Key Points
HMGB1 and DNA released from CLL cells induce nurse-like cell differentiation.
This differentiation appears TLR9/RAGE dependent.
Chronic lymphocytic leukemia (CLL) is a disease of an accumulation of mature B cells that are highly dependent on the microenvironment for maintenance and expansion. However, little is known regarding the mechanisms whereby CLL cells create their favorable microenvironment for survival. High-mobility group protein B-1 (HMGB1) is a highly conserved nuclear protein that can be actively secreted by innate immune cells and passively released by injured or dying cells. We found significantly increased HMGB1 levels in the plasma of CLL patients compared with healthy controls, and HMGB1 concentration is associated with absolute lymphocyte count. We therefore sought to determine potential roles of HMGB1 in modulating the CLL microenvironment. CLL cells passively released HMGB1, and the timing and concentrations of HMGB1 in the medium were associated with differentiation of nurse-like cells (NLCs). Higher CD68 expression in CLL lymph nodes, one of the markers for NLCs, was associated with shorter overall survival of CLL patients. HMGB1-mediated NLC differentiation involved internalization of both receptor for advanced glycation end products (RAGE) and Toll-like receptor-9 (TLR9). Differentiation of NLCs can be prevented by blocking the HMGB1-RAGE-TLR9 pathway. In conclusion, this study demonstrates for the first time that CLL cells might modulate their microenvironment by releasing HMGB1.
Introduction
Many cancers arise from sites of infection, chronic irritation, and inflammation. An inflammatory microenvironment is an important participant in the neoplastic process, fostering proliferation, survival, and migration for cancers, including chronic lymphocytic leukemia (CLL).1,,-4 Stressed, injured, or dying cells release damage-associated molecular patterns (DAMPs), which initiate noninfectious inflammatory responses.5,-7 The DAMP high-mobility group protein B1 (HMGB1) is a major player associating inflammation and cancer.8,9 HMGB1 is a nuclear protein that can be released passively by damaged or dead cells or actively by immune cells and stressed cancer cells.10,,,-14 HMGB1 regulates transcription factors but also behaves as a proinflammatory cytokine mediating inflammation.13,15,,-18 Nonprotein DAMPs, including DNA, RNA, and ATP, are also released by damaged or dying cells.6,7,19 DAMPs are associated with acute inflammatory responses, chronic inflammation, and wound healing, but are also important components of the disordered tumor microenvironment.8,20
HMGB1 is a DNA-binding protein, and increased serum concentrations of the HMGB1-DNA complex can activate the immune system and cause systemic autoimmune disease via the receptor for advanced glycation end products (RAGE) and toll-like receptor-9 (TLR9).21 Interactions of HMGB1-RAGE-TLR9 constitute a tripod that triggers nuclear factor κB (NF-κB) activation22 and promotes dendritic cell maturation.23 RAGE binds multiple ligands derived from a damaged cell environment including HMGB1 and S100 protein.13,24 RAGE is a critical mediator of pancreatic carcinogenesis through its ability to amplify interleukin (IL)-6–induced autophagic translocation of signal transducer and activator of transcription (STAT)3 to the mitochondria and enhance ATP production.25 Blockade of HMGB1 and RAGE suppressed tumor growth and metastasis in a murine model of lung cancer.26 As an intracellular receptor for DNA, TLR9 activation by an endogenous protein-nucleic acid complex plays an important role in autoimmune disease21,27 and also confers CLL cell resistance to fludarabine treatment.28
Tumor-associated macrophages (TAMs) are a significant component of inflammatory infiltrates in neoplastic tissues and are derived from peripheral blood (PB) CD14+ monocytes,4 attracted or recruited into the tumor from the local circulation in response to hypoxic/necrotic conditions and/or tumor-secreted chemokines.29,30 Factors inducing TAM differentiation could be potential therapeutic targets to control tumor progression, but TAM-inducing factors and their association with inflammation are poorly understood. IL-6 induces monocyte or immature dendritic cell in vitro differentiation to M2 TAMs when these cells were cultured in conditioned medium derived from tumor cell lines.31 Reactive oxygen species (ROS) production is critical for macrophage differentiation, and inhibition of superoxide production blocks M2 macrophages differentiation.32 However, it is unknown whether HMGB1 released by damaged tumor cells can promote TAM differentiation. CD14+ monocytes from CLL cells differentiate to nurse-like cells (NLCs) when cultured in vitro to express CD68, vimentin, and stromal derived factor-1.33,-35 Genetic screening and chemokinome identification demonstrate these NLCs belong to the M2 subset of TAMs.36,37 Numerous studies suggest that communication between CLL and stromal cells in the microenvironment support CLL cell survival and chemo-resistance.3,33,-35,38,39 However, little is known regarding the mechanisms whereby CLL cells create their protective microenvironment.
Here, we aimed to determine the factors that induce NLC differentiation in the CLL microenvironment. We report for the first time that CLL plasma contains high levels of circulating HMGB1, which promotes NLC differentiation through the RAGE/TLR9 signaling pathways.
Materials and methods
Cell separation and cell culture
Fresh primary CLL cells were obtained from CLL patients after obtaining written informed consent and ethical approval by the East London and the City Health Authority Local Research Ethics Committee 3 in accordance with the Declaration of Helsinki. CLL diagnosis was made by standard criteria. CLL PB samples from 60 untreated and previously treated patients were used for in vitro studies, and lymph node (LN) biopsies from 86 CLL/small lymphocytic leukemia patients were used in tissue microarrays (TMAs).
Plasma and mononuclear cells were isolated from whole blood by Ficoll-Paque PLUS (GE Healthcare) centrifugation. Plasma was stored at −80°C immediately after separation, and 5 × 106/mL of CLL cells were washed and resuspended in complete culture medium (RPMI-1640 medium supplemented with 10% heat-inactivated fetal calf serum, 25 mM N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid, and 2.0 mM l-glutamine). A Vi-Cell XR Cell Viability Analyzer (Beckman Coulter) was used to check cell numbers and viability. Cells were cultured in RPMI-1640 complete culture medium at 37°C in a 5% CO2 humidified incubator.
Reagents
Ethyl pyruvate (EP), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), and 4-well chambered slides were purchased from Sigma-Aldrich. ChargeSwitch gDNA 0.2-1 mL Serum Kit, 4% to 12% Bis-Tris NuPAGE gels, and 4-morpholinepropanesulfonic acid buffer were purchased from Invitrogen. HMGB1 enzyme-linked immunosorbent assay (ELISA) kit was from IBL International. G-type-inhibitory oligodeoxynucleotide (iODN) (class I) was from Enzo Life Science. NE-PER nuclear protein extraction kit was from Thermo-Scientific. Antibodies used in this study are listed in the supplemental Tables on the Blood Web site.
Determination of DNA and HMGB1 concentration in the plasma and conditioned medium
Conditioned cultured medium was collected from 5 × 106/mL CLL cells at different time points and spun down at 4000 rpm for 2 minutes to rid cell contamination. DNA was extracted from 200 µL of plasma using the ChargeSwitch gDNA 0.2-1 mL Serum Kit (Invitrogen), and concentration was determined using the NanoDrop 1000 spectrophotometer (Thermo Scientific). The HMGB1 concentration in human plasma or conditioned medium was determined using an HMGB1 ELISA kit (IBL International).
TMAs
LN biopsies from 86 patients in whom quality formalin-fixed paraffin-embedded (FFPE) tissue, clinical, and follow-up data were available and 58 reactive LN biopsies were used in the TMAs, which were constructed using an automated arraying system (TMABooster-Alphelys). After marking areas of characteristic morphology which were rich in malignant cells, triplicate 1-mm-diameter cores were taken as previously described.40 Optimally diluted primary antibodies (supplemental Table 1) were applied to slides for 40 minutes, and binding was visualized using a peroxidase-labeled system with 3,3′-diaminobenzidine (DAB) as the chromogen (Super-Sensitive Polymer-HRP IHC Detection System; BioGenex) and counterstained with Mayer hematoxylin. Appropriate positive and negative controls were always run. Slides were digitalized, image analysis was performed using a digital pathology system (Ariol; Lieca Microsystems), and results were manually and blinded reviewed and reported as an average of the triplicate cores.
Immunofluorescent staining
Fresh CLL PB cells or LN single cell suspension cells (5 × 106/mL) were cultured in 4-well chambered slides for 1 or 2 weeks. After removing suspension cells, slides were air dried. Cells on slides were fixed and permeabilized with Cytofix/Cytoperm solution (BD Biosciences) for 30 minutes, and nonspecific bindings were blocked for 30 minutes with a blocking buffer containing a mixture of 5% goat serum, 5% donkey serum, and 0.1% saponion in Tris-buffered saline (pH 7.4). Cells were then costained with 2 different isotype primary antibodies (supplemental Table 1; figure legends) for 1 hour at room temperature. After washing, slides were incubated in the dark with Alexa-Fluor–conjugated secondary antibodies (supplemental Table 2) at a 1:100 dilution. Slides were washed 3 times, stained with 50 ng/mL 4′,6-diamidino-2-phenylindole (DAPI), air-dried at 4°C in the dark, mounted in ProLong Gold anti-fade reagent (Invitrogen), and viewed under an Olympus BX40 fluorescence microscope (Artisan Scientific Corporation).
Flow cytometry
After a 2-week culture, suspension cells were removed by gentle aspiration, and adherent cells were removed from the bottom of cell culture plates. Cells were fixed/permeabilized for 30 minutes by mixing with CytoFix/CytoPerm (BD Biosciences). After washing, cells were incubated with 2% human anti-globulin antibody for 30 minutes on ice to block nonspecific bindings. To determine expression of NLC markers, cells were stained with anti–CD14-fluorescein isothiocyanate (FITC), anti–CD68-phycoerythrin (PE), anti–CD163-Allophycocyanin, and associated isotype controls for 30 minutes at room temperature in the dark. Cells were then washed with Phosphate Buffered Saline with Tween-20 (PBST) before analysis by flow cytometry.
MTT assay
The MTT assay is based on conversion of MTT into formazan crystals by living cells, and total mitochondrial activity is related to the number of viable cells. To determine numbers of NLCs, suspension cells and culture medium were removed. Adherent cells were then incubated with fresh medium containing 0.5 mg/mL MTT for 2 hours. the morphology of NLCs that contain large amounts of formazan was observed by phase-contrast microscopy. For quantification, formazom in the cells was dissolved by adding isopropanol (containing 0.4 N HCl) at a 1:1 dilution and mixed with pipetting. The optical density (OD) of soluble formazan was determined at a 570-nm wavelength by a plate reader.
Coimmunoprecipitation
Coimmunoprecipitation (Co-IP) was performed using Dynabeads protein A (Invitrogen). Dynabeads in 50 µL PBST were incubated with 4 µg mouse anti-TLR9 antibody or mouse IgG for 20 minutes at room temperature on a rotator. After 3 washes with PBST, Dynabeads protein A coated with antibody or IgG was mixed with 400 µg of proteins and incubated for 20 minutes at room temperature on a rotator. The protein complex was then eluted and mixed with NuPAGE LDS sample buffer mixed with reducing agent (Invitrogen).
Cellular fractionation and western blotting
Cytoplasmic and nuclear proteins were extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific). For whole cell analysis, proteins were extracted with CelLytic M cell Lysis Reagent (Sigma-Aldrich) supplied with protease inhibitor (Sigma-Aldrich). Proteins were subjected to 4% to 12% NuPAGE gels (Invitrogen) and transferred onto a polyvinylidene fluoride membrane (Sigma-Aldrich) at 20 V for 1 hour by semi-dry transfer. The polyvinylidene fluoride membrane was blocked with blocking buffer (5% polyvinyl pyrrolidone, 5% fetal calf serum, and 0.1% sodium azide in Tris-buffered saline containing 0.2% Tween-20) for 30 minutes and then incubated with primary antibodies overnight at 4°C against the primary antibodies (supplemental Table 2). Bound antibodies were detected using incubation with horseradish peroxidase–conjugated secondary antibodies (Santa Cruz Biotechnology) in Tris-buffered saline and Tween-20 containing 5% polyvinyl pyrrolidone and 5% fetal calf serum and visualized by a Luminescent image analyzer LAS-4000 (Fujifilm, Tokyo, Japan) after adding ECL plus (GE Healthcare Life Science), and the density of each band was analyzed with the Gelscan V5.1 program (BioSciTec, Frankfurt, Germany).
Statistical analysis
Statistical analysis was performed using GraphPad Prism software. Data are shown as either mean ± standard deviation or median with interquartile range when variation was high. Two group comparisons were performed using the Student t test. The Pearson product-moment correlation method was used to analyze the linear correlation between 2 groups. Categorical (cut point) data analysis was performed using the X-tile statistical package. Outcomes, measured from date of diagnosis to occurrence of event or date of last follow-up, were overall survival. Survival curves were analyzed using Kaplan-Meier survival fraction, and P values were generated by log-rank test.40 All P values <.05 were considered statistically significant. *, **, and *** indicate P < .05, .001, and .0001, respectively.
Results
CLL plasma contains higher levels of HMGB1 and DNA
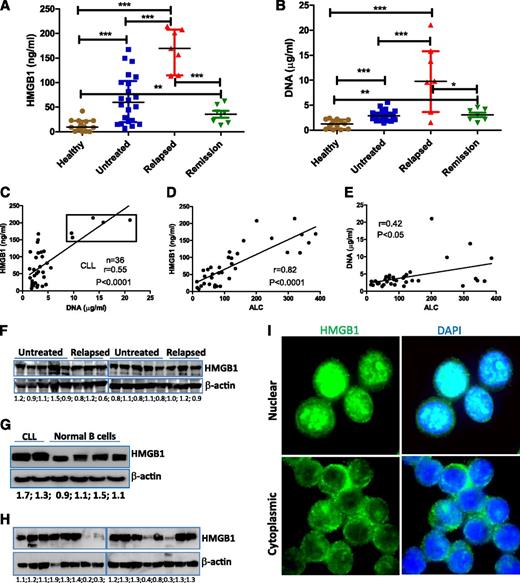
We first determined plasma concentrations of HMGB1 and DNA in 36 representative CLL patients (22 untreated and 14 previously treated) compared with 14 age-matched healthy controls. Both untreated and relapsed CLL had significantly increased levels of HMGB1 and DNA compared with donors (Figure 1A-B). HMGB1 and DNA concentrations appeared significantly correlated in both CLL and healthy controls (Figure 1C; supplemental Figure 1), and CLL patients with the highest plasma concentrations of both HMGB1 and DNA had progressive relapsed disease (supplemental Table 3). However, when these 5 highest cases (box in Figure 1C) were removed, the lower and intermediate levels of HMGB1 were not correlated with relative DNA concentrations (supplemental Figure 1B), suggesting that there may also be active release of HMGB1 without release of DNA. Plasma concentrations of HMGB1 (P < .0001) and DNA (P < .05) were strongly correlated with absolute lymphocyte count (ALC) of CLL samples (Figure 1D-E; supplemental Table 3), suggesting that HMGB1 and DNA are released by CLL cells. HMGB1 plasma concentration was not associated with HMGB1 intracellular expression, and HMGB1 protein is ubiquitously expressed in all CLL and normal B cells (Figure 1F-G). Fresh CLL cells were collected and stored at −80°C as dry pellets prior to western blotting. The freeze and thaw process leads to loss of intracellular HMGB1 protein, particularly in long-term cryopreserved CLL samples (Figure 1H; supplemental Figure 1C-D), suggesting this process can induce experimental necrosis and HMGB1 release.19 All fresh PB mononuclear cells express HMGB1, which can have nuclear and/or cytoplasmic localization (Figure 1I). HMGB1 is a nuclear protein, and its cytoplasmic localization may indicate HMGB1 prior to active or passive release.41 Taken together, we demonstrate that CLL cells and/or microenvironment macrophages/stromal cells release HMGB1 into the plasma, and the HMGB1 concentration in the plasma correlates with ALC.
Detection of extracellular and intracellular HMGB1 protein in CLL samples. (A) HMGB1 and (B) DNA concentrations were determined in the plasma of healthy (n = 14) and CLL samples (untreated, n = 22; relapsed, n = 7; partial remission, n = 7). Significantly increased HMGB1 and DNA concentrations in CLL samples were expressed as median ± interquartile range and differences analyzed by Student t test. (C) Correlation between HMGB1 and DNA concentration in 36 cases of CLL plasma. Dots in the box are 5 cases with the highest concentrations of both HMGB1 and DNA and represent the group of patients with the poorest prognosis. (D-E) Correlations between plasma (D) HMGB1 or (E) DNA concentration with ALC. ALC information was collected at the time of blood draw (supplemental Table 3). (F-H) Determination of HMGB1 expression in CLL and normal B cells by western blotting. (G) Representative results from 2 cases of CLL and 4 normal B-cell samples. (H) HMGB1 expression in long-term cryopreserved CLL samples. Fifty micrograms of proteins was loaded into each lane. Mouse anti-HMGB1 antibody (Sigma-Aldrich) was used at 1:3000 dilution. β-actin was used as a loading control. Numbers below each pair of bands are ratios of HMGB1 to β-actin. (I) Detection of HMGB1 intracellular localization in freshly isolated CLL cells by immunofluorescent staining. DAPI indicates nuclear localization. The isotype control images are demonstrated in supplemental Figure 1A-B.
Detection of extracellular and intracellular HMGB1 protein in CLL samples. (A) HMGB1 and (B) DNA concentrations were determined in the plasma of healthy (n = 14) and CLL samples (untreated, n = 22; relapsed, n = 7; partial remission, n = 7). Significantly increased HMGB1 and DNA concentrations in CLL samples were expressed as median ± interquartile range and differences analyzed by Student t test. (C) Correlation between HMGB1 and DNA concentration in 36 cases of CLL plasma. Dots in the box are 5 cases with the highest concentrations of both HMGB1 and DNA and represent the group of patients with the poorest prognosis. (D-E) Correlations between plasma (D) HMGB1 or (E) DNA concentration with ALC. ALC information was collected at the time of blood draw (supplemental Table 3). (F-H) Determination of HMGB1 expression in CLL and normal B cells by western blotting. (G) Representative results from 2 cases of CLL and 4 normal B-cell samples. (H) HMGB1 expression in long-term cryopreserved CLL samples. Fifty micrograms of proteins was loaded into each lane. Mouse anti-HMGB1 antibody (Sigma-Aldrich) was used at 1:3000 dilution. β-actin was used as a loading control. Numbers below each pair of bands are ratios of HMGB1 to β-actin. (I) Detection of HMGB1 intracellular localization in freshly isolated CLL cells by immunofluorescent staining. DAPI indicates nuclear localization. The isotype control images are demonstrated in supplemental Figure 1A-B.
Increased levels of cytoplasmic HMGB1 and CD68 in CLL-LN is associated with shorter overall survival
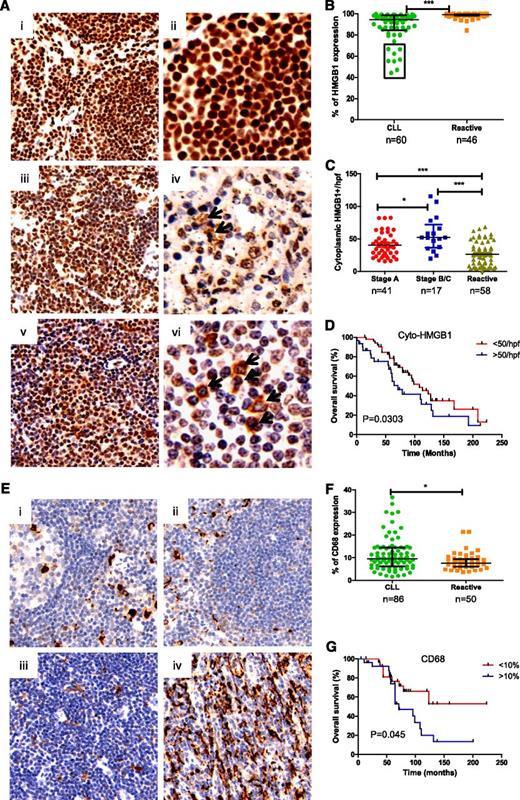
To determine whether CLL cells and macrophages in the LNs could release HMGB1 into their microenvironment, HMGB1 expression was evaluated in reactive (RA)-LN and CLL-LN TMAs. HMGB1 expression was mainly localized in the nucleus of RA-LN cells (Figure 2A, i-ii), whereas in CLL-LNs, there was nuclear and cytoplasmic localization and decreased HMGB1 nuclear expression (Figure 2A, iii-vi). CLL-LN showed significantly decreased overall HMGB1 expression compared with RA-LN (P < .0001; Figure 2B). Patients with reduced expression of HMGB1 (outlined in the box in Figure 2B) had a shorter time to first treatment and shorter survival. The finding of reduced overall HMGB1 expression in poor prognosis patients was initially surprising, but the number of cells expressing cytoplasmic HMGB1 was significantly increased in CLL-LN compared with RA-LN (P < .0001). Binet stages B/C showed higher cytoplasmic HMGB1 compared with stage A (P < .05; Figure 2C). A greater number of cells containing cytoplasmic HMGB1 in CLL-LN was significantly associated with shorter overall survival (P = .03; Figure 2D). We postulate that the decreased nuclear HMGB1 observed in CLL could result from cytoplasmic release and increased release into the microenvironment.
Determination of HMGB1 and CD68 expression in tissue microarrays. (A) Representative samples of HMGB1 expression in (i-ii) reactive and (iii-vi) CLL lymph nodes. Images were taken with a Leixa DM2500 microscope: i, iii, and v, original magnification ×200); ii, iv, and vi, original magnification ×400. (B) Overall HMGB1 expression. HMGB1 expression was determined by a computerized image analysis Ariol system using pathologist-trained visual parameters. All cores were reviewed manually before scoring, and only intact cores were used for scoring. Each datum represents an average of triplicate cores. There was a statistically significant decrease in expression of percentage of cells expressing HMGB1 in CLL compared with reactive LNs. (C) Cytoplasmic HMGB1 expression. Numbers of cytoplasmic HMGB1-positive cells were counted blindly by 2 reviewers in 4 high power fields (hpfs) and expressed as a mean value from triplicate cores. (D) Overall survival of CLL patients based on low (<50) and high (>50) numbers of cells containing cytoplasmic HMGB1 in 4 counted hpfs, with the cutoff determined using X-tiles software. (E) Expression of CD68 in (i-ii) reactive and CLL-LN with (iiii) lower and (iv) higher CD68 expression. (F) Statistical comparison of CD68 expression in CLL-LN and RA-LN. (G) Overall survival of CLL patients based on low (<10%) and high (>10%) expression of CD68.
Determination of HMGB1 and CD68 expression in tissue microarrays. (A) Representative samples of HMGB1 expression in (i-ii) reactive and (iii-vi) CLL lymph nodes. Images were taken with a Leixa DM2500 microscope: i, iii, and v, original magnification ×200); ii, iv, and vi, original magnification ×400. (B) Overall HMGB1 expression. HMGB1 expression was determined by a computerized image analysis Ariol system using pathologist-trained visual parameters. All cores were reviewed manually before scoring, and only intact cores were used for scoring. Each datum represents an average of triplicate cores. There was a statistically significant decrease in expression of percentage of cells expressing HMGB1 in CLL compared with reactive LNs. (C) Cytoplasmic HMGB1 expression. Numbers of cytoplasmic HMGB1-positive cells were counted blindly by 2 reviewers in 4 high power fields (hpfs) and expressed as a mean value from triplicate cores. (D) Overall survival of CLL patients based on low (<50) and high (>50) numbers of cells containing cytoplasmic HMGB1 in 4 counted hpfs, with the cutoff determined using X-tiles software. (E) Expression of CD68 in (i-ii) reactive and CLL-LN with (iiii) lower and (iv) higher CD68 expression. (F) Statistical comparison of CD68 expression in CLL-LN and RA-LN. (G) Overall survival of CLL patients based on low (<10%) and high (>10%) expression of CD68.
CLL-LN and RA-LN were stained with anti-CD68 to examine NLC number (Figure 2E). Significantly increased CD68 expression was detected in CLL-LN compared with RA-LN (P < .05). There was heterogeneity of CD68 expression among samples (Figure 2F), and higher CD68 expression in CLL-LN correlated with decreased overall survival (P = .045; Figure 2G). These results are in keeping with our hypothesis that HMGB1 passive or active release from CLL cells might be the cause of increased levels of HMGB1 in the plasma and contribute to increased numbers of NLCs in the CLL microenvironment.
NLC in vitro differentiation
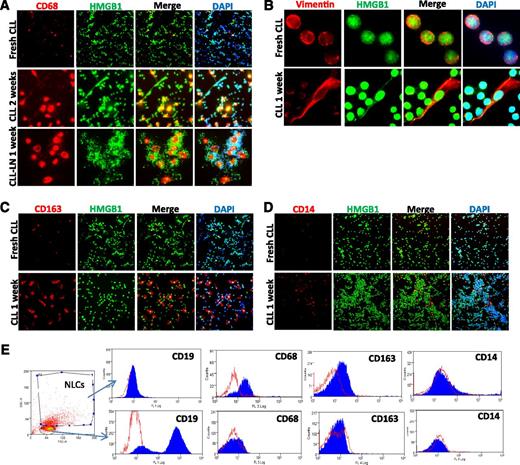
Monocytes in CLL PB and LN single suspension cells show the ability to develop stromal-like adherent cells that tightly contact with CLL cells (supplemental Figure 2A), defined as NLCs33 or TAMs,36,37 with an increase in stromal-like cells expressing CD68 and vimentin after 1 or 2 weeks of cell culture (Figure 3A-B), in agreement with previous studies.35 These NLCs also highly express CD163 (Figure 3C), a M2 polarized marker,42 although CD14 is expressed only weakly (Figure 3D). Expression of these markers was also confirmed by flow cytometry (Figure 3E), suggesting that in vitro differentiated NLCs are at least partly a M2-polarized subtype of TAMs.
NLC in vitro differentiation. Expression of (A) CD68, (B) vimentin, (C) CD163, and (D) CD14 of in vitro differentiated NLCs. Fresh CLL cells or CLL LN single cells were cultured in 4-well chambered slides for 1 to 2 weeks. Fresh CLL cell slides were fixed the day of separation from blood. Slides were costained with a rabbit anti-HMGB1 antibody (green) and DAPI. (E) Determination of NLC marker proteins by flow cytometry. CLL mononuclear cells were cultured for 2 weeks. Cells were fixed/permeabilized and then stained with anti–CD14-FITC, anti–CD68-PE, and anti–CD163-Allophycocyanin or relative isotype controls. The NLC population that is CD19-AF-488 negative and contains larger sizes (forward scatter) and high granulation (side scatter) was selected for marker protein analysis. Empty peaks were those stained with isotype controls and solid peaks were cells stained with specific antibodies. The flow cytometry profiles in the lower panel represent negative expression of 3 markers from selected small CD19-AF-488–positive CLL cells. Data shown are 1 of the typical CLL samples from 3 individual cases studied.
NLC in vitro differentiation. Expression of (A) CD68, (B) vimentin, (C) CD163, and (D) CD14 of in vitro differentiated NLCs. Fresh CLL cells or CLL LN single cells were cultured in 4-well chambered slides for 1 to 2 weeks. Fresh CLL cell slides were fixed the day of separation from blood. Slides were costained with a rabbit anti-HMGB1 antibody (green) and DAPI. (E) Determination of NLC marker proteins by flow cytometry. CLL mononuclear cells were cultured for 2 weeks. Cells were fixed/permeabilized and then stained with anti–CD14-FITC, anti–CD68-PE, and anti–CD163-Allophycocyanin or relative isotype controls. The NLC population that is CD19-AF-488 negative and contains larger sizes (forward scatter) and high granulation (side scatter) was selected for marker protein analysis. Empty peaks were those stained with isotype controls and solid peaks were cells stained with specific antibodies. The flow cytometry profiles in the lower panel represent negative expression of 3 markers from selected small CD19-AF-488–positive CLL cells. Data shown are 1 of the typical CLL samples from 3 individual cases studied.
NLC differentiation is associated with HMGB1 release from CLL cells
Differentiation of NLCs was not observed in all CLL samples in vitro, and samples with lower viability develop NLCs faster, but cells with higher viability failed to develop NLCs (Figure 4A). Of note, we observed no correlation between presence of high numbers of smudge cells in the blood film from these patients and their HMGB1 levels. Observation and quantification of NLCs were performed by phase-contrast microscopy (supplemental Figure 2A) and MTT staining (Figure 4B). We hypothesized that dead cells may release HMGB1 into the cell culture medium. HMGB1 passive release was first observed by immunofluorescent staining as early as 2 days of culture, shown as HMGB1 leaking from the nucleus (Figure 4C) and kinetically determined by ELISA (Figure 4D) and western blotting (Figure 4E,G). More rapid release of higher concentration of HMGB1 in CLL samples in vitro cultures was associated with earlier development of NLCs. HMGB1 release was accompanied by release of the cytosolic protein lactate dehydrogenase (LDH) (Figure 4E) and DNA (Figure 4F,H), suggesting these intracellular components were released passively by dead cells. A greater number of NLCs developing in 2-week culture was associated with higher concentrations of HMGB1 and DNA in conditioned medium (Figure 4G-H), suggesting that HMGB1/DNA plays important roles in NLC differentiation.
Association between HMGB1 release and NLC differentiation. (A) Association between cell viability and NLC differentiation. Cell viability of each sample was monitored every 2 or 3 days using a Cell Viability Analyzer (Beckman Coulter). The box represents the day on which NLCs were first identified. (B) Observation of NLCs by MTT staining. After 1 or 2 weeks of culture, the cells in suspension were removed. Cells were then cultured with fresh medium containing 0.5 mg/mL MTT for 2 hours. (C) HMGB1 passive release. CLL cells were cultured for 2 days on chambered slides. After fix/permeabilization, cells were costained with rabbit anti-COX IV (red) and mouse anti-HMGB1 (green) antibodies. Arrows indicate a typical phenomenon of HMGB1 passive release from the cells. (D-E) Time course of HMGB1 release determined by (D) ELISA and (E) western blotting in 2 representative cases of 6 independent cases examined. HMGB1 release into culture medium was monitored over 19 days. Conditioned medium was collected every 2 or 3 days and stored at −80°C. Ten or 40 µL of conditioned medium was used for ELISA assay or western blotting, respectively. Western blotting was probed with an anti-HMGB1 mouse antibody first and then reprobed with an anti-LDH rabbit antibody. (G) Determination of HMGB1 release by western blotting. Four CLL samples were cultured for 15 days, and conditioned medium was collected in every 5 days. Numbers of NLCs were quantified by MTT staining at the 15th day. The amounts of NLCs indicated as −, +, ++, +++, and ++++ were 0%, 10%, 25%, 50%, and >75% coverage of the well. (F,H) DNA concentration in the conditioned medium. Two hundred microliters of conditioned medium was used to determine DNA concentration. Boxes in A, D, E, and F indicate the time when NLCs were observed. Numbers in each chart are CLL sample IDs.
Association between HMGB1 release and NLC differentiation. (A) Association between cell viability and NLC differentiation. Cell viability of each sample was monitored every 2 or 3 days using a Cell Viability Analyzer (Beckman Coulter). The box represents the day on which NLCs were first identified. (B) Observation of NLCs by MTT staining. After 1 or 2 weeks of culture, the cells in suspension were removed. Cells were then cultured with fresh medium containing 0.5 mg/mL MTT for 2 hours. (C) HMGB1 passive release. CLL cells were cultured for 2 days on chambered slides. After fix/permeabilization, cells were costained with rabbit anti-COX IV (red) and mouse anti-HMGB1 (green) antibodies. Arrows indicate a typical phenomenon of HMGB1 passive release from the cells. (D-E) Time course of HMGB1 release determined by (D) ELISA and (E) western blotting in 2 representative cases of 6 independent cases examined. HMGB1 release into culture medium was monitored over 19 days. Conditioned medium was collected every 2 or 3 days and stored at −80°C. Ten or 40 µL of conditioned medium was used for ELISA assay or western blotting, respectively. Western blotting was probed with an anti-HMGB1 mouse antibody first and then reprobed with an anti-LDH rabbit antibody. (G) Determination of HMGB1 release by western blotting. Four CLL samples were cultured for 15 days, and conditioned medium was collected in every 5 days. Numbers of NLCs were quantified by MTT staining at the 15th day. The amounts of NLCs indicated as −, +, ++, +++, and ++++ were 0%, 10%, 25%, 50%, and >75% coverage of the well. (F,H) DNA concentration in the conditioned medium. Two hundred microliters of conditioned medium was used to determine DNA concentration. Boxes in A, D, E, and F indicate the time when NLCs were observed. Numbers in each chart are CLL sample IDs.
HMGB1 forms complexes with TLR9 and RAGE within intracellular vesicles
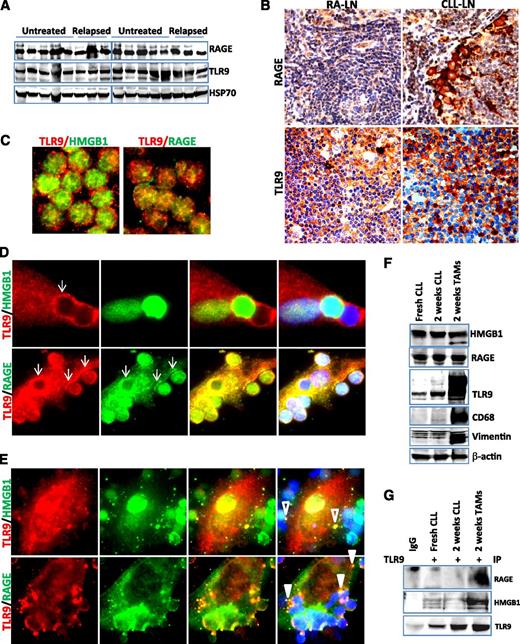
The putative cellular receptors for HMGB1 are TLR2, TLR4, TLR9, and RAGE.21,43 We hypothesized that extracellular HMGB1 and DNA may form a complex and stimulate immune cells via the TLR9 and/or RAGE pathway. RAGE and TLR9 are ubiquitously expressed in all CLL samples by western blotting (Figure 5A). Immunohistochemical staining showed that RAGE and TLR9 are strongly expressed in the microenvironment stromal cells in both CLL-LN and RA-LN and weakly expressed in lymphocytes (Figure 5B). Using fluorescent microscopy we observed that TLR9 and RAGE proteins were distributed in a dispersed pattern in fresh CLL cells and did not colocalize with each other or with HMGB1 (Figure 5C). TLR9, an intracellular receptor for the HMGB1/DNA complex, was recruited and aggregated at the contact sites with CLL cells after a 1-week culture and showed strong colocalization with the HMGB1 surface receptor RAGE (Figure 5D). After CLL cells were cultured for 2 weeks, HMGB1 and TLR9 formed complexes within distinct vesicles inside NLC and RAGE were internalized with TLR9 to endosomal-like structures (Figure 5E). Increased expression of complex proteins in NLCs was seen only for TLR9 but not RAGE (Figure 5F; supplemental Figure 3A). The interaction between RAGE and/or HMGB1 with TLR9 in NLCs was detected in protein complexes by co-IP using anti-TLR9 antibody (Figure 5G). These results demonstrate that the HMGB1-mediated RAGE/TLR9 signaling pathway is activated during differentiation and maturation of NLCs.
Activation of RAGE/TLR9 in the CLL microenvironment in vivo and in vitro. (A) Detection of RAGE and TLR9 expression in CLL cells by western blotting. Ten untreated and 6 relapsed CLL peripheral mononuclear cells were stored at −80°C and lysed directly from the pellets. Fifty micrograms of proteins was loaded onto each lane. Rabbit anti-RAGE antibody and mouse anti-TLR9 antibody were used at 1:3000 dilution. Hsp70 was used as a loading control. (B) Detection of RAGE and TLR9 expression in CLL-LN and RA-LN by immunohistochemical staining as described in the Materials and methods. Images were taken with a Leixa DM2500 microscope (original magnification, ×200). Immunofluorescent costaining of HMGB1/TLR9 and RAGE/TLR9 in (C) fresh CLL cells and differentiated NLCs after (D) 1 and (E) 2 weeks of in vitro culture. Costained primary antibodies used were mouse anti-TLR9 (red)/rabbit anti-HMGB1 (green) and mouse anti-TLR9 (red)/rabbit anti-RAGE (green) antibodies. Secondary antibodies for costaining were Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 546 donkey anti-mouse IgG. Arrows in D show TLR9 and RAGE aggregation at the contacting sites with CLL cells. Empty and solid triangles in E indicate specified vesicles containing HMGB1/TLR9 and RAGE/TLR9. (F) Expression of HMGB1, RAGE, and TLR9. Proteins were extracted from fresh and 2-week cultured CLL cells and NLCs. Protein expression was detected by western blotting using mouse anti-HMGB1, anti-TLR9, or a rabbit anti-RAGE antibody. CD68 and vimentin were used as markers for NLCs, and β-actin was used for a loading control. (G) Co-IP was performed using mouse anti-TLR9 antibody, and the blots were probed with a rabbit anti-RAGE, rabbit anti-HMGB1, and mouse anti-TLR9 antibodies, respectively. This is a typical result from 3 individual experiments performed.
Activation of RAGE/TLR9 in the CLL microenvironment in vivo and in vitro. (A) Detection of RAGE and TLR9 expression in CLL cells by western blotting. Ten untreated and 6 relapsed CLL peripheral mononuclear cells were stored at −80°C and lysed directly from the pellets. Fifty micrograms of proteins was loaded onto each lane. Rabbit anti-RAGE antibody and mouse anti-TLR9 antibody were used at 1:3000 dilution. Hsp70 was used as a loading control. (B) Detection of RAGE and TLR9 expression in CLL-LN and RA-LN by immunohistochemical staining as described in the Materials and methods. Images were taken with a Leixa DM2500 microscope (original magnification, ×200). Immunofluorescent costaining of HMGB1/TLR9 and RAGE/TLR9 in (C) fresh CLL cells and differentiated NLCs after (D) 1 and (E) 2 weeks of in vitro culture. Costained primary antibodies used were mouse anti-TLR9 (red)/rabbit anti-HMGB1 (green) and mouse anti-TLR9 (red)/rabbit anti-RAGE (green) antibodies. Secondary antibodies for costaining were Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 546 donkey anti-mouse IgG. Arrows in D show TLR9 and RAGE aggregation at the contacting sites with CLL cells. Empty and solid triangles in E indicate specified vesicles containing HMGB1/TLR9 and RAGE/TLR9. (F) Expression of HMGB1, RAGE, and TLR9. Proteins were extracted from fresh and 2-week cultured CLL cells and NLCs. Protein expression was detected by western blotting using mouse anti-HMGB1, anti-TLR9, or a rabbit anti-RAGE antibody. CD68 and vimentin were used as markers for NLCs, and β-actin was used for a loading control. (G) Co-IP was performed using mouse anti-TLR9 antibody, and the blots were probed with a rabbit anti-RAGE, rabbit anti-HMGB1, and mouse anti-TLR9 antibodies, respectively. This is a typical result from 3 individual experiments performed.
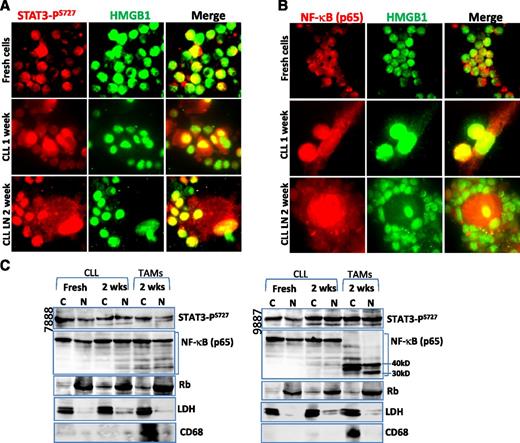
Activation of STAT3 and NF-κB in NLCs
Next, we determined whether activation of the RAGE/TLR9 pathway lea to activation of the transcription factors STAT3 and NF-κB. Immunofluorescent staining and cellular fractionation techniques demonstrate that STAT3-PS727 was expressed in the nucleus of NLCs, although CLL cells constitutively showed activated STAT3 (Figure 6A,C). Activation of STAT3-PY705 was not detected in NLCs (data not shown). There was no obviously increased STAT3 expression in both STAT3-PS727 and nonphosphorylated forms (supplemental Figure 3B). NF-κB (p65) nuclear translocation, an indicator for NF-κB activation, was detected in both CLL PB- and LN-derived NLCs (Figure 6B-C). NF-κB protein in NLCs was cleaved to 30 to 40 kD short length subunits in some cases (Figure 6C). STAT3-PS727 nuclear localization occurred in NLCs and CLL cells of all samples examined. NF-κB nuclear localization was found in all NLCs but only in 3 cases of CLL cells. These results indicate that both STAT3 and NF-κB transcription factors were activated in NLCs.
Nuclear translocation of STAT3 and NF-κB. (A-B) Immunofluorescent staining of (A) STAT3-PS727 and (B) NF-κB in cells before and after cell culture for 1 or 2 weeks. Slides were costained with mouse (A) STAT3-PS727 or (B) NF-κB antibody, showing the red color and a rabbit anti-HMGB1 antibody, showing green. Yellow color indicates nuclear colocalization between STAT3-PS727 or NF-κB with HMGB1. (C) Detection of STAT3 and NF-κB activation by cellular fractionation. Cytoplasmic (C) and nuclear (N) proteins were extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents. Fifty micrograms of proteins was loaded onto each lane. Mouse anti-STAT3-PS727 or NF-κB antibody was used for determination of nuclear translocation of each protein, and mouse anti-Rb and rabbit anti-LDH antibodies were used as marker for nucleus and cytoplasm, respectively. CD68 was used as a marker for NLCs.
Nuclear translocation of STAT3 and NF-κB. (A-B) Immunofluorescent staining of (A) STAT3-PS727 and (B) NF-κB in cells before and after cell culture for 1 or 2 weeks. Slides were costained with mouse (A) STAT3-PS727 or (B) NF-κB antibody, showing the red color and a rabbit anti-HMGB1 antibody, showing green. Yellow color indicates nuclear colocalization between STAT3-PS727 or NF-κB with HMGB1. (C) Detection of STAT3 and NF-κB activation by cellular fractionation. Cytoplasmic (C) and nuclear (N) proteins were extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents. Fifty micrograms of proteins was loaded onto each lane. Mouse anti-STAT3-PS727 or NF-κB antibody was used for determination of nuclear translocation of each protein, and mouse anti-Rb and rabbit anti-LDH antibodies were used as marker for nucleus and cytoplasm, respectively. CD68 was used as a marker for NLCs.
Blockage HMGB1-associated RAGE/TLR9 pathway prevented NLC differentiation
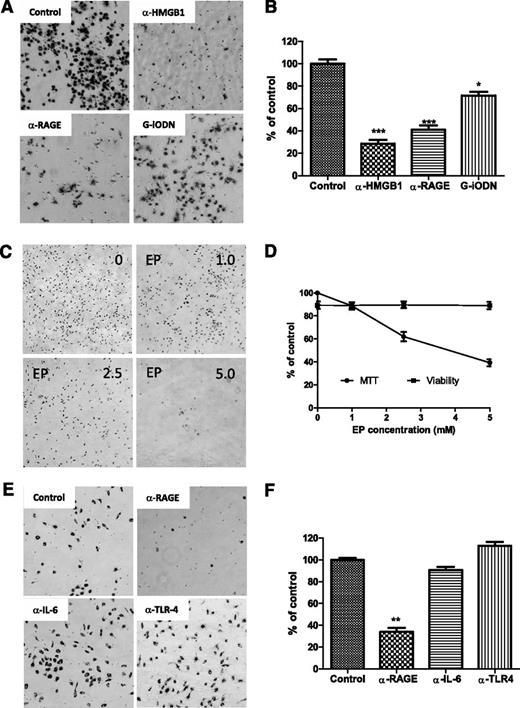
To determine whether NLC differentiation is dependent on HMGB1-mediated activation of the RAGE/TLR9 pathway, fresh CLL cells were incubated with anti-HMGB1 or RAGE antibody to either neutralize HMGB1 in the conditioned medium or block the surface receptor RAGE. TLR9, an intracellular receptor, was blocked by incubation with the TLR9 antagonist class I G-type iODN. After 1 week of culture, NLC differentiation was determined by MTT staining. Neutralizing HMGB1 or RAGE showed maximum blocking effect on NLC differentiation (Figure 7A-B). Control antibodies showed no effect on differentiation of NLCs (data not shown). The HMGB1 antagonist EP also prevented NLC differentiation in a dose-dependent manor (Figure 7C-D). Some but not all CLL cells expressed TLR4, another HMGB1 receptor (supplemental Figure 4), and CLL cells are capable of autocrine IL-6 (data not shown), but neutralizing IL-6 or TLR4 did not block NLC differentiation compared with the effect of neutralizing RAGE (Figure 7E-F). These results demonstrate that NLC differentiation in CLL appears to be HMGB1, RAGE, and TLR9 dependent.
Blocking differentiation of NLCs. (A,C,E) CLL cells were cultured for 1 week individually with different neutralizing antibodies or inhibitors. Suspending CLL cells were removed gently prior to adding the MTT solution. After incubation with MTT for 2 hours, a large amount of formazan was formed inside NLCs. Images of NLCs were taken under phase-contrast microscopy, which are displayed as larger dark cells on the bottom of plates. (B,D,F) To quantify the amount of NLCs, formazan in the cells was dissolved with isopropanol, and OD values were measured by spectrophotometry. Reduced numbers of NLCs were expressed as percentage of control. Data shown are mean ± standard deviation from 3 individual cases CLL patients with triplicate OD values. Significant changes were analyzed by the Student t test. (A-B) Blocking HMGB1, RAGE, or TLR9. Fresh CLL cells (5 × 106/mL) in 24-well plates were incubated with 10 µg/mL of anti-HMGB1 or anti-RAGE neutralizing antibody or 100 nM G-iODN for 1 week. (C-D) Inhibition of HMGB1 by EP. Cells were treated with 1.0, 2.5, and 5.0 mM of EP for 1 week. (D) Viability of CLL cells were determined by a Vi-Cell XR Cell Viability Analyzer, and MTT data represent the amount of NLCs before and after treatment with EP. (E-F) Blockade of RAGE, IL-6, or TLR4 by neutralizing antibodies. CLL cells were incubated with 10 µg/mL of anti-RAGE, anti–IL-6, or TLR4 neutralizing antibody for 1 week.
Blocking differentiation of NLCs. (A,C,E) CLL cells were cultured for 1 week individually with different neutralizing antibodies or inhibitors. Suspending CLL cells were removed gently prior to adding the MTT solution. After incubation with MTT for 2 hours, a large amount of formazan was formed inside NLCs. Images of NLCs were taken under phase-contrast microscopy, which are displayed as larger dark cells on the bottom of plates. (B,D,F) To quantify the amount of NLCs, formazan in the cells was dissolved with isopropanol, and OD values were measured by spectrophotometry. Reduced numbers of NLCs were expressed as percentage of control. Data shown are mean ± standard deviation from 3 individual cases CLL patients with triplicate OD values. Significant changes were analyzed by the Student t test. (A-B) Blocking HMGB1, RAGE, or TLR9. Fresh CLL cells (5 × 106/mL) in 24-well plates were incubated with 10 µg/mL of anti-HMGB1 or anti-RAGE neutralizing antibody or 100 nM G-iODN for 1 week. (C-D) Inhibition of HMGB1 by EP. Cells were treated with 1.0, 2.5, and 5.0 mM of EP for 1 week. (D) Viability of CLL cells were determined by a Vi-Cell XR Cell Viability Analyzer, and MTT data represent the amount of NLCs before and after treatment with EP. (E-F) Blockade of RAGE, IL-6, or TLR4 by neutralizing antibodies. CLL cells were incubated with 10 µg/mL of anti-RAGE, anti–IL-6, or TLR4 neutralizing antibody for 1 week.
Discussion
Here we demonstrate, for the first time, that CLL cells release HMGB1 protein and DNA into the plasma, and their concentrations appear correlated with tumor burden and adverse clinical outcome. HMGB1 demonstrates nuclear localization but is also cytoplasmic in CLL cells prior to its release. Lower nuclear and higher cytoplasmic HMGB1 expression in CLL-LN may be associated with poor outcome in CLL patients, but this should be verified in independent datasets. The CLL microenvironment contains increased numbers of NLCs, and NLC differentiation in vitro was associated with HMGB1 release from CLL cells, whereas blockade of the HMGB1-RAGE/TLR9 signaling pathway prevents NLC differentiation.
Increased plasma or serum levels of HMGB1 are observed in severe sepsis or disseminated intravascular coagulation, and higher HMGB1concentration is correlated with organ failure and mortality.44,45 Increased circulating HMGB1 is found in some cancers, including malignant peritoneal mesothelioma46 and myelodysplastic syndromes.20 Higher circulating HMGB1 is associated with worse clinical outcomes, and plasma samples with higher concentrations of HMGB1 (>150 ng/mL) in our study were from relapsed or progressive CLL patients and are correlated with circulating tumor bulk. In vitro culture experiments confirmed that HMGB1 passively released by dead CLL cells is a concurrent event with DNA and LDH release and occurred prior to NLC differentiation. HMGB1 translocation from the nucleus to the cytoplasm is required for its active release.17,41 Reduced nuclear and increased cytoplasmic HMGB1 expression were seen in CLL-LNs in both TAMs and CLL cells without necrotic cell death, suggesting active release of HMGB1 is also involved in the CLL tissue microenvironment.
CD68-positive TAMs are significantly increased in CLL-LNs compared with reactive controls, and higher CD68 expression was associated with shorter overall survival. We propose that extracellular HMGB1 in the CLL microenvironment induces NLC differentiation. This is prevented by incubation of CLL cells with HMGB1 neutralizing antibody, whereas human recombinant HMGB1 promotes NLC differentiation, enhanced by addition of DNA extracted from CLL patients’ plasma. HMGB1 induces dendritic cell maturation but not at the early phase of differentiation.23 ROS generation also induces TAM differentiation,32 and redox status has a significant impact on HMGB1 function.9 We propose that HMGB1 acts as a damage signal for cell repair and healing, and NLC differentiation is one of the responses and supports CLL cells to survive. Other factors likely also play roles in NLC differentiation.
In vitro differentiated NLCs express higher levels of CD68 but lower levels of CD14. The high expression of CD163 suggests these cells are M2-polarized TAMs,47 which also highly express the stromal cells marker vimentin.35 Activation of STAT3 and NF-κB was detected in both NLCs and CLL cells in keeping with a tumor-supporting function of NLCs.39,48
TLR4 is the most widely studied receptor for HMGB1-mediated inflammation.17,20 However, our results showed that most CLL samples have absent or weak TLR4 expression, and TLR4 blockade by the neutralizing antibody failed to prevent NLC differentiation. RAGE and TLR9 are ubiquitously expressed in CLL cells and are highly expressed in stromal-like cells in the CLL microenvironment. There is no specific pattern of distribution of these receptors in freshly isolated CLL cells. TLR9, an intracellular receptor for the HMGB1/DNA complex,16,21 recognizes DNA of invading pathogens and host self-DNA. It has been proposed that evolutionary pressure has exiled the DNA-recognizing TLR9 from the cell surface to intracellular compartments to avoid activation by extracellular host-derived DNA.49 We observed TLR9 resurfacing and aggregation at the contact sites with CLL cells, suggesting a recognition process of TLR9 to HMGB1/DNA complex released from damaged or dead CLL cells. Recruitment of HMGB1 by TLR9 to specific compartments was also observed by fluorescent microscopy. RAGE is a surface receptor for multiple ligands, including HMGB1 and S100, and HMGB1 and RAGE blockade suppresses tumor growth and metastasis in a murine model of lung cancer.13 HMGB1, TLR, and RAGE constitute a tripod that triggers NF-κB activation.22 There have been no reports on RAGE expression in CLL, but we found that RAGE is ubiquitously expressed in all CLL PB and LN samples. RAGE and TLR9 are both involved in HMGB1/DNA recognition and internalization into larger intracellular endosome-like compartments. Molecular interactions between TLR9-RAGE and TLR9-HMGB1 were detected by both immunostaining and immunoprecipitation, and blockade of HMGB1, RAGE, or TLR9 suppressed NLC differentiation.
In summary, this study suggests that extracellular HMGB1 is s a serologic biomarker for poor prognosis CLL. We demonstrate that HMGB1 induces NLC differentiation in vitro and probably also in vivo, and this is RAGE/TLR9 dependent. Increased numbers of NLCs in the CLL microenvironment are associated with shorter overall survival in CLL, although this will require verification in independent series. We propose extracellular HMGB1 is a promising therapeutic target for CLL treatment.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful for laboratory managing, technical, and statistical support from Drs Carol Jennings, Paul Greaves, Rita Coutinho, Tim Farren, Farideh Miraki-Moud, John Riches, William Day, and Guglielmo Rosignoli.
This project is supported by funding from program grant P01 CA81538 from the National Cancer Institute to the CLL Research Consortium (J.G.G.), and by a Cancer Research United Kingdom studentship (L.J.) for supporting A.M. We thank the Ian Hart summer studentship (2013) for supporting N.U.
Authorship
Contribution: L.J. and J.G.G. contributed project design and manuscript writing; L.J., A.C., F.-T.L., N.U., A.M., and E.H. performed experiments; L.J. and A.C. contributed data analysis; J.M. provided the clinical information for CLL patients; and C.D. and S.I. provided excellent tissue bank service.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Li Jia, Centre for Haemato-Oncology, Barts Cancer Institute, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK; e-mail: L.jia@qmul.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal