for RUNX1 downregulation. We generated expression constructs in which either the CXXC domain was deleted or contained point mutations that destroy the structure of CXXC zinc finger (C1155A, C1158A).37,38 Each of these constructs lacked the ability to decrease RUNX1 protein levels (Figure 3C), whereas a CXXC mutant of MLL-BP that maintains the CXXC zinc finger structure (E1165A37 ) retained the ability to downregulate RUNX1 (Figure 3C). We also cloned the CXXC domain and flanking regions from MLL1, MLL2/KMT2B, and CGBP (Figure 3A) to determine whether overexpression of only the CXXC domain and flanking regions could downregulate RUNX1/CBFβ. We found that minimal domains from both MLL1 and MLL2/KMT2B downregulate RUNX1/CBFβ significantly, whereas similar domains from CGBP had no effect (Figure 3D).
CXXC domain and flanking regions of MLL are responsible for RUNX1 downregulation. (A) Schematic representations of the major domains of MLL, the C-terminal truncation and internal deletion constructs, and the CXXC domain constructs of MLL1 (1137-1211 aa), MLL2/KMT2B (948-1022 aa), and CGBP (150-225 aa) generated for studying the MLL domains that were responsible for RUNX1 downregulation. (B-D) RUNX1 were cotransfected with blank vector, full-length MLL, or constructs indicated in panel A in 293T cells. At 40 hours’ posttransfection, whole cell lysates were analyzed for expression of RUNX1 and MLL or MLL deletions. β-Actin was used as a loading control. All of these experiments were repeated 3 times and representative data are shown.
CXXC domain and flanking regions of MLL are responsible for RUNX1 downregulation. (A) Schematic representations of the major domains of MLL, the C-terminal truncation and internal deletion constructs, and the CXXC domain constructs of MLL1 (1137-1211 aa), MLL2/KMT2B (948-1022 aa), and CGBP (150-225 aa) generated for studying the MLL domains that were responsible for RUNX1 downregulation. (B-D) RUNX1 were cotransfected with blank vector, full-length MLL, or constructs indicated in panel A in 293T cells. At 40 hours’ posttransfection, whole cell lysates were analyzed for expression of RUNX1 and MLL or MLL deletions. β-Actin was used as a loading control. All of these experiments were repeated 3 times and representative data are shown.
Runx1 and Cbfβ protein levels are downregulated in Mll-Af9 KI mice
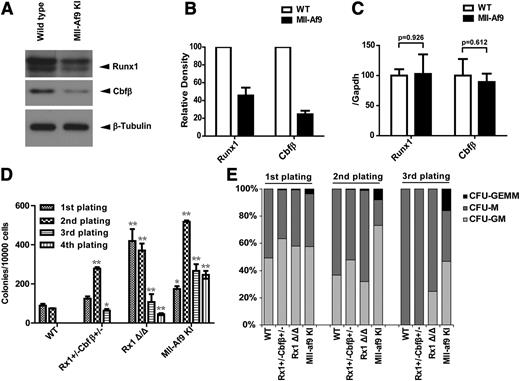
To extend our findings in vivo, LSK (Lin− c-kit+ sca1+) populations were isolated from the BM of wild-type or Mll-Af9 knock-in mice and the protein and mRNA expression levels of Runx1 and Cbfβ were determined. We found Runx1/Cbfβ protein levels were lower in Mll-Af9 knock-in LSK vs wild-type LSK (Figure 4A-B) with no significant change in Runx1/Cbfβ mRNA (Figure 4C), consistent with what we found in human AML cell lines.
Runx1 and Cbfβ are downregulated in Mll-Af9 knock-in mice. (A) Endogenous Runx1 and Cbfβ proteins were determined by western blotting in LSK (Lin− c-kit+ sca1+) populations selected by AutoMac from wild-type (WT) or Mll-Af9 knock-in C57 mice. (B) The band densities of Runx1 and Cbfβ in panel A were quantified relative to β-Actin band intensity. Each bar indicates data from 3 separate experiments. (C) Summary of relative Runx1 and Cbfβ mRNA levels in the mice LSK cells shown in panel A, which were measured by quantitative real-time polymerase chain reaction. All data were normalized to the expression of Gapdh. (D) BM cells from WT, Runx1+/−Cbfβ+/−, Rx1Δ/Δ (Runxflox/flox/Mx1-Cre+ with polyinosinic:polycytidylic acid injected), and Mll-Af9 knock-in mice were analyzed with M3434 methylcellulose-based medium that was formulated to support optimal growth of erythroid progenitors (burst forming unit-erythroid), granulocyte-macrophage (GM) progenitors (CFU-GM, CFU-M, CFU-G), and multipotential granulocyte, erythroid, macrophage, and megakaryocyte progenitors (CFU-GEMM). A total of 1 × 104 BM cells were plated in triplicate in M3434. Colony count scoring and replating were repeated every 7 days (*P < .05; **P < .01). (E) Proportion of CFUs in serial replatings in panel D.
Runx1 and Cbfβ are downregulated in Mll-Af9 knock-in mice. (A) Endogenous Runx1 and Cbfβ proteins were determined by western blotting in LSK (Lin− c-kit+ sca1+) populations selected by AutoMac from wild-type (WT) or Mll-Af9 knock-in C57 mice. (B) The band densities of Runx1 and Cbfβ in panel A were quantified relative to β-Actin band intensity. Each bar indicates data from 3 separate experiments. (C) Summary of relative Runx1 and Cbfβ mRNA levels in the mice LSK cells shown in panel A, which were measured by quantitative real-time polymerase chain reaction. All data were normalized to the expression of Gapdh. (D) BM cells from WT, Runx1+/−Cbfβ+/−, Rx1Δ/Δ (Runxflox/flox/Mx1-Cre+ with polyinosinic:polycytidylic acid injected), and Mll-Af9 knock-in mice were analyzed with M3434 methylcellulose-based medium that was formulated to support optimal growth of erythroid progenitors (burst forming unit-erythroid), granulocyte-macrophage (GM) progenitors (CFU-GM, CFU-M, CFU-G), and multipotential granulocyte, erythroid, macrophage, and megakaryocyte progenitors (CFU-GEMM). A total of 1 × 104 BM cells were plated in triplicate in M3434. Colony count scoring and replating were repeated every 7 days (*P < .05; **P < .01). (E) Proportion of CFUs in serial replatings in panel D.
BM cells with reduced expression of Runx1/Cbfβ exhibit enhanced colony-forming activity
As expected, Mll-Af9 knock-in BM cells or splenocytes can replate in vitro and rapidly develop AML in vivo. In contrast, BM or spleen cells from polyinosinic:polycytidylic acid injected Runx1flox/floxMx1-Cre+ (Runx1Δ/Δ) mice show enhanced replating activity in vitro, but do not develop spontaneous AML.34 To determine whether intermediate levels of Runx1/Cbfβ expression impact colony-replating activity, BM cells from Runx1+/−Cbfβ+/− mice were analyzed in CFU assays (Figure 4D-E). In fact, Runx1+/−Cbfβ+/− BM cells produced significantly more colonies in the second and third plating compared with wild-type BM cells. Runx1Δ/Δ and Mll-Af9 knock-in BM cells also produced significantly more colonies than wild-type BM cells. These data indicate that Runx1+/−Cbfβ+/− BM cells can maintain an immature phenotype more easily than wild-type BM cells.
Runx1 and Cbfβ hypomorph phenotype results in HSPC expansion
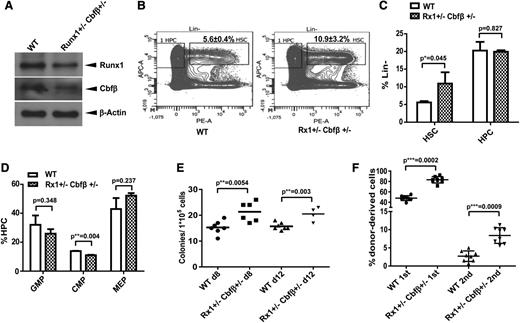
We have demonstrated that MLL translocations result in decreased levels of RUNX1/CBFβ, a hypomorph condition; however, the significance of reduced RUNX1/CBFβ levels has not been examined thus far. We generated Runx1+/−Cbfβ+/− compound heterozygous mice as a Runx1/Cbfβ hypomorph model (Figure 5A). We analyzed hematopoiesis in Runx1+/−Cbfβ+/− mice with a specific focus on the function of HSPC as previously described.39 BM cells from Runx1+/−Cbfβ+/− mice exhibited a higher percentage of LSK cells in the Lin− population as compared with LSK from wild-type mice (Figure 5B-C) and statistically fewer CMP within the hematopoietic cell (HPC, Lin− c-kit+ sca1+) population (P < .01, Figure 5D). The percentage of Lin− c-kit+ sca1− (LK, HPC) cells in the Lin− population as well as the Lin−c-Kit+Sca1−CD34+CD16/32+ and (Lin−c-Kit+Sca1−CD34−CD16/32− percentages in the HPC populations (Figure 5C-D) were comparable between the 2 genotypes.
Runx1 and Cbfβ hypomorph results in HSC expansion. (A) Endogenous Runx1 and Cbfβ proteins were determined by western blotting in LSK populations selected by AutoMac from wild-type or Runx1+/−Cbfβ+/− mice. (B) Flow cytometric analysis of HSC and HPC compartments in WT and Runx1+/−Cbfβ+/− mice. n = 4 mice/group. (C) Summary of HSC (Lin−c-Kit+Sca-1+) and HPC (Lin−c-Kit+Sca-1−) population percentages in Lin− populations in the genotypes described in panels A and B. (D) Summary of GMP (Lin−c-Kit+Sca1−CD34+CD16/32+), CMP (Lin−c-Kit+Sca1−CD34+CD16/32−), and MEP (Lin−c-Kit+Sca1−CD34−CD16/32−) percentages in HPC (lin−c-Kit+Sca-1−) populations in the genotypes described in panels A and B. (E) BM cells from WT or Runx1+/−Cbfβ+/− mice were transplanted into lethally irradiated mouse recipients. The spleens from each group were harvested and homogenized 8 or 12 days after transplantation and placed in Tellesniczky fixative to visualize the colonies. (F) The fractions of WT or Runx1+/−Cbfβ+/− (CD45.2+) BM cells were transplanted into lethally irradiated CD45.1+/CD45.2+ WT recipient mice along with 1 × 105 WT (CD45.1+) helper cells. Engraftment was assessed 16 weeks after transplantation. The mean ± standard deviation percentage of donor-derived wild-type or Runx1+/−Cbfβ+/− (CD45.2+) in recipients’ BM is shown. APC-A, Anti-c-kit-APC; PE-A, Anti-Sca1-PE.
Runx1 and Cbfβ hypomorph results in HSC expansion. (A) Endogenous Runx1 and Cbfβ proteins were determined by western blotting in LSK populations selected by AutoMac from wild-type or Runx1+/−Cbfβ+/− mice. (B) Flow cytometric analysis of HSC and HPC compartments in WT and Runx1+/−Cbfβ+/− mice. n = 4 mice/group. (C) Summary of HSC (Lin−c-Kit+Sca-1+) and HPC (Lin−c-Kit+Sca-1−) population percentages in Lin− populations in the genotypes described in panels A and B. (D) Summary of GMP (Lin−c-Kit+Sca1−CD34+CD16/32+), CMP (Lin−c-Kit+Sca1−CD34+CD16/32−), and MEP (Lin−c-Kit+Sca1−CD34−CD16/32−) percentages in HPC (lin−c-Kit+Sca-1−) populations in the genotypes described in panels A and B. (E) BM cells from WT or Runx1+/−Cbfβ+/− mice were transplanted into lethally irradiated mouse recipients. The spleens from each group were harvested and homogenized 8 or 12 days after transplantation and placed in Tellesniczky fixative to visualize the colonies. (F) The fractions of WT or Runx1+/−Cbfβ+/− (CD45.2+) BM cells were transplanted into lethally irradiated CD45.1+/CD45.2+ WT recipient mice along with 1 × 105 WT (CD45.1+) helper cells. Engraftment was assessed 16 weeks after transplantation. The mean ± standard deviation percentage of donor-derived wild-type or Runx1+/−Cbfβ+/− (CD45.2+) in recipients’ BM is shown. APC-A, Anti-c-kit-APC; PE-A, Anti-Sca1-PE.
We also performed standard CFU-spleen assays to dissect the function of Runx1+/−Cbfβ+/− HSPCs. On days 8 and 12 of a CFU-spleen assay, we found a 39% and 30% increase in CFUs from Runx1+/−Cbfβ+/− BM compared with wild-type BM, respectively (P < .01, Figure 5E). These data indicate hematopoietic stem cell (HSC) expansion within the Runx1+/−Cbfβ+/− BM. We further analyzed the in vivo reconstituting ability of BM cells obtained from Runx1+/−Cbfβ+/− mice. We found these BM cells have greater engraftment potential and long-term reconstitution ability than the control wild-type BM cells in both first and second BMT assays (Figure 5F). These data show that Runx1+/−Cbfβ+/− mice have a significant expansion of phenotypic and functional HSPCs compared with their wild-type littermates. Thus, downregulation of Runx1/Cbfβ could contribute to MLL fusion leukemogenesis not only through blocking terminal differentiation, but also through expanding self-renewal within the HSPC pool, potentiating further accumulation of cooperative mutations and progression to full-blown leukemias.
Restoration of RUNX1 expression inhibits MLL and induces differentiation
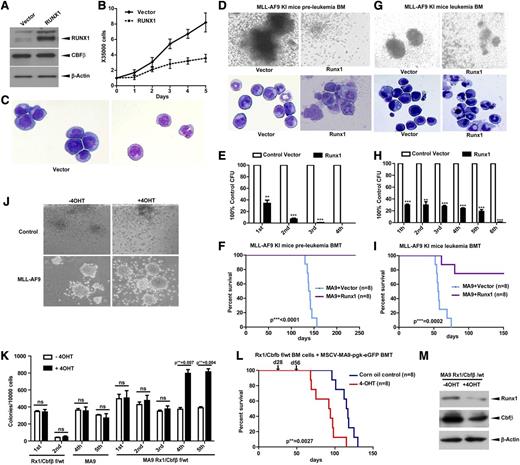
Given that MLLs have low RUNX1 levels, to understand whether restoration of RUNX1 can impair the MLL phenotype, we overexpressed RUNX1 in MV4-11 cells and found cell growth arrest and morphological differentiation of sorted stable transfectants (Figure 6A-C). To validate this finding in an in vivo BMT assay, we used Mll-Af9 knock-in mice and overexpressed RUNX1 in BM cells from preleukemic (Figure 6D-F) or leukemic (Figure 6G-I) mice. Under both conditions, overexpression of RUNX1 resulted in terminal differentiation (Figure 6D,G) and reduced colony-forming ability (Figure 6E,H). More importantly, RUNX1 overexpression in Mll-Af9 knock-in HSPCs completely blocked their leukemic potential in BMT assays (Figure 6F,I).
RUNX1 inhibits AML caused by MLL fusion proteins. (A) The immunoblot of RUNX1 and CBFβ in the MV4-11 cell line infected with recombinant amphotropic retroviruses carrying either pMY blank vector alone or with RUNX1. eGFP+ cells were sorted after infection and used for western blotting. (B) Growth curves of cells obtained in panel A. (C) Morphology of cells obtained in panel A by cytospin. BM cells from preleukemia (D-F) or leukemia (G-I) staged Mll-Af9 knock-in mice were infected with retroviruses carrying either pMY blank vector alone or with pMY-RUNX1. The eGFP+ cells were used for CFU replating assays or transplanted into lethally irradiated mouse recipients. Colony formation of each plating (E,H) and the representative image of colonies (D,G, upper panels) and cytospin (D,G, lower panels) of the first round of plating are shown (**P < .01; ***P < .001). (F,I) The survival curve of transplanted mice. (J-L) Low-density BM cells from Runx1flox/wtCbfβflox/wt/Rosa-Cre-ERT2 mice were infected with retroviruses carrying MLL-AF9, and eGFP+ cells were sorted for CFU plating assays with or without tamoxifen (4-OHT) (J-K) or for BMT (L). Arrows in panel L indicate the time points when 4-OHT or corn oil was injected into the treatment group or control group separately. (M) Endogenous Runx1 and Cbfβ proteins were measured by immunoblotting with leukemic cells in panel J. ns, not significant.
RUNX1 inhibits AML caused by MLL fusion proteins. (A) The immunoblot of RUNX1 and CBFβ in the MV4-11 cell line infected with recombinant amphotropic retroviruses carrying either pMY blank vector alone or with RUNX1. eGFP+ cells were sorted after infection and used for western blotting. (B) Growth curves of cells obtained in panel A. (C) Morphology of cells obtained in panel A by cytospin. BM cells from preleukemia (D-F) or leukemia (G-I) staged Mll-Af9 knock-in mice were infected with retroviruses carrying either pMY blank vector alone or with pMY-RUNX1. The eGFP+ cells were used for CFU replating assays or transplanted into lethally irradiated mouse recipients. Colony formation of each plating (E,H) and the representative image of colonies (D,G, upper panels) and cytospin (D,G, lower panels) of the first round of plating are shown (**P < .01; ***P < .001). (F,I) The survival curve of transplanted mice. (J-L) Low-density BM cells from Runx1flox/wtCbfβflox/wt/Rosa-Cre-ERT2 mice were infected with retroviruses carrying MLL-AF9, and eGFP+ cells were sorted for CFU plating assays with or without tamoxifen (4-OHT) (J-K) or for BMT (L). Arrows in panel L indicate the time points when 4-OHT or corn oil was injected into the treatment group or control group separately. (M) Endogenous Runx1 and Cbfβ proteins were measured by immunoblotting with leukemic cells in panel J. ns, not significant.
Although MLLs have low RUNX1/CBFβ levels, further reducing the level by Runx1 deletion in an MLL-ENL model accelerated the leukemia.40 However, residual RUNX1/CBFβ levels are absolutely required for MLL because complete deletion of both Runx1/Cbfβ blocked MLL formation.41 To understand the gene dosage effect in the context of MLL, we also took advantage of BM cells from the Runx1flox/wt/Cbfβflox/wt/Rosa26-Cre-ERT2 mouse model. HSPCs were transduced with MLL-AF9, sorted for eGFP+ cells, and plated in the presence or absence of 4-hydroxytamoxifen (4-OHT) to induce hypomorphic Runx1/Cbfβ function. Deletion of 1 allele of Runx1 and Cbfβ resulted in significantly more colonies upon replating of MLL-AF9 cells (Figure 6J-K). We also performed BMT assays with the sorted eGFP+ cells. After confirming eGFP+ cell reconstitution in recipient mice, we injected 1 group of mice with tamoxifen and the other with corn oil, and found accelerated AML development in the tamoxifen-injected group (Figure 6L). We confirmed Runx1/Cbfβ gene deletion and protein level changes (supplemental Figures 5 and 6M).
Discussion
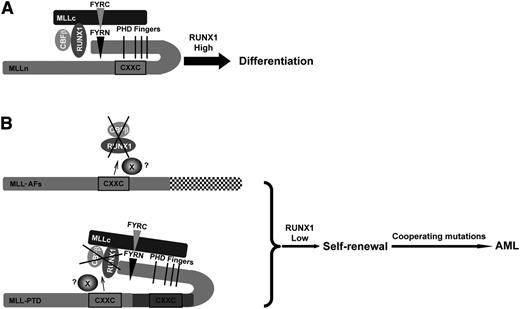
Our study suggests a working model (Figure 7) for wild-type MLL cells, in which the downregulation of RUNX1/CBFβ proteins mediated by the CXXC domain is inhibited by the presence of the PHD finger domains; this results in a normal level of RUNX1/CBFβ that then allows HSC differentiation (Figure 7A). In the presence of MLL fusion proteins or MLL-PTD (Figure 7B), the loss of MLL-AFs or duplication of the CXXC domain (MLL-PTD) leads to downregulation of RUNX1/CBFβ, which promotes self-renewal and differentiation blockade. Additional mutations can then cooperate to cause leukemia.
Working model. (A) The effect of wild-type MLL on the stability of RUNX1/CBFβ results in HSC differentiation. In the state of wild-type MLL, the downregulation of RUNX1/CBFβ mediated by the CXXC domain is intrinsically inhibited by PHD finger domains. High levels of RUNX1 will induce HSC differentiation. (B) MLL oncoproteins lose the ability to inhibit the CXXC domain via the PHD finger domains and the RUNX1/CBFβ protein complex is constitutively downregulated. X, the possible protein or complex that mediates the effect of CXXC domain and flanking region to RUNX1/CBFβ. (Upper panel) In MLL fusion proteins, the PHD finger domains can no longer inhibit the CXXC domain. (Lower panel) MLL-PTD has only a single PHD finger domain, yet still contains 2 CXXC domains, leaving 1 CXXC domain free to downregulate RUNX1/CBFβ constitutively and promote HSC self-renewal. Subsequent mutations then cooperate to lead to AML development.
Working model. (A) The effect of wild-type MLL on the stability of RUNX1/CBFβ results in HSC differentiation. In the state of wild-type MLL, the downregulation of RUNX1/CBFβ mediated by the CXXC domain is intrinsically inhibited by PHD finger domains. High levels of RUNX1 will induce HSC differentiation. (B) MLL oncoproteins lose the ability to inhibit the CXXC domain via the PHD finger domains and the RUNX1/CBFβ protein complex is constitutively downregulated. X, the possible protein or complex that mediates the effect of CXXC domain and flanking region to RUNX1/CBFβ. (Upper panel) In MLL fusion proteins, the PHD finger domains can no longer inhibit the CXXC domain. (Lower panel) MLL-PTD has only a single PHD finger domain, yet still contains 2 CXXC domains, leaving 1 CXXC domain free to downregulate RUNX1/CBFβ constitutively and promote HSC self-renewal. Subsequent mutations then cooperate to lead to AML development.
The leukemogenic 11q23 translocations fuse the MLL N terminus with a wide variety of fusion partners, ranging from nuclear factors to cytoplasmic proteins.16,29,30 More than 70 MLL fusion partners have been identified, and the number is increasing. Many previous studies on MLL fusion proteins have focused on the loss of the MLL C terminus and the “gain of function” provided by the different fusion partners. Our study reveals an unreported function of MLL fusion proteins, mediated by a common protein domain within the NH2 terminus of MLL, that downregulates RUNX1/CBFβ. This property contrasts with our previous demonstration that full-length MLL prevents ubiquitin-proteasome–mediated degradation of RUNX1 by reducing poly-ubiquitination of RUNX1.33
All MLL translocations occur in the BCR of MLL that spans exons 9-11, indicating that all MLL fusion proteins retain the CXXC domain and the adjacent RD2 region while deleting the PHD finger domain and the MLL C terminus including the SET domain. MLL-PTD is another mutant form of MLL, which contains a partial tandem duplication of the MLL N terminus, with the duplicated breakpoints matching the BCR found in MLL translocations (exons 9-11).22,27
Our study indicates that the CXXC domain is the part of the MLL N terminus responsible for the downregulation of RUNX1/CBFβ. The CXXC domain of MLL is a cysteine-rich sequence containing 2 CGXCXXC repeats in its core region (1154-1194 aa), which adopts an extended crescent-like structure coordinating 2 zinc ions.37,42 The CXXC domain of MLL plays a crucial role in myeloid cell transformation, which correlates with its recognition of nonmethylated CpG dinucleotides.37,38 Although the carboxyl-flanking region of the MLL CXXC domain (the “post-CxxC” moiety of MLL, which is rich in basic aa) is not required for binding to CpG sites,37 it also contributes to MLL-associated myeloid transformation.37,43 We found that the MLL deletion construct lacking the CXXC carboxy-flanking region (MLL 1-1194, Figure 3B) could downregulate RUNX1, although not as efficiently as the MLL deletion construct that retains both the core CXXC motif and the post-CXXC region (MLL 1-1211, Figure 3B), suggesting that the CXXC flanking region may play a synergistic role with the core CXXC motif in RUNX1 downregulation either through stabilization of the structure or through a protein–protein interaction.
Recently, the polymerase-associated factor complex (PAFc) has been found to interact with the CXXC-RD2 region in MLL.44 PAFc stimulates MLL and MLL fusion protein—mediated transcriptional activation of HOXA9; disrupting the MLL-PAFc interaction selectively inhibits the growth of MLL-fusion leukemic cells.45 Whether PAFc and its associated complexes and factors (eg, super elongation complex,46 BRE1/RAD6 complex47 ) are also involved in regulating RUNX1/CBFβ will be important to dissect in future studies.
The MLL PHD finger domains region (MLL 1406-2666, shown in Figure 3A as the “inhibitory region” of RUNX1 downregulation) is lost in all MLL fusion proteins. Insertion of PHD finger domains into MLL-ENL or MLL-AF9 suppresses MLL fusion protein–induced HSC immortalization, indicating that the loss of the PHD finger domains in MLL fusion proteins is necessary for leukemogenesis.48,49 In our study, the 300-kDa fragment of MLL generated by taspase cleavage (MLL 1-2666), which retains the CXXC domain and all the PHD finger domains, showed no effect on RUNX1 downregulation (Figure 3B). Thus, the mechanism by which the PHD finger domains block the CXXC effects on RUNX1/CBFβ will be an interesting question for future study.
The downregulation of CBF (RUNX1/CBFβ) is involved in leukemogenesis. It has been shown that depletion of Aml1/Runx1 accelerates the development of MLL-ENL in a mouse BMT model.40 We found that introduction of MLL-BP into wild-type BM cells conferred better replating potential (Figure 1I) and that the hypomorph condition of Runx1/Cbfβ in Runx1+/−Cbfβ+/− mice resulted in HSPC expansion (Figure 5). Conversely, RUNX1/CBFβ upregulation could be a potential therapeutic target for MLLs as an alternative therapy approach.
In our study, overexpression of RUNX1 inhibited the AML development of Mll-Af9 knock-in HSPCs (Figure 6A-I); consistent with this, further reducing of Runx1/Cbfβ levels accelerated Mll-Af9–mediated AML in BMT assays (Figure 6J-M). These results are fully consistent with the tumor suppressor role of Runx1/Cbfβ. At the same time, we recently showed that a certain level of RUNX1 is required for the growth and survival of MLL-AF9 cells,41 and a recent report showed that RUNX1 expression is important for the growth of t(4;11) leukemia cell lines, in which it plays a role in the activation of specific target genes by direct interaction with AF4-MLL.21,50 Taken together, these data argue that RUNX1 level is tightly controlled and low RUNX1/CBFβ activity is critical for leukemogenesis. Therefore, modulating RUNX1/CBFβ dosage, either by overexpression or by further inhibition, could offer a therapeutic strategy for the treatment of MLL.
In summary, we have demonstrated a new mechanism of leukemogenesis mediated by MLL translocations. The common portion of the MLL fusion proteins (the approximately 1400-aa region of the MLL N terminus) can downregulate RUNX1/CBFβ protein; this function is dependent on its CXXC domain. Losing the PHD finger domains exposes this negative regulatory activity. RUNX1/CBFβ hypomorphic mice have HSPC expansion, suggesting that decreased RUNX1/CBFβ dosage could potentially contribute to the initiation and maintenance of AML. Modulating RUNX1 protein levels or RUNX1/CBFβ activity, either up or down, is a new therapeutic strategy for leukemias bearing rearrangements of chromosome 11q23.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr H. Leighton Grimes and Ashish R. Kumar for careful reading and giving of advice on experiments and the manuscript.
This work was supported by grants from Ohio Cancer Research Associates, Cancer Free Kids, Leukemia Research Foundation and Pilot Research Grant of the State Key Laboratory of Experimental Hematology (Tianjin, China) (G.H.), Leukemia Lymphoma Society SCOR grant (S.D.N.), and the National Natural Science Foundation of China grants 81200356 (X.Z.) and 81000220 (F.H.).
Authorship
Contribution: X.Z., X.Y., and G.H. designed the research; X.Z., A.C., X.Y., Y.Z., F.H., Y.H., Y.D., Y.R., B.L., R.M.C., S.E.E., N.H., and G.H. performed research; X.Z., Y.Z., F.H., and G.H. analyzed data; A.M.-D., J.Z., Z.X., W.T., D.G.T., Q.W., W.C., J.C.M., and S.D.N. contributed vital new reagents; and X.Z. and G.H. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gang Huang, Cincinnati Children's Hospital Medical Center, 3333 Burnet Ave, Room S7.607, MLC 7013, Cincinnati, OH 45229-3039; e-mail: gang.huang@cchmc.org.
References
Author notes
X.Z. and A.C. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal