Key Points
Quantitative proteomics identifies BRG as the main ATPase of BAF complexes expressed in leukemia.
BRG is essential for the proliferation of leukemic cells.
In mammals, combinatorial assembly of alternative families of subunits confers functional specificity to adenosine triphosphate (ATP)-dependent SWI/SNF-like Brg/Brm-associated factor (BAF) chromatin remodeling complexes by creating distinct polymorphic surfaces for interaction with regulatory elements and DNA-binding factors. Although redundant in terms of biochemical activity, the core ATPase subunits, BRG/SMARCA4 and BRM/SMARCA2, are functionally distinct and may contribute to complex specificity. Here we show using quantitative proteomics that BAF complexes expressed in leukemia are specifically assembled around the BRG ATPase. Moreover, using a mouse model of acute myeloid leukemia, we demonstrate that BRG is essential for leukemia maintenance, as leukemic cells lacking BRG rapidly undergo cell-cycle arrest and apoptosis. Most importantly, we show that BRG is dispensable for the maintenance of immunophenotypic long-term repopulating hematopoietic stem cells, suggesting that adroit targeting of BRG in leukemia may have potent and specific therapeutic effects.
Introduction
The cancer stem cell (CSC) theory proposes that many cancers are organized as a hierarchy with a rare population of neoplastic cells at the apex that have self-renewal capacity.1 Because self-renewal provides CSCs with capability for long-term clonal maintenance, the model predicts that cure must depend on CSCs eradication in a tumor. Although the validity and universality of the CSC model remains to be established, emerging evidence from acute myeloid leukemia (AML) supports the idea that leukemia stem cell (LSC) properties may be prognostic. For example, recent correlative studies have linked clinical outcome with specific LSC properties, such as cell-surface expression of LSC-specific markers or their capacity to initiate the disease in xenograft models.2 Although, LSCs share common regulatory mechanisms with normal hematopoietic stem cells (HSCs),3,,-6 several of these genes and pathways are often mutated, activated or aberrantly expressed in leukemia. Thus, a therapeutic window may exist, whereby interfering with these mechanisms might ablate LSCs while sparing the normal HSC compartment.
The mouse genome encodes 29 different SWI2/SNF2-like adenosine triphosphatases (ATPases), several of which are assembled into SWI/SNF-like chromatin remodeling complexes. Two of these ATPases, Brahma/Smarca2 (Brm) and Brg/Smarca4, are alternative subunits in a subfamily of 1.5 to 2 MDa complexes termed BAF (Brg/Brm-associated factor) or mSWI/SNF.7 Increasing evidence indicates that combinatorial assembly of alternative families of subunits confers functional specificity to BAF complexes in different tissues and cell-types. Specialized assemblies of BAF complexes have been observed in embryonic stem cells (esBAF), neural progenitors (npBAF) and postmitotic neurons (nBAF),8,,-11 and recent studies have suggested that the choice of alternative ATPases within BAF complexes12 represents a key determinant of complex specificity. Brg null mice die at pre- or peri-implantation stage,13 whereas Brm null animals are viable and fertile,14 indicating nonredundant roles in embryonic development. Distinct functions for BRM- and BRG-based complexes have been observed in smooth muscle development,15 osteoblast differentiation,16 as well as cellular proliferation.14,17,,,,-22 BRM depletion is essential for neoplastic transformation of mouse fibroblasts by various oncogenes and its overexpression is sufficient to revert the RasV12 transformed phenotype.17 In contrast to BRM, BRG is essential for oncogenic transformation of mouse fibroblasts and tumor formation in BAF47 null mice.17,19 Importantly, BRG is not a general proliferation nor survival factor, as BRG is dispensable for embryonic epidermis formation,23 glia proliferation,11 and mouse embryonic fibroblast proliferation and viability.13
Increasing evidence suggests that BAF subunits also play nonredundant and dosage-sensitive roles in normal and leukemic hemopoiesis.24,,,,-29 Based on these findings, we hypothesized that specialized assemblies of BAF complexes may regulate normal and leukemic stem cell activity. Here we show that BAF complexes expressed in leukemia are specifically assembled around the BRG ATPase and that BRG is required for leukemia propagation in a mouse model of AML. Altogether, these studies suggest that manipulating Brg function or downstream effectors may be of therapeutic benefit in AML.
Methods
Mouse strains and genotyping
B6.SJL-Ptprca Pep3b/BoyJ (Pep3B) and C57Bl/6J congenic mice (Jackson Labs) were bred in a specific pathogen-free animal facility at the Institute for Research in Immunology and Cancer (IRIC) in accordance with institutional guidelines. Brgfl/fl mice (backcross 3 in C57Bl/6J background; kindly provided by Dr Pierre Chambon) were crossed with C57Bl/6J Mx1-Cre transgenics (Jackson Labs). Genomic probes and primers used for genotyping are described in the supplemental Data on the Blood Web site.
Generation and in vivo expansion of FLA2 and FLB1 mouse primary leukemias
Generation and characterization of FLA2 and FLB1 leukemias is described in Thorsteinsdottir et al30 and Wilhelm et al.31 Briefly, KLS cells sorted from C57Bl/6J:Pep3B E14.5 fetal liver cells were plated at limiting dilution in 24-well plates containing murine stem cell virus Hoxa9-IRES-Meis1a-pgk-Neo GP+E retroviral producers. After infection, the content of each well was either transferred into methylcellulose cultures to assess leukemia colony forming cell (L-CFC) frequencies or transplanted at limiting dilution into sublethally irradiated congenic recipients to evaluate LSC frequencies, which was estimated to be 1 per 1.4 cells in FLA2 and 1 per 347 in FLB1 leukemia. Beside their different LSC frequencies, FLA2 and FLB1 leukemias did not reveal any significant difference in morphology, chromosomal integrity, cell cycle distribution, immunophenotype, percentage of leukemic blasts (varied between 95% to 98%), and dissemination characteristics (ie, spleen infiltration).31 LSC frequencies remained stable during successive transplantations and on multiple freeze/thaw procedures. For in vivo expansion, FLA2/FLB1 cells were thawed and washed twice in 2% fetal bovine serum (Invitrogen) in phosphate-buffered saline. A total of 2 × 105 cells/mouse was injected into the tail vein of sublethally irradiated (700 cGy) C57Bl/6J mice. Recipient mice were euthanized when moribund (approximately 4 weeks after injection) and leukemic cells collected from femurs and tibias.
Affinity purification of BAF complexes and preparation for two-dimensional liquid chromatography (2DLC)-MS/MS
Affinity purification of BAF complexes was performed as described10 using J1 polyclonal anti-BRG/BRM antibodies, which recognize both ATPases with equal efficiency. Peptide identifications and tandem mass spectrometry (MS/MS) MS/MS spectra using Mascot 2.2.0 (Matrix Science) are available online in ProteoConnections32 (http://www.thibault.iric.ca/proteoconnections/index.php?view=14;project=137;idfview=142). All protein identifications are shown in supplemental Table 1.
Glycerol gradient cosedimentation experiments
FLA2 nuclear extracts were fractionated on a 10% to 30% glycerol gradient as described.10
Generation of hematopoietic chimeras and retroviruses
Production of vesicular stomatitis virus-pseudotyped and ecotropic retroviruses was done as described.4 E14.5 fetal liver cells (Ly5.2+) were infected for 2 days on Hoxa9-IRES-Meis1a-pgk-Neo GP+E retroviral producers and 5 × 105 cells transplanted into the tail veins of sublethally irradiated congenic Pep3B/Ly5.1+ recipients (700 cGy; 5 days before transplantation) together with 5 × 104 Pep3B helper BM cells. Mice were euthanized when moribund and leukemic blasts isolated from the femurs and tibias were either transplanted into secondary hosts (see as follows) or cultured for 48 hours on Cre-green fluorescent protein (GFP) or GFP (control) retroviral producers before plating in methylcellulose to assess leukemic CFC frequencies and generate highly proliferative clones (HPCs). To generate secondary leukemias, 5 × 105 leukemic cells were transplanted into irradiated (700 cGy; 5 days before transplantation) Pep3B mice together with 5 × 104 Pep3B helper BM cells. At day 5 posttransplantation, animals were treated (or not) with polyI:polyC (pIpC) (GE Healthcare Life Sciences) to induce Brg deletion. There was 7 µg pIpC/g body weight injected intraperitoneally (in 200 µL phosphate-buffered saline) followed by 3 injections every 2 days of 6 µg pIpC/g body weight.
Flow cytometry
Flow cytometry acquisitions were performed on an LSRII or fluorescence-activated cell sorter (FACS) Canto II cytometer and sorting experiments were performed on an FACSAria cell sorter (Becton Dickinson). Data were analyzed using FACSDiva (Becton Dickinson) and ModFit LT (Verity Software House) software.
Colony assays and generation of HPCs
Methylcellulose cultures of mouse myeloid clonogenic progenitors were done as described.4 For generating HPCs, myeloid progenitors derived from methylcellulose cultures of leukemic bone marrow cells isolated from primary recipients were plucked on day 6 and expended in vitro in liquid culture media containing Dulbecco’s modified Eagle medium (Invitrogen), 15% fetal calf serum (pre-selected; Invitrogen), 6 ng/mL Interleukin (IL)-3, 10 ng/mL IL-6, 100 ng/mL Steel factor and 100 ng/mL IL-11 (all generated and titrated at IRIC from COS cell supernatants) and 1 × 10−5 M β-Mercaptoethanol. Clones that could generate more than 1010 progeny cells are referred to as HPCs.
RNA-sequencing (RNA-seq)
Human bone marrow and blood samples were collected by the Quebec Leukemia Cell Bank with the approval the Research Ethics Board of the Maisonneuve-Rosemont Hospital affiliated with Université de Montréal. Informed consent was provided according to the Declaration of Helsinki. Total RNA from ∼5 million cells was isolated using Trizol (Roche). Paired-end sequencing was performed on an Illumina HiSeq2000 running TruSeq v3 chemistry. Numerical values are shown in supplemental Tables 3-4.
Quantitative real-time polymerase chain reaction (RT-PCR) analyses
Total RNA was isolated from 20 000 freshly sorted cells using Trizol (Roche). Reactions were done in triplicate on a high-throughput ABI 7900HT Fast Real-Time PCR System (Applied Biosystems). Specificity of all oligos used in these studies was tested on complementary DNAs encoding family members. Oligo sequences and numerical values are shown in supplemental Tables 6-7.
Statistical analyses
The significance of differences was determined by a one-tailed Student t test. Statistical significance is indicated by P < .05 and P < .005.
Results
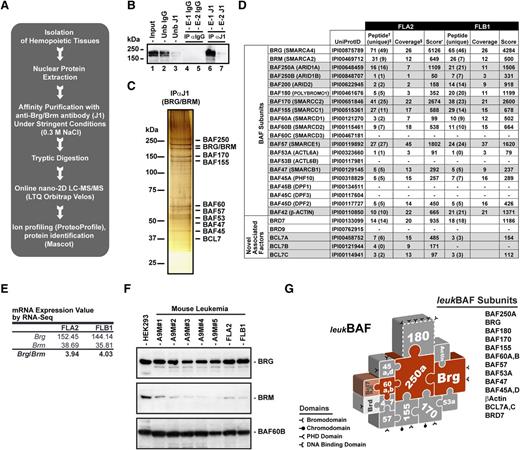
To determine the subunit composition of BAF complexes expressed in leukemia, label-free quantitative proteomics analyses32 of affinity-purified BAF complexes were performed in 2 Hoxa9 and Meis1a (A9M)-induced mouse primary leukemias4,30 exhibiting different leukemia-initiating cell frequencies, ranging from 1 in 1.4 (∼71%) for FLA2 to 1 in 347 (∼0.3%) for FLB132 (Figure 1 and supplemental Tables 1-2). Anti-BRG/BRM (J1) immunoprecipitates isolated from freshly isolated (in vivo expanded) FLA2 and FLB1 leukemias were analyzed by nano two-dimensional liquid chromatography tandem mass spectrometry (nano-2D LC-MS/MS; Figure 1A-C). Comparative MS analysis yielded 78 unambiguously identified proteins with a false discovery rate <2% that included most (16 of 20) family members of the 10 core subunits of BAF complexes (Figure 1D and supplemental Table 1). In both types of leukemia, complexes are mainly assembled on BRG ATPase, as shown by the relative number of unique peptides identified for BRG vs BRM (Figure 1D). No peptides from the BAF45B/DPF1, BAF45C/DPF3, BAF53B/ACTL6B, and BAF60C/SMARCD3 proteins were identified, indicating that leukemic BAF complexes specifically lack these family subunits (Figure 1D). Label-free quantitative proteomics analyses indicated that nonalternative subunits (BAF47/SMARCB1 and BAF57/SMARCE1) were present at similar levels in FLA2 and FLB1 complexes, whereas the relative abundance of alternative subunits varied less than 4-fold (supplemental Figure 1). Glycerol gradient cosedimentations of FLA2 nuclear extracts confirmed the subunit composition of leukemic BAF complexes identified by mass spectrometry, including the prevalence of BRG ATPase over BRM (supplemental Figure 2). Western blot analyses of a panel of A9M-induced mouse AMLs and transcriptomics studies of FLA2 and FLB1 leukemia32 confirmed the relative abundance of BRG ATPase over BRM in leukemic cells (Figure 1E-F). Altogether, these studies indicated that BAF complexes expressed in leukemia are assembled around the BRG ATPase and have a unique subunit composition that is not seen in other normal hematopoietic lineages (supplemental Figure 3), cell types (brain, embryonic stem cells, and mouse embryonic fibroblasts),8,10 or cell lines.7 Major subunit differences included undetectable levels of BAF45B/DPF1, BAF45C/DPF3, and BAF53B/ACTL6B that appear to be specifically expressed in postmitotic neurons.8 For simplicity, we refer to SWI/SNF-like complexes expressed in acute myeloid leukemia as leukBAF complexes (Figure 1G).
Leukemic BAF complexes are assembled around BRG ATPase. (A) Strategy for purification and sequencing of BAF complexes expressed in leukemia. Highly stringent conditions were used to isolate only the core components of the complexes and tightly associated factors. See supplemental Table 1 for all identifications. (B) Immunoprecipitation and western blot analysis of BRG/BRM ATPase subunits in nuclear extracts isolated from FLA2 leukemia using anti-BRG/BRM (J1) antibodies, which recognize both ATPases with equal efficiency. (C) Silver-stain analysis of immunopurified (anti-BRG/BRM) BAF complexes in nuclear extracts isolated from FLA2 leukemia. (D) Subunit composition of leukemic BAF (leukBAF) complexes identified by a proteomics approach in FLA2 and FLB1 leukemia. Frequencies of leukemia-initiating cells are 1 in 1.4 and 1 in 347, respectively. †Total number of different peptides for each protein entry. ‡Number of specific peptides corresponding to a unique protein entry in IPI mouse database. §Sequence coverage (%). •Mascot score with false discovery rate <2%. (E) Relative abundance of Brg and Brm messenger RNAs (mRNAs) in FLA2 and FLB1 leukemia based on adjusted reads by coverage obtained by RNA-seq.32 (F) Relative abundance of BRG and BRM ATPases in nuclear extracts isolated from various mouse A9M-derived leukemias using anti-BRG and BRM specific antibodies. BAF60B is a loading control. HEK293 cells were used as a positive control for BRM expression. (G) Schematic representation of leukBAF complexes assembled on BRG ATPase. Complexes are drawn in a jigsaw puzzle configuration to denote the apparent fit of the subunits within the complexes. These positions have not been experimentally defined, except for actin and BAF53, which contact the catalytic domain of BRG. Subunits shown in dashed outline are inconstant components of the complexes. Domains that bind DNA or modified histones and hence could target the complexes to specific loci independent of (or in cooperation with) transcription factors are shown. 2DLC-MS/MS, two-dimensional liquid chromatography-tandem mass spectrometry; Ub, unbound fraction; IgG, immunoglobulin; E-1 and −2, elution 1 and 2, respectively. See also supplemental Figures 1-3 and supplemental Tables 1-2.
Leukemic BAF complexes are assembled around BRG ATPase. (A) Strategy for purification and sequencing of BAF complexes expressed in leukemia. Highly stringent conditions were used to isolate only the core components of the complexes and tightly associated factors. See supplemental Table 1 for all identifications. (B) Immunoprecipitation and western blot analysis of BRG/BRM ATPase subunits in nuclear extracts isolated from FLA2 leukemia using anti-BRG/BRM (J1) antibodies, which recognize both ATPases with equal efficiency. (C) Silver-stain analysis of immunopurified (anti-BRG/BRM) BAF complexes in nuclear extracts isolated from FLA2 leukemia. (D) Subunit composition of leukemic BAF (leukBAF) complexes identified by a proteomics approach in FLA2 and FLB1 leukemia. Frequencies of leukemia-initiating cells are 1 in 1.4 and 1 in 347, respectively. †Total number of different peptides for each protein entry. ‡Number of specific peptides corresponding to a unique protein entry in IPI mouse database. §Sequence coverage (%). •Mascot score with false discovery rate <2%. (E) Relative abundance of Brg and Brm messenger RNAs (mRNAs) in FLA2 and FLB1 leukemia based on adjusted reads by coverage obtained by RNA-seq.32 (F) Relative abundance of BRG and BRM ATPases in nuclear extracts isolated from various mouse A9M-derived leukemias using anti-BRG and BRM specific antibodies. BAF60B is a loading control. HEK293 cells were used as a positive control for BRM expression. (G) Schematic representation of leukBAF complexes assembled on BRG ATPase. Complexes are drawn in a jigsaw puzzle configuration to denote the apparent fit of the subunits within the complexes. These positions have not been experimentally defined, except for actin and BAF53, which contact the catalytic domain of BRG. Subunits shown in dashed outline are inconstant components of the complexes. Domains that bind DNA or modified histones and hence could target the complexes to specific loci independent of (or in cooperation with) transcription factors are shown. 2DLC-MS/MS, two-dimensional liquid chromatography-tandem mass spectrometry; Ub, unbound fraction; IgG, immunoglobulin; E-1 and −2, elution 1 and 2, respectively. See also supplemental Figures 1-3 and supplemental Tables 1-2.
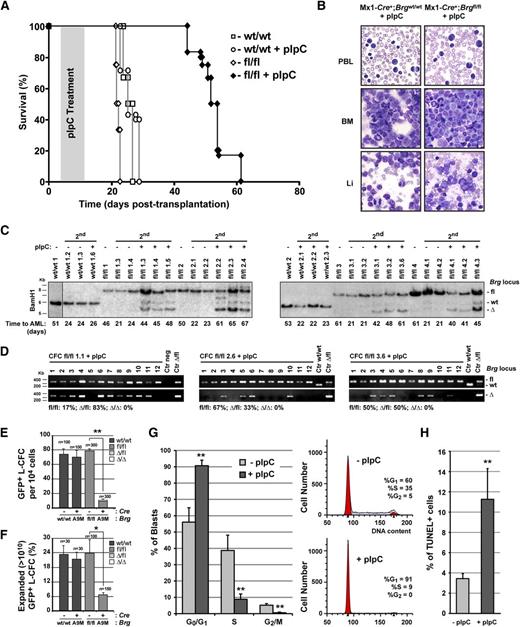
To address the importance of BRG ATPase in leukemia, we generated Brg conditional (Mx1-Cre+;Brgfl/fl) A9M-leukemia in E14.5 fetal liver cells and treated recipient mice with pIpC to induce Brg deletion (supplemental Figures 4 and 5A). Although control (treated or not; groups A and C, respectively) and untreated Mx1-Cre+;Brgfl/flA9M (group B) mice came down with AML within 22 ± 1 days posttransplantation, pIpC-treated Mx1-Cre+;Brgfl/flA9M (group D) mice survived as long as 70 days posttransplantation, but eventually also came down with AML (Figure 2A and supplemental Figure 5B-C). Cytological analyses of leukemic cells isolated from all 4 groups of mice at time of death indicated that the leukemias were morphologically indistinguishable (Figure 2B). In all cases, we observed massive infiltration of leukemic blasts in all hematopoietic and nonhematopoietic organs analyzed that showed strong expression of the myeloid Mac-1 (95%) and Gr-1 (11% to 35%) surface antigens (Figure 2B and data not shown). Western blot analyses revealed near identical levels of BRG in nuclear extracts isolated from pIpC-treated Mx1-Cre+;Brgfl/fl, and control (untreated) A9M animals (supplemental Figure 5D). This observation prompted us to investigate the possibility that Brg deletion is not permissive for leukemia development. To first address the kinetics of Brg allele recombination upon pIpC treatment, we performed a time-course experiment in Mx1-Cre+;Brgfl/flA9M secondary animals (supplemental Figure 5E). In untreated animals, the leukemias rapidly developed after transplantation, as shown by the relative abundance of the Brgfl allele (donor/leukemia) over Brgwt (recipient) in their BM at time of sacrifice. In pIpC-treated animals, maximal recombination of the Brgfl allele was achieved as early as 24 hours after the first injection. The leukemias remained in the BrgΔ configuration up to 7 days posttreatment, but then progressively became Brgfl. The progressive accumulation of the Brgwt allele during the pIpC treatment period (8 days) is indicative of the rapid regression of the leukemias, probably due to a proliferative disadvantage of the Brg-deficient leukemic cells (supplemental Figure 5E). Leukemia regression was also evident from the ratio of Ly5.2+ (donor/leukemia) over Ly5.1+ (recipient) BM cells determined by cytofluorometric analyses at similar time points (data not shown).
BRG is essential for the proliferation of leukemic cells. (A) Survival curves of secondary leukemic Mx1-Cre+;Brgfl/fl and Mx1-Cre;Brgwt/wt (control) A9M mice treated or not with polyI:polyC (pIpC). (B) Cytological preparations of peripheral blood leukocytes (PBL), bone marrow (BM), and liver (Li) of representative secondary leukemic mice at time of death. (C) Southern blot analysis of Brg locus configuration in primary and secondary Mx1-Cre+;Brgfl/fl and control A9M leukemias at time of death. The vertical line indicates a repositioned gel lane. (D) Brg locus configuration of leukemic colony-forming cells (L-CFCs) derived from 3 independent pIpC-treated Mx1-Cre+;Brgfl/fl (group D) primary leukemias at time of death as determined by polymerase chain reaction. (E) Frequency of L-CFCs derived from Cre-GFP and GFP (control) transduced Brgfl/fl primary leukemia isolated at time of death (n = 50 to 100 GFP+ L-CFC per leukemia were scored; n = 2 to 3 leukemias per group). Mean ± SD. *P < .05. (F) Frequency of highly proliferative clones (HPCs) derived from Brgfl/fl L-CFCs infected with Cre-GFP or GFP (control) (n = 15-75 HPCs analyzed per leukemia; n = 2 primary leukemias per group). Mean ± SD. *P < .05. (G) Cell cycle analysis of donor-derived (Ly5.2+) Mx1-Cre+;Brgfl/fl primary leukemias isolated on days 2 to 4 after pIpC or mock treatment using propidium iodide (PI) staining (n = 3 to 5 mice per group). Mean ± SD. **P < .005. (H) Apoptotic index of donor-derived (Ly5.2+) Mx1-Cre+;Brgfl/fl primary leukemias isolated on days 2 to 4 after pIpC or mock treatment as determined by terminal deoxynucleotidyltransferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL) assay (n = 3 to 5 mice per group). Mean ± SD. **P < .005. 2nd, secondary leukemia; L-CFC, leukemic colony-forming-cell; HPC, highly proliferative clone. See also supplemental Figures 4-8.
BRG is essential for the proliferation of leukemic cells. (A) Survival curves of secondary leukemic Mx1-Cre+;Brgfl/fl and Mx1-Cre;Brgwt/wt (control) A9M mice treated or not with polyI:polyC (pIpC). (B) Cytological preparations of peripheral blood leukocytes (PBL), bone marrow (BM), and liver (Li) of representative secondary leukemic mice at time of death. (C) Southern blot analysis of Brg locus configuration in primary and secondary Mx1-Cre+;Brgfl/fl and control A9M leukemias at time of death. The vertical line indicates a repositioned gel lane. (D) Brg locus configuration of leukemic colony-forming cells (L-CFCs) derived from 3 independent pIpC-treated Mx1-Cre+;Brgfl/fl (group D) primary leukemias at time of death as determined by polymerase chain reaction. (E) Frequency of L-CFCs derived from Cre-GFP and GFP (control) transduced Brgfl/fl primary leukemia isolated at time of death (n = 50 to 100 GFP+ L-CFC per leukemia were scored; n = 2 to 3 leukemias per group). Mean ± SD. *P < .05. (F) Frequency of highly proliferative clones (HPCs) derived from Brgfl/fl L-CFCs infected with Cre-GFP or GFP (control) (n = 15-75 HPCs analyzed per leukemia; n = 2 primary leukemias per group). Mean ± SD. *P < .05. (G) Cell cycle analysis of donor-derived (Ly5.2+) Mx1-Cre+;Brgfl/fl primary leukemias isolated on days 2 to 4 after pIpC or mock treatment using propidium iodide (PI) staining (n = 3 to 5 mice per group). Mean ± SD. **P < .005. (H) Apoptotic index of donor-derived (Ly5.2+) Mx1-Cre+;Brgfl/fl primary leukemias isolated on days 2 to 4 after pIpC or mock treatment as determined by terminal deoxynucleotidyltransferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL) assay (n = 3 to 5 mice per group). Mean ± SD. **P < .005. 2nd, secondary leukemia; L-CFC, leukemic colony-forming-cell; HPC, highly proliferative clone. See also supplemental Figures 4-8.
To investigate the possibility that Brg deletion is incompatible with leukemia development, we examined the Brg loci configuration of the leukemias at the time of death. In all 4 groups of mice, both primary and secondary leukemias were derived from 2 to 4 leukemic clones (supplemental Figure 5C). However, the relative abundance of the BrgΔ allele was significantly weaker than the Brgfl allele in pIpC-treated Mx1-Cre+;Brgfl/flA9M (group D) secondary animals, suggesting that the leukemias were mainly derived from nondeleted Brgfl/fl cells (Figure 2C). Clonal analyses performed on leukemic colony-forming cells (L-CFCs) isolated from pIpC-treated group D animals at time of death indicated that all leukemic clones were either in the Brgfl/fl or Brgfl/Δ configuration and not a single BrgΔ/Δ clone was identified (Figure 2D). Although we did not observe a direct correlation between the Brgfl/BrgΔ allelic ratio of the leukemias and the time to AML, we identified clones that rapidly disappeared in group D animals upon pIpC-treatment, likely corresponding to BrgΔ/Δ clones (see supplemental Figure 5C). Altogether, these studies suggested that Brg is absolutely required for the maintenance of leukemia.
To explain the apparent exhaustion of the BrgΔ/Δ leukemic clones, we monitored their survival and growth characteristics in vitro and in vivo, by infecting Brgfl/fl A9M primary leukemias with retroviruses expressing either Cre-GFP or GFP (control) (supplemental Figures 5A and 6). The cellularity, survival, and L-CFC content of Cre-GFP-infected cultures was significantly reduced compared with control cultures and BrgΔ/Δ L-CFCs could not be derived (Figure 2E and supplemental Figure 6). To further investigate the proliferative impairment of Brg-deficient leukemia, we attempted to derive highly proliferative clones (HPCs; more than 1010 cells) from Cre-GFP infected Brgfl/fl L-CFCs. Although 15% of control (GFP+) L-CFCs generated HPCs, it was impossible to derive a single BrgΔ/Δ HPC from Cre-GFP infected L-CFCs (Figure 2F). DNA content analysis of leukemias isolated from Mx1-Cre+;Brgfl/flA9M (group D) animals revealed an accumulation of cells in the G0/G1 phases of the cell cycle, and a concomitant decrease in the percentage of S and G2/M cells relative to control (group B) leukemias (Figure 2G). Furthermore, the proportion of apoptotic cells was significantly increased in leukemias isolated from Mx1-Cre+;Brgfl/flA9M (group D) animals compared with controls (Figure 2H). Together, these studies indicated that Brg is essential for cell cycle progression and survival of leukemic cells. Moreover, using a short hairpin RNA-mediated knockdown strategy, we showed that BRG is required for the proliferation and survival of 3 human leukemia cell lines (both AML and acute lymphoblastic leukemias subtypes) originating from distinct oncogenic events, suggesting that BRG-dependency may be generalized to other types of leukemia (supplemental Figure 7).
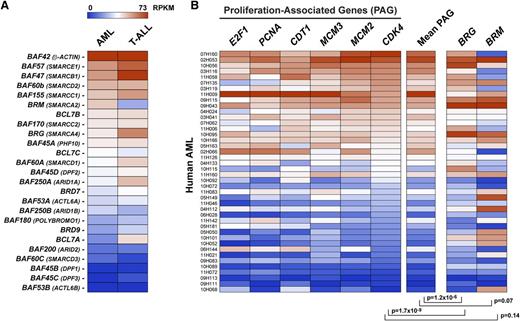
To investigate the evolutionarily conservation of leukBAF complexes, we next analyzed BAF gene expression in 40 human-derived acute myeloid leukemias (38 of 40 displaying a normal karyotype [NK]-AMLs) and 12 human T-cell lymphoblastic leukemias by an RNA-seq approach (supplemental Tables 3-4). Consistent with our findings in mouse models of AMLs (Figure 1), expression of BAF45B/C, BAF53B, BAF60C, and BAF200/ARID2 was low to undetectable in both types of human leukemia, indicating that human leukBAF complexes specifically lack these subunits (Figure 3A). Strikingly, BRG mRNA levels correlated with the proliferative activity of the human AMLs, as evaluated from the expression of several proliferation-associated genes (Figure 3B; P < 1.2 × 10−6).
BRG levels correlate with the proliferative activity of human acute myeloid leukemias (AMLs). (A) BAF gene expression in human AMLs and T-cell acute lympholastic leukemias (T-ALLs) based on adjusted reads by coverage obtained by RNA-seq. Mean values of 40 AMLs and 12 T-ALLs are shown. (B) BRG levels correlate with the proliferative activity of human AMLs based on adjusted reads by coverage obtained by RNA-seq. AMLs were ordered according to CDK4 expression levels. E2F1, PCNA, CDT1, and MCM2/3 proliferation-associated genes (PAGs) are used as comparative controls. Note that all AMLs had normal karyotypes, except leukemias identified as 07H160 and 10H068, which had monosomy 7 and 11p deletion, respectively. See also supplemental Tables 3-4.
BRG levels correlate with the proliferative activity of human acute myeloid leukemias (AMLs). (A) BAF gene expression in human AMLs and T-cell acute lympholastic leukemias (T-ALLs) based on adjusted reads by coverage obtained by RNA-seq. Mean values of 40 AMLs and 12 T-ALLs are shown. (B) BRG levels correlate with the proliferative activity of human AMLs based on adjusted reads by coverage obtained by RNA-seq. AMLs were ordered according to CDK4 expression levels. E2F1, PCNA, CDT1, and MCM2/3 proliferation-associated genes (PAGs) are used as comparative controls. Note that all AMLs had normal karyotypes, except leukemias identified as 07H160 and 10H068, which had monosomy 7 and 11p deletion, respectively. See also supplemental Tables 3-4.
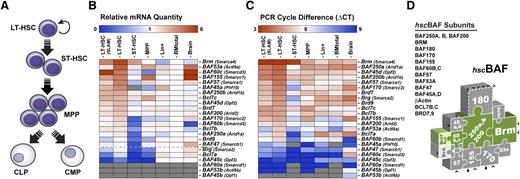
Several studies have demonstrated the potential importance of ablating LSCs when treating leukemia. The unique characteristics of LSCs that differentiate them from their normal counterparts can be exploited to specifically target the malignant population.33,34 To get insights into the composition of BAF complexes expressed in the mainly quiescent long-term repopulating hemopoietic stem cells (LT-HSCs),35 we measured BAF mRNA levels in purified BM populations by quantitative RT-PCR (Figure 4 and supplemental Tables 5-7). In contrast to leukBAF complexes in which Brg is the dominant ATPase (Figures 1 and 3A), quiescent LT-HSCs show highest levels of the alternative Brm ATPase (both in term of relative quantity and ΔCt values), with close to a 6-fold enrichment over total BM cells (Figure 4B-C). Brm mRNA levels progressively decrease in short-term HSCs (ST-HSCs), multipotent progenitors and lineage positive cells and are barely detectable in FLA2 and FLB1 leukemia (Figure 4B-C and data not shown). Quiescent LT-HSCs also show high levels of BAF45a/Phf10, BAF45d/Dpf2, BAF53a/Actl6a, BAF60b/Smarcd2, BAF60c/Smarcd3, and Bcl7b/c, whereas expression of the alternative homologous BAF45b/Dpf1, BAF45c/Dpf3, BAF53b/Actl6b, BAF60a/Smarcd1, and Bcl7a genes is weak to undetectable (Figure 4B-C). These studies indicated that quiescent LT-HSCs expressed distinctive SWI/SNF-like BAF complexes defined by the presence of BRM, BAF45A/D, BAF53A, and BAF60B/C subunits, and the absence of the alternative BRG, BAF45B/C, BAF53B, and BAF60A family members. For simplicity, we refer to BAF complexes expressed in quiescent LT-HSCs as hscBAF complexes (Figure 4D).
BAF complexes expressed in quiescent LT-HSCs are assembled around the BRAHMA (BRM) ATPase. (A) Developmental hierarchy of the (B-C) BM populations analyzed. (B) BAF gene expression in purified populations of mouse BM cells, as determined by quantitative reverse-transcription polymerase chain reaction. Relative quantity (RQ) values represent the relative expression of each gene in the different populations analyzed over total BM cells after GAPDH normalization. BAF genes were ordered according to their expression level in quiescent LT-HSCs. Oligo specificity was tested on complementary DNAs encoding alternative family members. Gray boxes: undetectable expression. Brain: mouse embryonic brain (E14.5). Purity of all sorted population was evaluated to 98% to 99% upon reanalysis. Frequency of all sorted long-term repopulating hemopoietic stem cells (LT-HSC) populations was 1 in 4. (C) ΔCT values for each gene were determined relative to GAPDH in each population (done in triplicate). (D) Schematic representation of BAF complexes expressed in quiescent LT-HSCs (hscBAF) assembled around the BRM ATPase. See also supplemental Tables 5-7. BM, bone marrow; δCT, delta cycle threshold; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
BAF complexes expressed in quiescent LT-HSCs are assembled around the BRAHMA (BRM) ATPase. (A) Developmental hierarchy of the (B-C) BM populations analyzed. (B) BAF gene expression in purified populations of mouse BM cells, as determined by quantitative reverse-transcription polymerase chain reaction. Relative quantity (RQ) values represent the relative expression of each gene in the different populations analyzed over total BM cells after GAPDH normalization. BAF genes were ordered according to their expression level in quiescent LT-HSCs. Oligo specificity was tested on complementary DNAs encoding alternative family members. Gray boxes: undetectable expression. Brain: mouse embryonic brain (E14.5). Purity of all sorted population was evaluated to 98% to 99% upon reanalysis. Frequency of all sorted long-term repopulating hemopoietic stem cells (LT-HSC) populations was 1 in 4. (C) ΔCT values for each gene were determined relative to GAPDH in each population (done in triplicate). (D) Schematic representation of BAF complexes expressed in quiescent LT-HSCs (hscBAF) assembled around the BRM ATPase. See also supplemental Tables 5-7. BM, bone marrow; δCT, delta cycle threshold; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
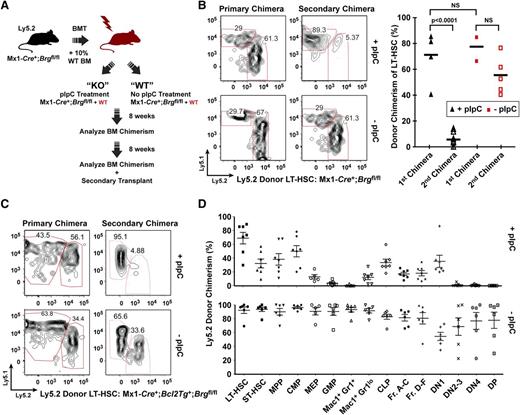
To verify whether Brg is dispensable for the function of the mainly quiescent35 LT-HSCs, we generated Mx1-Cre+;Brgfl/fl, and control bone marrow chimeras, and we deleted Brg using pIpC treatment (Figure 5A and supplemental Figure 8A-G). We chose this approach because constitutive loss of Brg is embryonic lethal13 and its acute deletion in Mx1-Cre+;Brgfl/fl primary animals leads to BM failure within 2 weeks post-pIpC treatment (data not shown). As shown in Figure 5, the frequency of Brg-deficient LT-HSCs analyzed 12 to 16 weeks after deletion remained unchanged, indicating that Brg does not impact the frequency of immunophenotypic quiescent LT-HSCs (Figure 5B,D). The frequencies of ST-HSCs, multipotent progenitors and common myeloid and lymphoid progenitors were reduced between 2- and 3-fold upon deletion of Brg, whereas more mature myeloid and lymphoid progenitors were severely affected (Figure 5D and supplemental Figures 9-10). Interestingly, Brg-deficient LT-HSCs and downstream progenitors were significantly reduced when forced to proliferate after their transplantation into myeloablated secondary mice (Figure 5B), even upon overexpression of the Bcl2 anti-apopototic gene (Figure 5C and supplemental Figure 9C), indicating that Brg is dispensable for the maintenance of LT-HSCs, but essential for their full proliferative activity and multipotentiality. More broadly, these studies suggest an unanticipated degree of functional specialization of BAF complexes in quiescent vs proliferative HSCs (either normal or leukemic) linked with the choice of alternative ATPases (Figure 6).
Brg is dispensable for LT-HSC maintenance but is required for the full proliferative activity of LT-HSCs and downstream progenitors. (A) Schematic of transplant strategy used to achieve bone-marrow-specific deletion of Brg using a Mx1-Cre transgene. Donor and recipient bone marrow is distinguished by the expression of different alleles of the cell-surface marker Ly5. (B) Frequency of donor-derived (Ly5.2+) LT-HSCs in polyI:polyC (pIpC)-treated Mx1-Cre+;Brgfl/fl and control hematopoietic chimeras as analyzed 16 weeks (primary) and 12 weeks (secondary) after transplantation. Representative fluorescence-activated cell sorter (FACS) profiles are shown (left). LT-HSCs were CD34- Flk2- Sca1+ Lin- c-kit+. (C) Donor bone marrow containing the H2k-Bcl2 transgene show a similar impairment in LT-HSC to short-term hematopoietic stem cell (ST-HSC) transition after Brg deletion 16 weeks (primary) and 12 weeks (secondary) post-pIpC treatment. Representative FACS profiles are shown. (D) Donor (Ly5.2) versus host (Ly5.1) chimerism of bone marrow HSC and progenitor populations 18 weeks after pIpC-induced deletion of Brg (upper panel) or mock treatment (lower panel) (n = 5 to 6 mice per group). CLP, common lymphoid progenitor; CMP, common myeloid progenitor; DN, double negative; DP, double positive; Fr, fraction; GMP, granulocyte-macrophage progenitor; LT-HSC, long-term hemopoietic stem cells; MEP, megakaryocyte-erythrocyte progenitor; MPP, multipotent progenitor; NS, nonsignificant. See also supplemental Figures 8-10.
Brg is dispensable for LT-HSC maintenance but is required for the full proliferative activity of LT-HSCs and downstream progenitors. (A) Schematic of transplant strategy used to achieve bone-marrow-specific deletion of Brg using a Mx1-Cre transgene. Donor and recipient bone marrow is distinguished by the expression of different alleles of the cell-surface marker Ly5. (B) Frequency of donor-derived (Ly5.2+) LT-HSCs in polyI:polyC (pIpC)-treated Mx1-Cre+;Brgfl/fl and control hematopoietic chimeras as analyzed 16 weeks (primary) and 12 weeks (secondary) after transplantation. Representative fluorescence-activated cell sorter (FACS) profiles are shown (left). LT-HSCs were CD34- Flk2- Sca1+ Lin- c-kit+. (C) Donor bone marrow containing the H2k-Bcl2 transgene show a similar impairment in LT-HSC to short-term hematopoietic stem cell (ST-HSC) transition after Brg deletion 16 weeks (primary) and 12 weeks (secondary) post-pIpC treatment. Representative FACS profiles are shown. (D) Donor (Ly5.2) versus host (Ly5.1) chimerism of bone marrow HSC and progenitor populations 18 weeks after pIpC-induced deletion of Brg (upper panel) or mock treatment (lower panel) (n = 5 to 6 mice per group). CLP, common lymphoid progenitor; CMP, common myeloid progenitor; DN, double negative; DP, double positive; Fr, fraction; GMP, granulocyte-macrophage progenitor; LT-HSC, long-term hemopoietic stem cells; MEP, megakaryocyte-erythrocyte progenitor; MPP, multipotent progenitor; NS, nonsignificant. See also supplemental Figures 8-10.
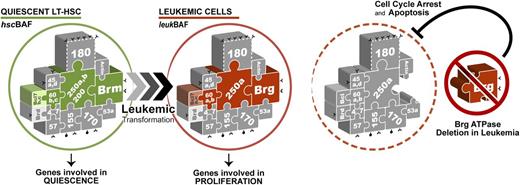
Proposed model for Brg ATPase function in leukemia. BAF complexes expressed in quiescent long-term hematopoietic stem cells (LT-HSCs) (hscBAF) are assembled around the BRM ATPase, whereas leukemic BAF complexes (leukBAF) specifically contain the BRG ATPase. BRG is dispensable for long-term HSC maintenance, but is required for the full proliferative activity of both normal and leukemic HSCs.
Proposed model for Brg ATPase function in leukemia. BAF complexes expressed in quiescent long-term hematopoietic stem cells (LT-HSCs) (hscBAF) are assembled around the BRM ATPase, whereas leukemic BAF complexes (leukBAF) specifically contain the BRG ATPase. BRG is dispensable for long-term HSC maintenance, but is required for the full proliferative activity of both normal and leukemic HSCs.
Discussion
In these studies, we demonstrate that SWI/SNF-like BAF complexes expressed in leukemia are specifically assembled around the BRG ATPase, whereas BRM is the main ATPase expressed in quiescent long-term repopulating HSCs. Consistent with these findings, we provide evidence that BRG is essential for leukemia propagation using a mouse model of AML. Moreover, we demonstrate that BRG is dispensable for the maintenance of quiescent LT-HSCs, but essential for their full proliferative activity and multipotentiality.
The findings described above are consistent with earlier reports describing antagonistic roles for the BRM and BRG ATPases in cellular proliferation.14,17,,,,-22,36,37 BRG is absolutely required for oncogenic transformation of NIH3T317 and elevated levels of Brg were found in several types of human cancers.38,-40 In contrast to Brg,Brm expression is often silenced in primary and tumor-derived cell lines41,42 and BRM overexpression can revert the RasV12 transformed phenotype.17,18 Although redundant in terms of ATPase activity,12 the divergent roles of Brg and Brm in cellular proliferation may be explained, at least in part, by a unique ability of Brg to constrain p53-mediated growth suppression,20 through a proline-rich motif not present in Brm.21 Consistently, Brg, but not Brm, is uniformly expressed in cancer cells that harbor wild-type p53, whereas Brg-deficient tumor cells contain p53 mutations or do not express p53.21 Differential affinity for regulatory elements and/or DNA-bound factors leads to preferential recruitment of Brm-based complexes to genes essential for the quiescent state of cells and Brg-complexes to cell cycle entry and proliferation genes22 (Figure 6). The divergent roles of Brg and Brm in cellular proliferation are also consistent with the notion that the BAF genes do not function equally in cancer. Notably, differences in cancer types have been associated with the loss of distinct subunits in mice13,25,43,,-46 and opposing roles in cell cycle for BAF complexes that differed by a single variant subunit (BAF250) have been observed. Although the exact mechanisms remain to be investigated, these observations are reminiscent of the functional antagonism between Polycomb and SWI/SNF-like BAF subunits in cellular proliferation47,48 or the changes in DNA compaction and gene expression that are associated with the assembly of distinct histone variants.49
Unlike genetic mutations, epigenetic modifications are usually reversible and thus represent attractive targets for therapeutic intervention. The apparent difficulty of bypassing the requirement for Brg in leukemic stem/progenitor cells suggests that its adroit molecular targeting might have potent and specific therapeutic effects. Because elevated levels of BRG were found in gastric,38 prostate,39 and skin40 cancer, and correlate with tumor progression,38,39 it will be interesting to determine whether the findings reported here might also extend to other types of cancer stem cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank P. Chambon for providing the Brg targeting vector, E. Bonneil for mass spectrometry analyses, M.-A. Bernard for help with hematopoietic cell transplantations, J. Krosl for providing growth factors, D. Gagné from the IRIC flow cytometry platform for cell sorting, P. Chagnon and R. Lambert from the IRIC genomic platform for help with quantitative RT-PCR, members of the Leucegene project for RNA-seq analyses and Sebastien Lemieux for statistical analyses.
This work was supported by a grant from the Canadian Cancer Society (CCS) and a career development award from the Human Frontiers Science Program Organization (HFSP) to J.L., a grant from Genome-Quebec to G.S., J.H., and L.H. for the Leucegene project, and a grant from the Fonds de Recherche du Québec-Santé (FRQS) Cancer Research Network in support of the Leukemia Cell Bank of Quebec (LCBQ). J.L. holds a Canada Research Chair in Molecular Genetics of Stem Cells Hematopoiesis and PT a Canada Research Chair in Proteomics and Bioanalytical Spectrometry. IRIC is supported in part by the Canadian Center of Excellence in Commercialization and Research (CECR), the Canada Foundation for Innovation (CFI) and the FRQS.
Authorship
Contribution: M.B. and V.K. performed the proteomics, molecular, and cellular biology and in vivo studies; C.S. analyzed RNA-seq data and participated in biochemistry studies; L.H. performed most of the studies of Brg function in normal HSCs in G.R.C. laboratory; J.H. provided human leukemia samples; B.W. performed RNA-seq analyses; J.A.L., P.T., G.S., G.R.C., and M.B. designed the studies; J.A.L. and M.B. wrote the paper; and all authors discussed and commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Julie A. Lessard, the Institute for Research in Immunology and Cancer, C.P. 6128 Succ., Downtown, Montreal, H3C 3J7, Canada; e-mail: j.lessard.1@umontreal.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal