Abstract
Chronic myeloid leukemia in chronic phase (CML-CP) is induced by BCR-ABL1 oncogenic tyrosine kinase. Tyrosine kinase inhibitors eliminate the bulk of CML-CP cells, but fail to eradicate leukemia stem cells (LSCs) and leukemia progenitor cells (LPCs) displaying innate and acquired resistance, respectively. These cells may accumulate genomic instability, leading to disease relapse and/or malignant progression to a fatal blast phase. In the present study, we show that Rac2 GTPase alters mitochondrial membrane potential and electron flow through the mitochondrial respiratory chain complex III (MRC-cIII), thereby generating high levels of reactive oxygen species (ROS) in CML-CP LSCs and primitive LPCs. MRC-cIII–generated ROS promote oxidative DNA damage to trigger genomic instability, resulting in an accumulation of chromosomal aberrations and tyrosine kinase inhibitor–resistant BCR-ABL1 mutants. JAK2(V617F) and FLT3(ITD)–positive polycythemia vera cells and acute myeloid leukemia cells also produce ROS via MRC-cIII. In the present study, inhibition of Rac2 by genetic deletion or a small-molecule inhibitor and down-regulation of mitochondrial ROS by disruption of MRC-cIII, expression of mitochondria-targeted catalase, or addition of ROS-scavenging mitochondria-targeted peptide aptamer reduced genomic instability. We postulate that the Rac2-MRC-cIII pathway triggers ROS-mediated genomic instability in LSCs and primitive LPCs, which could be targeted to prevent the relapse and malignant progression of CML.
Introduction
Genomic instability is one of the most common hallmarks of cancer and can be responsible for the accumulation of mutations affecting tumor cell malignant properties and response to therapies.1 The mechanisms and consequences of genomic instability may be substantially different in cancer stem cells (CSCs) and cancer progenitor cells (CPCs). Genetic aberrations in CSCs may not cause problems if acquired in quiescent CSCs, but when these cells eventually divide or the aberrations induce proliferation or appear in CSCs that are already cycling, they may generate drug-resistant and/or more malignant clones. Conversely, genomic instability in CPCs must induce the acquisition of CSC properties to prevent mutations from disappearing before undergoing terminal maturation.
BCR-ABL1+ chronic myeloid leukemia (CML) has served for decades as a paradigm for understanding the stepwise process of carcinogenesis, which involves CSCs and CPCs responsible for the initiation and/or maintenance of the disease.2 CML is initiated by a BCR-ABL1 tyrosine kinase that transforms hematopoietic stem cells (HSCs) to leukemia stem cells (LSCs) to induce CML in chronic phase (CML-CP). Deregulated growth of LSC-derived leukemia progenitor cells (LPCs) leads to manifestation of the disease. ABL tyrosine kinase inhibitors (TKIs) such as imatinib, dasatinib, and nilotinib frequently induce complete cytogenetic or major molecular responses, but LSCs are intrinsically insensitive to TKIs despite inhibition of BCR-ABL1 kinase.3 CML-CP cells may at some stage acquire additional genetic changes that confer TKI resistance and induce the more aggressive blast phase (CML-BP).4
Genomic instability usually results from an aberrant cellular response to enhanced DNA damage.5 One of the leading causes of DNA damage is reactive oxygen species (ROS). The first ROS molecule produced is the superoxide anion (·O2−), which is a moderately stable free radical. Dismutation of ·O2− by superoxide dismutase (SOD) results in the production of hydrogen peroxide (H2O2), which may be converted by Fe2+-driven cleavage to the highly reactive hydroxyl group (·OH). ROS can damage DNA bases to produce 7,8-dihydro-8-oxo-2′-deoxyguanosine (8-oxoG) and other oxo-derivatives that lead to point mutations. ROS also induce spontaneous DNA double-strand breaks (DSBs) the unsuccessful repair of which can result in chromosomal aberrations.
We and others have demonstrated previously that leukemia cell lines expressing BCR-ABL1 kinase and other oncogenic tyrosine kinases (OTKs) such as TEL-ABL1, TEL-JAK2, TEL-PDGFβR, JAK2(V617F), and FLT3-ITD accumulate ROS and oxidative DNA damage (8-oxoG and DSBs), resulting in genomic instability.6-9 However, the mechanisms and consequences of genomic instability may be different in various subpopulations of leukemia cells, so it is critical to determine whether this process originates in LSCs or LPCs and which molecular mechanisms are involved.
Methods
Human cells
For patient specimens, freshly isolated or frozen BM and peripheral blood samples from anonymous CML-CP patients at diagnosis (90%-100% Philadelphia chromosome positive by FISH) were obtained from the Institute of Hematology and Blood Transfusion, Warsaw, Poland; Medical University of Warsaw, Warsaw, Poland; University of Glasgow, Glasgow, United Kingdom; British Columbia Cancer Agency, Vancouver, BC; Medical University of Vienna & Ludwig-Boltzmann Cluster Oncology, Vienna, Austria; Jena University Hospital, Jena, Germany; University of Adelaide, Adelaide, Australia; and University of Tuebingen, Stuttgart, Germany. Acute myeloid leukemia (AML) cells were from Dana-Farber Cancer Institute. Polycythemia vera (PV) cells were received from Dr Ross Levine (Memorial Sloan-Kettering Cancer Center, NY). Healthy donor samples were purchased from Cambrex BioScience. Lin−CD34+ cells were obtained from Ficoll-separated samples by magnetic sorting using the EasySep negative selection human progenitor cell enrichment cocktail, followed by human CD34+ selection cocktail (StemCell Technologies). CD34+CD38− and CD34+CD38+ cells were sorted after staining with anti–human CD38 Ab conjugated with PE-Cy7 (BD Pharmingen). FACS-sorted cells were suspended in StemSpanSFEM (StemCell Technologies, PeproTech, and R&D Systems) supplemented with 10% FBS in the presence of growth factors (StemCell Technologies) at concentrations similar to that found in stroma-conditioned medium in long-term BM cultures (200 pg/mL of SCF, 200 pg/mL of GM-CSF, 1 ng/mL of G-CSF, 50 pg/mL of LIF, 200 pg/mL of MIP-1α, and 1 ng/mL of IL-6). Some Lin−CD34+CD38− samples were further characterized by detection of the Philadelphia chromosome. For quiescent and proliferating cells, CD34+ cells were stained with CellTrace CFSE (Invitrogen) or CellTrace Violet (CTV; Invitrogen), incubated for 3-5 days in StemSpanSFEM supplemented with a cocktail of growth factors (100 ng/mL of SCF, 20 ng/mL of IL-3, 100 ng/mL of Flt-3 ligand, 20 ng/mL of G-CSF, and 20 ng/mL of IL-6), and sorted. The studies were approved by the appropriate institutional review boards.
Murine cells
32Dcl3 murine myeloid precursor cells and derivatives expressing nonmutated BCR-ABL1 kinase, the indicated TKI-resistant BCR-ABL1 kinase domain mutants (P-loop Y253H, the site of a hydrogen bond with imatinib, T315I; activation loop hinge, M351T; and activation loop, H396P) and TEL-ABL1, TEL-JAK2, TEL-PDGFβR, TEL-TRKC(L), and BCR-FGFR1 were maintained in IMDM plus 10% FBS and 1.5% WEHI-conditioned medium as a source of IL-3. E9, FG23-1, and A22 cell lines were derived from L929 cells. FG23-1 harbors the ND613887iC frameshift mutation abolishing the synthesis of the mitochondrial respiratory chain (MRC) complex I subunit; A22 contains the CYTbG15263A mutation causing an E373K amino acid substitution, which is deleterious for the MRC complex III (MRC-cIII); and E9 is an isogenic control.10 These cells were transfected with pMIG-BCR-ABL1-IRES-GFP or pMIG-IRES-GFP retroviral vectors (kindly obtained from Dr W. Pear, University of Pennsylvania, Philadelphia, PA) and green fluorescent protein–positive (GFP+) cells were used for experiments. 32Dcl3 cells were transfected with expression plasmids encoding the mitochondria matrix-targeted circularly permuted yellow fluorescence protein (mt-cpYFP),11 followed by p210BCR-ABL1 retroviral constructs, and BCR-ABL1+ clones were selected by Western blotting. Murine BM cells (muBMCs) from 5-fluorouracil–pretreated C57Bl/6 (Cre-Tg+);Rac1flox/flox (Rac1Δ/Δ), Rac2−/−, Rac3−/−, Rac1Δ/ΔRac2−/−, and +/+ mice were transfected with retroviral constructs. Briefly, helper-free retroviruses were generated by transiently transfecting retroviral vectors into the amphotropic packaging cell line by the calcium-phosphate/chloroquine method using pMIG-BCR-ABL1-IRES-GFP retroviral construct encoding p210BCR-ABL1 and GFP or with pMIG-IRES-GFP. Culture supernatants containing viral particles were centrifuged for 3 hours at 36 100g. Mononuclear BM cells were infected with retroviral supernatants in the presence of SCF, IL-3, and polybrene. GFP+ cells were sorted and used for experiments. Puromycin-resistant BCR-ABL1-32Dcl3 cells expressing the Rac(T17N) mutant have been described previously.12 GFP+ 32Dcl3 cells expressing hemagglutinin-tagged Rac(T17N) and control cells were obtained after transfection with pMIG-HA-Rac(T17N)-IRES-GFP and pMIG-IRES-GFP retroviral constructs, respectively. GFP+ 32Dcl3 cells expressing mitochondrial-targeted catalase (MitCat) and a control clone were obtained from Dr Joel Greenberger (University of Pittsburgh Cancer Institute, Pittsburgh, PA). Mitochondrial localization of catalase was achieved by adding the mitochondrial localization sequence of superoxide dismutase 2 (MnSOD). BCR-ABL1-32Dcl3-MitCat cells were obtained after electroporation with pBABE-puro retroviral construct encoding for p210BCR-ABL1 and selection in puromycin. The studies involving animals were approved by the appropriate institutional animal care and use committees.
ROS, oxidative DNA damage, and genomic instability
Cells were incubated for 48 hours at 37°C in normoxia (17% O2 in 5% CO2 and 95% N2) or hypoxia (1% O2, 5% CO2, and 94% N2) as described previously.13 Fluorochromes displaying preferred sensitivity to ·O2−, H2O2, and ·OH, such as dihydroethidium (DHE), Mitosox Red (MSR), RedoxSensor Red (CC1), and chloromethyl-dichlorodihydrofluorescein diacetate (DCFDA; all from Invitrogen), were used to detect various ROS, as described previously.7 mt-cpYFP fluorescence, an indicator of mitochondrial ·O2−, was measured as described previously.11 8-oxoG and γ-H2AX nuclear foci were detected as described previously.7,8 In addition, FITC-avidin conjugates (avidin binds with high affinity to 8-oxoG in DNA) were used in flow cytometry analyses to detect 8-oxoG in LSCs according to the instructions in the OxyDNA fluorimetric kit (Argutus Medical). TKI-resistant clones expressing BCR-ABL1 kinase mutants were detected in methylcellulose in the presence of 1μM imatinib, as described previously.7 Chromosomal aberrations were detected by spectral karyotyping analysis and analyzed with Applied Spectral Imaging SKY 2.6 software as described previously.14
Rho0 cells
MitDNA-depleted Rho0 cells were generated as described previously.15 Briefly, cells were cultured in medium supplemented with growth factors and ethidium bromide (EB; 250 ng/mL), uridine (50 μg/mL), and sodium pyruvate (100 ng/mL) for 10 days. Control cells were cultured without EB. Rho0 cells were washed free of EB before being used in the experiments. The depletion of mitochondrial DNA was verified by examination of the expression of cytochrome oxidase II (Cox II) mRNA, coded by mitochondrial DNA, by RT-PCR analysis using mouse Cox II–specific upstream (5′-TGCATGTGGCTGTGGATGTCATCAA-3′) and downstream (5′-CACTAAGACAGACCCGTCATCTCCA-3′) primers and human Cox II–specific upstream (5′-ACAATAGCTAAGACCCAAACTGGG-3′) and downstream (5′-GCCCATGAGGTGGCAAGAAATGGG-3′) primers. As a control, the expression of GAPDH mRNA, coded by chromosomal DNA, was also analyzed by RT-PCR using mouse-specific upstream (5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′) and downstream (5′-CATGTGGGCCATGAGGTCCACCAC-3′) primers and human-specific upstream (5′-ACCACAGTCCATGCCATCAC-3′) and downstream (5′-TGCACCACCCTGTTGCTGTA-3′) primers. Rho0 cells displayed stable loss of CoxII mRNA and their survival depended on uridine and pyruvate.
Mitochondrial functions
For the measurement of mitochondria count, cells were stained with 200nM Mitotracker in culture medium, washed with PBS, and mounted on slides, followed by DNA staining with 4′,6′diamedino-2-phenylindole (DAPI). Specific staining was visualized using an inverted Olympus IX70 fluorescence microscope equipped with a 100× UPlan Apo lens (numeric aperture, 1.35), and a Sensicam QE camera (Cooke). At least 50 individual cells were analyzed per experimental group. Images were acquired with Slidebook Version 3.0 software (Intelligent Imaging Innovations) combined with de-convolution technology. For the measurement of mitochondrial membrane potential, cells were stained with 1-10μM JC-1 fluorescent probe in the presence or absence of a 50μM concentration of the depolarizing agent CCCP for 30 minutes at 37°C in the dark. Cells were then washed once with warm PBS and fluorescence was read using a 488-nm blue argon laser flow cytometer. Increased potential was detected as the increased shift of JC-1 fluorescence from 529-590 nm and is expressed as the red/green ratio. For the measurement of electron flow through MRC, mitochondrial complexes I-III and II-III electron flow activities were examined as described previously.16 Briefly, 1-2 × 108 cells were placed in ice-cold homogenization buffer containing 0.23M mannitol, 70mM sucrose, 10mM EGTA with 0.1% BSA, pH 7.4. Cells were homogenized in a dounce homogenizer and the homogenate was put through 3 freeze-thaw cycles. For complex I-III activity, 500 μg of whole-cell lysate was thawed and added to a cuvette containing 50μM oxidized cytochrome c and 1mM KCN with or without 1μM rotenone in 100mM potassium phosphate (pH 7.2) in a thermostated spectrophotometer at 30°C. A baseline absorbance at λ = 550 nm was recorded and the reaction was initiated with 0.2mM NADH. The increase in absorbance, corresponding to cytochrome c reduction, was recorded continuously for 60 seconds. For complex II-III, the electron flow activity (succinate-cytochrome c reductase) was determined using 2 mg/mL of lysate in 100mM potassium phosphate buffer containing 50μM oxidized cytochrome c, 1mM KCN, and 1μM rotenone, and was initiated by the addition of 6mM succinate. The activity of complexes I-III and II-III was expressed as the concentration (nanomolar) of reduced cytochrome c per minute per milligram (ε550 = 21.1nM−1cm−1).
Inhibitors of MRC complexes
Cells were incubated with or without specific inhibitors of MRC complexes (all from Sigma-Aldrich): 1mM rotenone (complex I), 4μM malonate (complex II), 2μM stigmatellin, 1μM myxothiazol, 1μM antimycin A (complex III), and 100μM KCN (complex IV) for 3 and 48 hours in the presence of uridine and sodium pyruvate before ROS and oxidative DNA damage, respectively, were examined. The inhibitors were not toxic for cells under these experimental conditions.
Mitochondria-targeted antioxidants
Cells were treated for 24 hours with 1μM mitochondria-targeted ubiquinone (obtained from Dr R. A. J. Smith, University of Otago, Dunedin, New Zealand). SS31 (d-Arg-Dmt-Lys-Phe-NH2; Dmt = 2′,6′-dimethyltyrosine) and SS20 (Phe-d-Arg-Phe-Lys-NH2) peptides were purchased from Genemed Synthesis. Cells were incubated with 1μM SS20 and SS31 for 24 hours (under pretested conditions described previously17 ). ICR-SCID female mice (Taconic) were total-body irradiated with 3 Gy from biologic irradiator RS 2000 (Rad Source Technologies) and IV injected with GFP+ muBMCs transfected with pMIG1-p210BCR-ABL1-IRES-GFP. Mice received daily IP treatment with 1.5 mg/kg of SS20 or SS31 or a combination of imatinib (50 mg/kg every morning and 100 mg/kg every evening by gavage) and SS20 or SS31.17,18 CD34+ CML-CP cells were cultured for 6 weeks with 1μM imatinib and 1μM SS20 or SS31 in medium supplemented with growth factors.
Rac activity and inhibitors
Cells were incubated with 25μM NSC23766 or 10μM EHT1864 (Calbiochem). Inhibition of Rac was confirmed in the PAK-binding domain pull-down assay according to the manufacturer's protocol (Upstate Biotechnology). Densitometric analysis was done using Quantity One Version 4.6.5 software (Bio-Rad). The hemagglutinin-tagged Rac(T17N) dominant-negative mutant [HA-Rac(T17N)] cDNA was described previously12 and was cloned in the LXSP-puro and pMIG-IRES-GFP retroviral constructs. CD34+ CML-CP cells and BCR-ABL1-32Dcl3 cells were infected with pMIG-HA-Rac(T17N)-IRES-GFP or LXSP-HA-Rac(T17N)-puro or with empty retroviruses as described previously.19 Expression of HA-Rac(T17N) in GFP+ and puromycin-resistant cells was confirmed by Western analysis. BCR-ABL1-32Dcl3 cells were transfected with pGFP-V-RS retroviral constructs encoding Rac2-specific or scrambled shRNA (Origene). Down-regulation of Rac2 in GFP+ cells was confirmed by Western analysis.
Western analyses
Proteins were detected in total cell lysates by SDS-PAGE, followed by Western blot analysis using the Abs recognizing ABL1, actin, and tubulin (Oncogene Science), Rac2 (Santa Cruz Biotechnology), catalase (Calbiochem), and hemagglutinin (Abcam).
Statistics
Results are presented as means ± SD and were analyzed by 2-tailed paired or unpaired Student t test and the Mann-Whitney rank-sum test when appropriate. The frequency of TKI-resistant mutations was calculated using the Luria-Delbruck fluctuation test with modifications to exclude the potential impact of proliferation rates.20
Results
Primitive CML-CP cells exhibit high levels of ROS and oxidative DNA damage
Lin−CD34+CD38− (CD34+CD38−) cells contain HSCs and LSCs, some of which are quiescent (CFSEmax or CTVmax).21 The Lin−CD34+CD38+ (CD34+CD38+) immunophenotype characterizes the populations of an early hematopoietic progenitor cells and LPCs, which show a greater proliferative activity (CFSElow or CTVlow).
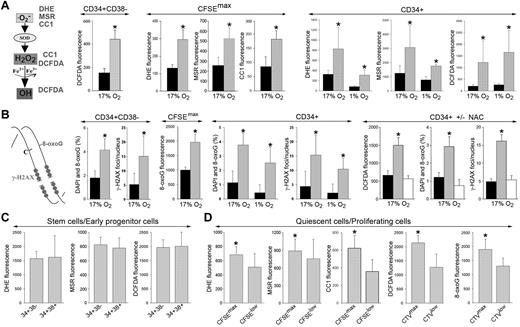
O2−, H2O2, and ·OH display different capabilities to cause oxidative DNA damage: ·O2− < H2O2 < ·OH.22 Using fluorescent reagents that are selectively sensitive to these distinct ROS (Figure 1A left), we detected a 2- to 2.5-fold increase in the levels of ·O2−, H2O2 and ·OH in the most primitive CD34+CD38− and quiescent CD34+CFSEmax CML-CP cells compared with their normal counterparts (Figure 1A CD34+CD38− and CFSEmax). Moreover, a 2- to 6-fold elevation of ROS was observed in primitive CD34+ CML-CP cells in normoxia (17% O2), which mimics arterial blood (> 10% O2), and also in hypoxia (1% O2; Figure 1A CD34+), which mimics the BM niche.
Elevated levels of ROS and oxidative DNA damage in primitive CML-CP CD34+ subsets. (A) DHE, MSR, CC1, and DCFDA were used to detect various ROS with different DNA damaging capabilities (left). ROS was measured in CD34+CD38−, quiescent CD34+CFSEmax (CFSEmax), and total CD34+ cells from healthy donors (black bars) and CML-CP patients (gray bars) in 17% and 1% O2, as indicated. (B) Diagram showing oxidative DNA lesions: 8-oxoG and DSBs (stained by γ-H2AX; left). 8-oxoG and γ-H2AX were detected by specific fluorescence in CD34+CD38−, quiescent CD34+CFSEmax (CFSEmax), and total CD34+ cells from healthy donors (black bars) and CML-CP patients (gray bars) maintained in 17% and 1% O2, as indicated (CD34+ ± NAC). CD34+ cells from healthy donors (black bars) and those from CML-CP patients were incubated (white bars) or not (gray bars) with 50μM N-acetyl-L-cysteine for 48 hours. ROS (DCFDA fluorescence), 8-oxoG, and γ-H2AX foci were detected. (C-D) ROS (DHE, MSR, CC1, and DCFDA fluorescence) and/or 8-oxoG was detected in CD34+CD38− and CD34+CD38+ CML-CP cells (C) and in CD34+(CFSEmax or CTVmax) quiescent and CD34+(CFSElow or CTVlow) proliferating CML-CP cells (D). Results represent means of 3-20 samples/group ± SD. *P < .05 compared with healthy counterparts (A-B), these treated with NAC (B), and CFSElow or CTVlow (D).
Elevated levels of ROS and oxidative DNA damage in primitive CML-CP CD34+ subsets. (A) DHE, MSR, CC1, and DCFDA were used to detect various ROS with different DNA damaging capabilities (left). ROS was measured in CD34+CD38−, quiescent CD34+CFSEmax (CFSEmax), and total CD34+ cells from healthy donors (black bars) and CML-CP patients (gray bars) in 17% and 1% O2, as indicated. (B) Diagram showing oxidative DNA lesions: 8-oxoG and DSBs (stained by γ-H2AX; left). 8-oxoG and γ-H2AX were detected by specific fluorescence in CD34+CD38−, quiescent CD34+CFSEmax (CFSEmax), and total CD34+ cells from healthy donors (black bars) and CML-CP patients (gray bars) maintained in 17% and 1% O2, as indicated (CD34+ ± NAC). CD34+ cells from healthy donors (black bars) and those from CML-CP patients were incubated (white bars) or not (gray bars) with 50μM N-acetyl-L-cysteine for 48 hours. ROS (DCFDA fluorescence), 8-oxoG, and γ-H2AX foci were detected. (C-D) ROS (DHE, MSR, CC1, and DCFDA fluorescence) and/or 8-oxoG was detected in CD34+CD38− and CD34+CD38+ CML-CP cells (C) and in CD34+(CFSEmax or CTVmax) quiescent and CD34+(CFSElow or CTVlow) proliferating CML-CP cells (D). Results represent means of 3-20 samples/group ± SD. *P < .05 compared with healthy counterparts (A-B), these treated with NAC (B), and CFSElow or CTVlow (D).
In addition, 3-5 times more DNA lesions, such as 8-oxoG and DSBs marked by γ-H2AX (Figure 1B left), were detected in the total CD34+ CML-CP cells in normoxia and hypoxia, and a 2- to 3-fold increase was also observed in the most primitive CD34+CD38− and quiescent CD34+CFSEmax CML-CP cells (Figure 1B). This effect depended on ROS, because N-acetyl-L-cysteine (NAC), a widely used antioxidant, diminished DNA damage in leukemia cells by 2.5- to 3.5-fold (Figure 1B CD34+ ± NAC).
CD34+CD38− and CD34+CD38+ CML-CP cells displayed similarly high levels of various ROS moieties (Figure 1C). Interestingly, the most primitive quiescent CD34+CFSEmax/CTVmax CML-CP cells accumulated 1.4-1.8 times more ROS (except mitochondrial ·O2−) and approximately 1.4 times more 8-oxoG compared with proliferating pools (Figure 1D).
On the basis of these observations and the studies in murine hematopoietic 32Dcl3 cells expressing TKI-resistant BCR-ABL1 kinase variants (supplemental Figure 1A-D, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), we predicted that primitive CML-CP cells carrying these mutations would also contain high levels of ROS and oxidative DNA damage. Moreover, cells transformed with TEL-ABL1, TEL-JAK2, TEL/PDGFβR, TEL-TRKC(L), BCR-FGFR1, JAK2(V617F), and FLT3-ITD also stimulate ROS, suggesting a broad effect of OTKs on the production of ROS (supplemental Figure 1E).6
Metabolic alterations in mitochondria are associated with enhanced production of ROS in primitive CML-CP cells
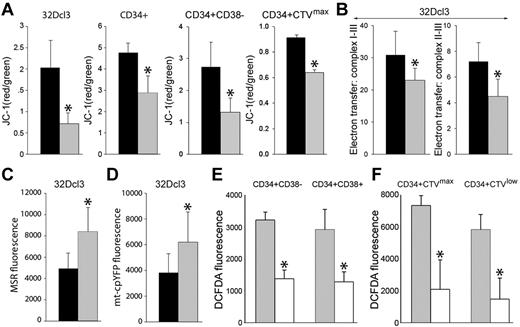
Results with MSR selectively detecting mitochondrial ·O2− suggested that primitive CML-CP cells generate > 2 times more mitochondrial ROS than their normal counterparts (Figure 1A CFSEmax MSR and CD34+ MSR). CML-CP cells and their normal counterparts contain similar numbers of mitochondria detected by MitoTracker Red (40 ± 18 and 34 ± 19, respectively). Using JC-1, a voltage-sensitive green cationic dye in which mitochondrial uptake and red fluorescence is directly related to the magnitude of mitochondrial membrane potential (ΔΨm),23 we found that the mitochondrial membrane was depolarized in BCR-ABL1-32Dcl3 cells, in total CD34+ CML-CP cells, and in most primitive CD34+CD38− and quiescent CD34+CTVmax CML-CP cells compared with their normal counterparts (Figure 2A). Abnormal ΔΨm in BCR-ABL1–32Dcl3 cells was associated with a 33%-67% reduction of the electron flow rate between MRC complexes I-III and II-III (Figure 2B). These biophysical alterations in BCR-ABL1–32Dcl3 cells were associated with approximately 1.6-fold enhanced production of mitochondrial ·O2− measured by MSR (Figure 2C) and by mt-cpYFP (Figure 2D), an ultrasensitive biosensor of mitochondrial ·O2−.11
Biochemical alterations in the mitochondria in BCR-ABL1 leukemia cells. (A) BCR-ABL1-32Dcl3 cells (gray bars) and 32Dcl3 cells (black bars), and CD34+, CD34+CD38− and CD34+CTVmax cells from CML-CP patients (gray bars) and healthy donors (black bars) were stained with JC-1 and the red/green fluorescence ratio is shown. (B) 32Dcl3 cells (black bars) and BCR-ABL1-32Dcl3 cells (gray bars) were used to measure electron transfer activity between MRC complexes I-III (left panel) and II-III (right panel). (C-D) Mitochondrial ·O2− was measured in 32Dcl3 cells (black bars) and BCR-ABL1-32Dcl3 cells (gray bars) by MSR (C) and mt-cpYFP fluorescence (D). (E-F) ROS were measured by DCFDA in CML-CP CD34+CD38− and CD34+CD38+ subpopulations (E) and in CD34+CTVmax quiescent and CD34+CTVlow proliferative cells incubated (white bars) or not (gray bars) with mitochondria-targeted ubiquinone (F). Results represent mean values of a minimum of 3 measurements/group ± SD. *P < .05 compared with normal/parental (A-D) and untreated (E-F) cells.
Biochemical alterations in the mitochondria in BCR-ABL1 leukemia cells. (A) BCR-ABL1-32Dcl3 cells (gray bars) and 32Dcl3 cells (black bars), and CD34+, CD34+CD38− and CD34+CTVmax cells from CML-CP patients (gray bars) and healthy donors (black bars) were stained with JC-1 and the red/green fluorescence ratio is shown. (B) 32Dcl3 cells (black bars) and BCR-ABL1-32Dcl3 cells (gray bars) were used to measure electron transfer activity between MRC complexes I-III (left panel) and II-III (right panel). (C-D) Mitochondrial ·O2− was measured in 32Dcl3 cells (black bars) and BCR-ABL1-32Dcl3 cells (gray bars) by MSR (C) and mt-cpYFP fluorescence (D). (E-F) ROS were measured by DCFDA in CML-CP CD34+CD38− and CD34+CD38+ subpopulations (E) and in CD34+CTVmax quiescent and CD34+CTVlow proliferative cells incubated (white bars) or not (gray bars) with mitochondria-targeted ubiquinone (F). Results represent mean values of a minimum of 3 measurements/group ± SD. *P < .05 compared with normal/parental (A-D) and untreated (E-F) cells.
We next sought to investigate the contribution of mitochondria to total ROS in primitive CML-CP cells. Mitochondria-targeted ubiquinone, which converts mitochondrial H2O2 to H2O and O2,24 reduced ROS by 2- to 3-fold in most primitive CD34+CD38− and quiescent CD34+CTVmax cells and also in CD34+CD38+ progenitor and CD34+CTVlow proliferative cells (Figure 2E-F).
MRC-cIII is responsible for overproduction of ROS and excessive oxidative DNA damage in primitive CML-CP cells
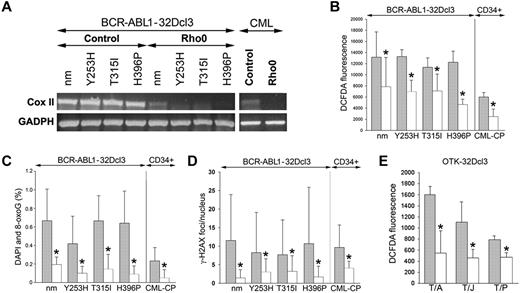
The MRC consists of 87 proteins, 13 of which are encoded by mitochondrial DNA (mtDNA). Cells devoid of mtDNA, and therefore lacking functional MRC (Rho0 cells), were generated after exposure to EB, which intercalates preferentially with mtDNA, thereby interfering with replication enzymes to generate cells stably depleted of mtDNA.15 Loss of mtDNA in Rho0 cells was confirmed by RT-PCR showing down-regulation of CoxII after EB treatment, which persisted throughout the experiment (Figure 3A). Various Rho0 cells (32Dcl3 cells transformed by BCR-ABL1 kinase or the TKI-resistant mutants and CD34+ CML-CP cells) accumulated 1.6-2.5 times less ROS and 2.5-12 times less oxidative DNA damage (8-oxoG and γ-H2AX) than their MRC-proficient counterparts (Figure 3B-D, respectively). At the time of analysis, Rho0 cells and MRC-proficient cells displayed a similar cell-cycle profile (data not shown), thus excluding a possible impact of cell proliferation on ROS and oxidative DNA damage. In addition, Rho0 cells transformed by other OTKs such as TEL-ABL1, TEL-JAK2 and TEL-PDGFβR also demonstrated a 1.6- to 3.0-fold reduction of ROS, implicating a broad role of MRC in the production of ROS in leukemias (Figure 3E).
MRC-depleted BCR-ABL1 leukemia cells display reduced ROS and oxidative DNA damage. (A-D) BCR-ABL1-32Dcl3 cells expressing nonmutated BCR-ABL1 kinase (nm) and indicated TKI-resistant (TKIR) mutants and CD34+ cells from CML-CP patients (control cells, gray bars) were depleted of mtDNA (Rho0 cells, white bars). (A) Expression of Cox II and GADPH mRNA by RT-PCR. ROS were detected by DCFDA (B) and 8-oxoG (C) and γ-H2AX (D) foci were analyzed by immunofluorescence. (E) ROS were detected by DCFDA in 32Dcl3 cells expressing TEL-ABL1 (T/A), TEL-JAK2 (T/J), and TEL-PDGFβR (T/P; gray bars) and in corresponding Rho0 cells (white bars). Results represent mean values ± SD from a minimum of 2 experiments/group (A,B,E) and from 25-55 cells/group (C-D). *P < .05 compared with control cells.
MRC-depleted BCR-ABL1 leukemia cells display reduced ROS and oxidative DNA damage. (A-D) BCR-ABL1-32Dcl3 cells expressing nonmutated BCR-ABL1 kinase (nm) and indicated TKI-resistant (TKIR) mutants and CD34+ cells from CML-CP patients (control cells, gray bars) were depleted of mtDNA (Rho0 cells, white bars). (A) Expression of Cox II and GADPH mRNA by RT-PCR. ROS were detected by DCFDA (B) and 8-oxoG (C) and γ-H2AX (D) foci were analyzed by immunofluorescence. (E) ROS were detected by DCFDA in 32Dcl3 cells expressing TEL-ABL1 (T/A), TEL-JAK2 (T/J), and TEL-PDGFβR (T/P; gray bars) and in corresponding Rho0 cells (white bars). Results represent mean values ± SD from a minimum of 2 experiments/group (A,B,E) and from 25-55 cells/group (C-D). *P < .05 compared with control cells.
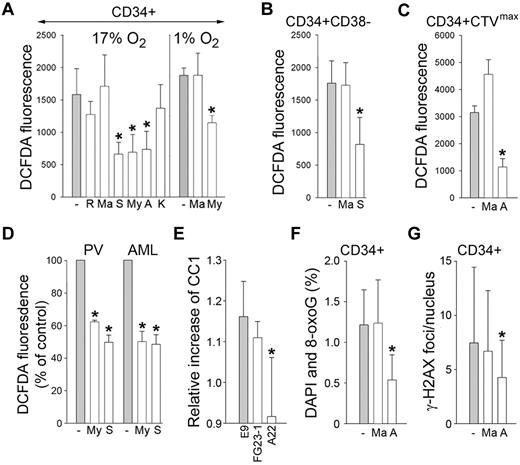
To determine which MRC complex might be contributing to the overproduction of DNA-damaging ROS (H2O2 and ·OH), CD34+ cells from CML-CP patients were incubated with specific inhibitors of complex I (rotenone), complex II (malonate), complex III (myxothiazol, stigmatellin, and antimycin A), and complex IV (KCN).25 MRC-cIII inhibitors, but not others, reduced the levels of H2O2 and ·OH in normoxia (17% O2) and hypoxia (1% O2) by 1.6- to 2.4-fold (Figure 4A). Moreover, antimycin A, but not malonate, reduced these ROS in the most primitive CD34+CD38− and quiescent CD34+CTVmax CML-CP subsets by 2.2-2.9 times (Figure 4B-C, respectively). MRC-cIII inhibitors also diminished the levels of H2O2 and ·OH by 40%-50% in primary FLT3(ITD)–positive AML and JAK2(V617F)–PV vera cells (Figure 4D), and this was associated with reduced O2 consumption (supplemental Figure 2). MRC-cIII inhibitors did not affect SOD, which catalyzes the dismutation of ·O2− into H2O2 and O2, nor catalase, which degrades H2O2 (supplemental Figure 3), thus confirming their specificity.
MRC-cIII is responsible for ROS-induced oxidative DNA damage in primitive CML-CP CD34+ subsets and in PV and AML cells. CD34+ (A), CD34+CD38− (B), and quiescent CD34+CTVmax (C) cells from CML-CP patients were incubated without (gray bars) or with (white bars) the indicated inhibitors (R indicates rotenone; Ma, malonate; S, stigmatellin; My, myxothiazol; A, antimycin; and K, KCN) for 3 hours in 17% or 1% O2 (A) and in 17% O2 (B-C). ROS were measured by DCFDA. (D) ROS were also measured by DCFDA in primary FLT3(ITD)–positive AML cells and in JAK2(V617F)–positive PV cells untreated (−) or treated with myxothiazol (My) or stigmatellin (S). Results show relative ROS levels compared with untreated cells. (E) Control L929 cells (E9) and clones containing a defective mutation in complex I (FG23-1) and complex III (A22) were cotransfected with BCR-ABL1–IRES-GFP or IRES-GFP expression plasmid. ROS was measured 72 hours later in GFP+ cells with CC1. Results represent the relative increase of ROS in BCR-ABL1+ cells. 8-oxoG (F) and γ-H2AX (G) DNA lesions were detected in CD34+ CML-CP cells incubated with malonate, antimycin A, or diluent (−) for 48 hours. Results represent mean values of a minimum of 3 measurements/group ± SD. *P < .05 compared with the untreated cells (A-D,F-G) and with E9 (E).
MRC-cIII is responsible for ROS-induced oxidative DNA damage in primitive CML-CP CD34+ subsets and in PV and AML cells. CD34+ (A), CD34+CD38− (B), and quiescent CD34+CTVmax (C) cells from CML-CP patients were incubated without (gray bars) or with (white bars) the indicated inhibitors (R indicates rotenone; Ma, malonate; S, stigmatellin; My, myxothiazol; A, antimycin; and K, KCN) for 3 hours in 17% or 1% O2 (A) and in 17% O2 (B-C). ROS were measured by DCFDA. (D) ROS were also measured by DCFDA in primary FLT3(ITD)–positive AML cells and in JAK2(V617F)–positive PV cells untreated (−) or treated with myxothiazol (My) or stigmatellin (S). Results show relative ROS levels compared with untreated cells. (E) Control L929 cells (E9) and clones containing a defective mutation in complex I (FG23-1) and complex III (A22) were cotransfected with BCR-ABL1–IRES-GFP or IRES-GFP expression plasmid. ROS was measured 72 hours later in GFP+ cells with CC1. Results represent the relative increase of ROS in BCR-ABL1+ cells. 8-oxoG (F) and γ-H2AX (G) DNA lesions were detected in CD34+ CML-CP cells incubated with malonate, antimycin A, or diluent (−) for 48 hours. Results represent mean values of a minimum of 3 measurements/group ± SD. *P < .05 compared with the untreated cells (A-D,F-G) and with E9 (E).
Furthermore, BCR-ABL1 kinase failed to induce ROS in the L929 clone lacking the assembled complex III (A22), whereas it elevated ROS in the clone lacking complex I (FG23-1) and in E9 control cells (Figure 4E), supporting the role of MRC-cIII in the generation of ROS in primitive leukemia cells.
In addition, antimycin A, but not malonate, diminished 8-oxoG and DSBs by 1.8- to 2.4-fold in CD34+ CML-CP cells (Figure 4F-G, respectively), indicating that MRC-cIII plays a significant role in the induction of oxidative DNA damage in very primitive CML cells.
Rac2-GTP–dependent production of mitochondrial ROS and oxidative DNA damage in primitive CML-CP cells
Among numerous signaling proteins activated in CML cells, Rac GTPases were potential candidates to regulate the mitochondrial production of ROS.12,26 The Rac(T17N) dominant-negative mutant was used to examine the role of Rac-GTP in the induction of mitochondrial ROS, oxidative DNA damage, and genomic instability.12 Expression of HA-Rac(T17N) in BCR-ABL1–32Dcl3 cells was confirmed by Western blotting (Figure 5A Western blots), and cells expressing untagged Rac(T17N) have been described previously.12 Rac(T17N) increased ΔΨm by approximately 1.8 times (Figure 5A JC-1). HA-Rac(T17N) accelerated electron transfer between MRC complexes I-III and II-III by 25%-40% (Figure 5A electron transfer) and diminished mitochondrial ·O2− and cellular ·O2− and H2O2 by approximately 3.8-fold (Figure 5A MSR and CC1, respectively) and decreased 8-oxoG and γ-H2AX by 1.5- and 5-fold, respectively (Figure 5A 8-oxoG and γ-H2AX). HA-Rac(T17N) also inhibited mitochondrial and cellular ROS in CD34+ CML-CP cells by 2.8- and 2.0-fold, respectively (Figure 5B).
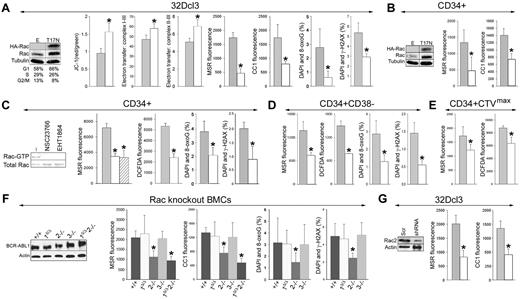
Rac2 induces mitochondrial ROS, oxidative DNA damage, and genomic instability. (A) BCR-ABL1-32Dcl3 cells were transfected with pMIG-HA-Rac(T17N)–IRES-GFP or LXSP-Rac(T17N) (T17N) and pMIG-IRES-GFP or LXSP (E) retroviruses. Expression of HA-tagged Rac(T17N) mutant and total Rac was detected by Western blot analysis. Results from cells expressing E and T17N are represented by gray and white bars, respectively. ΔΨm (JC-1 red/green fluorescence ratio), electron transport between MRC complexes I-III and II-III, ROS (MSR and CC1 fluorescence), 8-oxoG, and DSBs (marked by γ-H2AX) were assessed (respective panels). (B) CD34+ CML-CP cells were transfected with HA-Rac(T17N)–IRES-GFP (T17N) or IRES-GFP (E) retroviruses. Western blot analyses of the expression of HA-tagged Rac(T17N) mutant and total Rac in GFP+ cells. ROS (bar panels) were measured with MSR and CC1. CD34+ (C), CD34+CD38− (D), and quiescent CD34+CTVmax (E) CML-CP cells were untreated (−) or treated with NSC23766 or EHT1864. (C) Western blot. Rac activation assay: reaction samples (topbox) and total cell lysates (bottom box) were analyzed for Rac-GTP and total Rac protein content, respectively. (C-E bar panels) ROS (MRC and DCFDA fluorescence), 8-oxoG, and γ-H2AX were measured in untreated cells (gray bars) and in cells treated with NSC23766 (white bars) and EHT1864 (striped bars). (F) BCR-ABL1 detected by Western analysis in double-positive, Rac1Δ/Δ, Rac2−/−, Rac3−/−, and Rac1Δ/ΔRac2−/− muBMCs. ROS (MSR and CC1 fluorescence) and oxidative DNA lesions (8-oxoG) and DSBs (γ-H2AX) were detected (bar panels). (G) BCR-ABL1-32Dcl3 cells were transfected with pGFP-V-RS retroviral vector encoding Rac2-specific shRNA (shRNA) or scrambled RNA (Scr). Down-regulation of Rac2 in GFP+ cells was detected by Western blot analysis. ROS (MRC and CC1 fluorescence) were measured in GFP+ cells expressing Scr (gray bars) and shRNA (white bars). Results represent mean values of a minimum of 3 measurements/group ± SD (A-E,G) and mean values of 3-14 muBMC samples/group ± SD (F). *P < .05 compared with empty/Scr plasmid (A,B,G), untreated cells (C-E), and double-positive cells (F).
Rac2 induces mitochondrial ROS, oxidative DNA damage, and genomic instability. (A) BCR-ABL1-32Dcl3 cells were transfected with pMIG-HA-Rac(T17N)–IRES-GFP or LXSP-Rac(T17N) (T17N) and pMIG-IRES-GFP or LXSP (E) retroviruses. Expression of HA-tagged Rac(T17N) mutant and total Rac was detected by Western blot analysis. Results from cells expressing E and T17N are represented by gray and white bars, respectively. ΔΨm (JC-1 red/green fluorescence ratio), electron transport between MRC complexes I-III and II-III, ROS (MSR and CC1 fluorescence), 8-oxoG, and DSBs (marked by γ-H2AX) were assessed (respective panels). (B) CD34+ CML-CP cells were transfected with HA-Rac(T17N)–IRES-GFP (T17N) or IRES-GFP (E) retroviruses. Western blot analyses of the expression of HA-tagged Rac(T17N) mutant and total Rac in GFP+ cells. ROS (bar panels) were measured with MSR and CC1. CD34+ (C), CD34+CD38− (D), and quiescent CD34+CTVmax (E) CML-CP cells were untreated (−) or treated with NSC23766 or EHT1864. (C) Western blot. Rac activation assay: reaction samples (topbox) and total cell lysates (bottom box) were analyzed for Rac-GTP and total Rac protein content, respectively. (C-E bar panels) ROS (MRC and DCFDA fluorescence), 8-oxoG, and γ-H2AX were measured in untreated cells (gray bars) and in cells treated with NSC23766 (white bars) and EHT1864 (striped bars). (F) BCR-ABL1 detected by Western analysis in double-positive, Rac1Δ/Δ, Rac2−/−, Rac3−/−, and Rac1Δ/ΔRac2−/− muBMCs. ROS (MSR and CC1 fluorescence) and oxidative DNA lesions (8-oxoG) and DSBs (γ-H2AX) were detected (bar panels). (G) BCR-ABL1-32Dcl3 cells were transfected with pGFP-V-RS retroviral vector encoding Rac2-specific shRNA (shRNA) or scrambled RNA (Scr). Down-regulation of Rac2 in GFP+ cells was detected by Western blot analysis. ROS (MRC and CC1 fluorescence) were measured in GFP+ cells expressing Scr (gray bars) and shRNA (white bars). Results represent mean values of a minimum of 3 measurements/group ± SD (A-E,G) and mean values of 3-14 muBMC samples/group ± SD (F). *P < .05 compared with empty/Scr plasmid (A,B,G), untreated cells (C-E), and double-positive cells (F).
To investigate the role of Rac in the generation of ROS in primitive CML-CP cells, we used 2 Rac-specific inhibitors: NSC23766, which inhibits the guanine nucleotide exchange factor–dependent activation of Rac, and EHT1864, which impairs Rac-GTP formation and prevents the activation of Rac downstream effectors.27,28 Treatment of CD34+ CML-CP cells with NSC23766 or EHT1864 down-regulated Rac-GTP (Figure 5C Western blots) and also inhibited mitochondrial ·O2− (Figure 5C MSR) and cellular levels of H2O2 and ·OH by 2.0- to 2.2-fold (Figure 5C DCFDA), thereby reducing oxidative DNA damage by 1.9-2.2 times (Figure 5C 8-oxoG and γ-H2AX). Moreover, NSC23766 down-regulated mitochondrial ·O2−, cellular H2O2, and ·OH and oxidative DNA damage in the most primitive CD34+CD38− cells (Figure 5D) and ROS levels in quiescent CD34+CTVmax (Figure 5E) CML-CP cells by 2.0-2.8 times. Consistent with our previous results,12 NSC23766 did not affect the cell-cycle progression of CD34+ CML-CP cells in the presence of growth factors significantly (supplemental Figure 4). In addition, NSC23766 reduced ROS in G1, S, and G2/M cell-cycle phases, indicating that the inhibitor exerts its activity via the cell cycle.
There are 3 members of the Rac family: Rac1, Rac2, and Rac3, which exert unique and overlapping roles in hematopoietic cells.29 To determine which Rac stimulates the production of mitochondrial ROS in leukemia cells, BCR-ABL1 was expressed in muBMCs lacking specific Rac (Rac1Δ/Δ, Rac2−/−, Rac3−/−, and Rac1Δ/ΔRac2−/−) and also in their +/+ counterparts (Figure 5F Western blot). Production of mitochondrial ·O2− and total cellular H2O2 and ·O2− was impaired in BCR-ABL1+Rac2−/− and Rac1Δ/ΔRac2−/− cells compared with BCR-ABL1-positive cells by 2.0- to 2.7-fold, whereas it was not affected in Rac1Δ/Δ and Rac3−/− cells (Figure 5F MSR and CC1). BCR-ABL1+Rac2−/− cells also displayed approximately 2.1 times less oxidative DNA damage (Figure 5F 8-oxoG and γ-H2AX).
To support a key role of Rac2 in the stimulation of ROS production, BCR-ABL1-32Dcl3 cells were transfected with Rac2-specific shRNA. Down-regulation of Rac2 was associated with an approximately 2.5-fold reduction in the levels of mitochondrial ·O2− and total cellular H2O2 and ·O2− (Figure 5G MSR and CC1, respectively). Down-regulation of Rac2 also diminished mitochondrial ·O2− in 32Dcl3 cells expressing TEL-ABL1, TEL-JAK2, and TEL-PDGFβR (supplemental Figure 5).
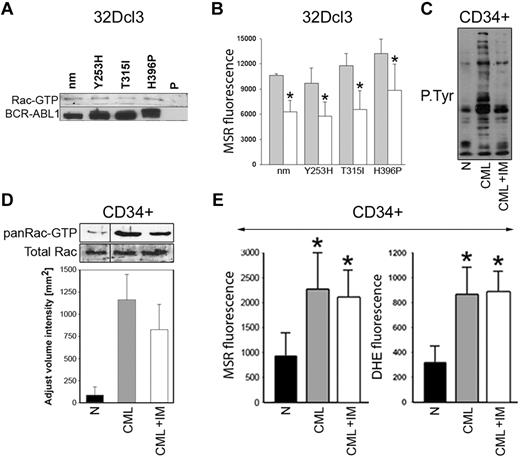
Rac-GTP was stimulated not only in leukemia cells expressing TKI-sensitive BCR-ABL1 kinase, but also in cells containing TKI-resistant BCR-ABL1 mutants (Figure 6A). The expression of Rac(T17N) reduced mitochondrial ·O2− by approximately 1.5- to 1.9-fold in these cells (Figure 6B). Moreover, despite almost complete inhibition of BCR-ABL1 kinase–mediated tyrosine phosphorylation (Figure 6C), approximately 73% of Rac remained active in CML-CP CD34+ cells treated with imatinib (Figure 6D), and this was associated with elevated levels of the cellular and mitochondrial ·O2− in these cells (Figure 6E).
Role of Rac in the induction of mitochondrial ROS in TKI-resistant BCR-ABL1 cells and CD34+ CML-CP cells treated with imatinib. (A) Rac activation was examined in 32Dcl3 parental cells (P) and clones expressing nonmutated BCR-ABL1 (nm) and the indicated TKI-resistant mutants (Y253H, T315I, and H396P) using a PAK-binding domain pull-down assay; top box shows the Rac-GTP protein content, bottom box shows the expression of BCR-ABL1 protein variants. (B) The indicated cells were transfected with HA-Rac(T17N)-IRES-GFP (white bars) or IRES-GFP (gray bars) retroviral particles and mitochondrial ·O2− was measured with MSR in GFP+ cells. (C-E) CD34+ cells from healthy donors (N) and from CML-CP patients were incubated with 1μM imatinib (CML + IM) or placebo (CML) for 12 hours in the presence of growth factors. (C) Tyrosine phosphorylation (P.Tyr) of total cellular proteins was analyzed by Western blot using anti-phosphotyrosine Ab. (D) Top panel is a representative Western blot analysis of the Rac-GTP active form detected using a PAK-binding domain pull-down assay; bottom panel shows the total Rac protein in cell lysates by densitometric analysis of Rac-GTP. (E) Total cellular and mitochondrial ·O2− was measured with the use of DHE and MSR, respectively. Results represent mean values of 2 or 3 measurements/group ± SD. *P < .05 compared with cells transfected with an empty plasmid (B) or normal counterparts (D).
Role of Rac in the induction of mitochondrial ROS in TKI-resistant BCR-ABL1 cells and CD34+ CML-CP cells treated with imatinib. (A) Rac activation was examined in 32Dcl3 parental cells (P) and clones expressing nonmutated BCR-ABL1 (nm) and the indicated TKI-resistant mutants (Y253H, T315I, and H396P) using a PAK-binding domain pull-down assay; top box shows the Rac-GTP protein content, bottom box shows the expression of BCR-ABL1 protein variants. (B) The indicated cells were transfected with HA-Rac(T17N)-IRES-GFP (white bars) or IRES-GFP (gray bars) retroviral particles and mitochondrial ·O2− was measured with MSR in GFP+ cells. (C-E) CD34+ cells from healthy donors (N) and from CML-CP patients were incubated with 1μM imatinib (CML + IM) or placebo (CML) for 12 hours in the presence of growth factors. (C) Tyrosine phosphorylation (P.Tyr) of total cellular proteins was analyzed by Western blot using anti-phosphotyrosine Ab. (D) Top panel is a representative Western blot analysis of the Rac-GTP active form detected using a PAK-binding domain pull-down assay; bottom panel shows the total Rac protein in cell lysates by densitometric analysis of Rac-GTP. (E) Total cellular and mitochondrial ·O2− was measured with the use of DHE and MSR, respectively. Results represent mean values of 2 or 3 measurements/group ± SD. *P < .05 compared with cells transfected with an empty plasmid (B) or normal counterparts (D).
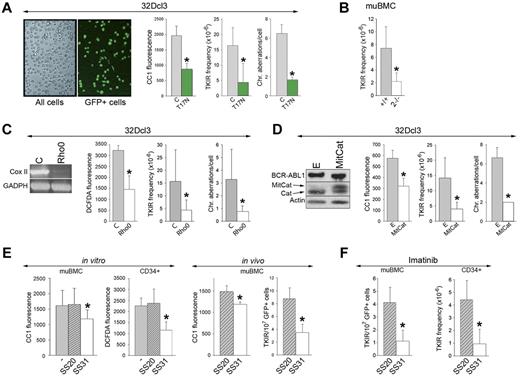
Targeting the Rac2 mitochondrial ROS pathway prevents genomic instability in BCR-ABL1 leukemia
GFP+ BCR-ABL1-32Dcl3 cells cultured for 12 weeks or harvested from SCID mice after 6-8 weeks accumulated TKI-resistant BCR-ABL1 kinase mutants and chromosomal aberrations (ie, translocations, deletions, and additions), as described previously.7,14 Expression of HA-Rac(T17N) in GFP+ in BCR-ABL1-32Dcl3 cells down-regulated ROS by approximately 2.4-fold (Figure 7A left), reduced the frequency of TKI-resistant clones by 4-fold, and inhibited the accumulation of chromosomal aberrations by approximately 2.2-fold after 12 weeks of in vitro culture (Figure 7A right). In addition, after 8 weeks of in vitro culture, the frequency of TKI-resistant clones in BCR-ABL1+Rac2−/− cells was diminished by approximately 3.5 times compared with their BCR-ABL1–positive counterparts (Figure 7B).
Targeting the Rac-mitochondrial ROS pathway reduces genomic instability in BCR-ABL1 leukemia cells. (A) GFP+ BCR-ABL1-32Dcl3 cells expressing Rac(T17N) and GFP- BCR-ABL1-32Dcl3 control cells (C) as illustrated. BCR-ABL1+/+ and Rac2−/− muBMCs are described in Figure 5F (B); BCR-ABL1-32Dcl3 Rho0 cells (Rho0) and BCR-ABL1-32Dcl3 MRC-proficient cells (C) as confirmed by RT-PCR–detected down-regulation of expression of Cox II in Rho0 cells; and BCR-ABL1-32Dcl3 cells expressing MitCat (D), or empty plasmid (E) as documented by Western analysis showing the expression of BCR-ABL1, MitCat, and endogenous catalase (Cat), were maintained for 8 (B) or 12 (A,C,D) weeks in liquid culture. ROS were measured using CC1 and DCFDA fluorescence. The frequency of TKI-resistant (TKIR) clones and/or spectral karyotyping analysis of chromosomal aberrations were determined; results represent mean values of a minimum of 3 experiments ± SD. (E) In the in vitro experiment, BCR-ABL1 muBMCs and CML-CP CD34+ cells were cultured either without (−) or with SS20 or SS31 peptides. ROS were measured using CC1 and DCFDA fluorescence; results represent mean values of a minimum of 3 experiments ± SD. In the in vivo experiment, 5 SCID mice/group bearing GFP+ BCR-ABL1 muBMCs were treated with SS20 or SS31 for 6-8 weeks. ROS (CC1 fluorescence) and the number of TKIR clones in GFP+ muBMCs were determined. (F) Left panel shows 5 SCID mice/group bearing GFP+ BCR-ABL1 muBMCs treated with imatinib and SS20 or SS31 for 8 weeks. TKIR clones were detected in GFP+ cells from BM and spleen. Right panel shows CD34+ cells from 2 CML-CP patients cultured for 6 weeks with imatinib and SS20 or SS31 in medium supplemented with growth factors, after which time the frequency of TKIR clones was determined. *P < .05 compared with control, untreated, and SS20-treated cells or animals.
Targeting the Rac-mitochondrial ROS pathway reduces genomic instability in BCR-ABL1 leukemia cells. (A) GFP+ BCR-ABL1-32Dcl3 cells expressing Rac(T17N) and GFP- BCR-ABL1-32Dcl3 control cells (C) as illustrated. BCR-ABL1+/+ and Rac2−/− muBMCs are described in Figure 5F (B); BCR-ABL1-32Dcl3 Rho0 cells (Rho0) and BCR-ABL1-32Dcl3 MRC-proficient cells (C) as confirmed by RT-PCR–detected down-regulation of expression of Cox II in Rho0 cells; and BCR-ABL1-32Dcl3 cells expressing MitCat (D), or empty plasmid (E) as documented by Western analysis showing the expression of BCR-ABL1, MitCat, and endogenous catalase (Cat), were maintained for 8 (B) or 12 (A,C,D) weeks in liquid culture. ROS were measured using CC1 and DCFDA fluorescence. The frequency of TKI-resistant (TKIR) clones and/or spectral karyotyping analysis of chromosomal aberrations were determined; results represent mean values of a minimum of 3 experiments ± SD. (E) In the in vitro experiment, BCR-ABL1 muBMCs and CML-CP CD34+ cells were cultured either without (−) or with SS20 or SS31 peptides. ROS were measured using CC1 and DCFDA fluorescence; results represent mean values of a minimum of 3 experiments ± SD. In the in vivo experiment, 5 SCID mice/group bearing GFP+ BCR-ABL1 muBMCs were treated with SS20 or SS31 for 6-8 weeks. ROS (CC1 fluorescence) and the number of TKIR clones in GFP+ muBMCs were determined. (F) Left panel shows 5 SCID mice/group bearing GFP+ BCR-ABL1 muBMCs treated with imatinib and SS20 or SS31 for 8 weeks. TKIR clones were detected in GFP+ cells from BM and spleen. Right panel shows CD34+ cells from 2 CML-CP patients cultured for 6 weeks with imatinib and SS20 or SS31 in medium supplemented with growth factors, after which time the frequency of TKIR clones was determined. *P < .05 compared with control, untreated, and SS20-treated cells or animals.
To determine the impact of MRC-generated ROS on genomic instability, Rho0 cells displaying down-regulation of CoxII mRNA and of ROS and their control MRC-proficient counterparts (Figure 7C left) were cultured in vitro for 12 weeks. The frequency of TKI-resistant clones and the accumulation of chromosomal aberrations were reduced 3.8 and 4.6 times, respectively, in Rho0 cells compared with MRC-proficient cells (Figure 7C right).
We also used MitCat, which is located in close proximity to MRC and degrades H2O2.30 Expression of MitCat in BCR-ABL1–32Dcl3 cells reduced ROS levels by approximately 1.9 times (Figure 7D left) and inhibited the accumulation of TKI-resistant clones and chromosomal aberrations by 4.7- and 3.3-fold, respectively, in an in vitro mutagenic test (Figure 7D right).
We also applied a mitochondrial-targeted ROS-scavenging peptide as a “druggable” approach to preventing the accumulation of TKI-resistant clones. SS31 tetrapeptide concentrates > 1000-fold in the inner mitochondrial membrane independently of ΔΨm, followed by scavenging of H2O2 and ·OH by the Dmt analog from the peptide. CD34+ CML-CP cells and GFP+ BCR-ABL1 muBMCs incubated with the antioxidant SS31 peptide displayed 1.3-2.0 times fewer ROS than cells exposed to control SS20 peptide and their untreated counterparts (Figure 7E in vitro); these peptides did not affect cell proliferation (supplemental Figure 6A). In addition, SCID mice injected with GFP+ BCR-ABL1 muBMCs (TKI-resistant clones not detected at the time of injection) were treated with SS31 or SS20. Whereas this treatment did not affect the survival of injected animals (supplemental Figure 6B), GFP+ leukemia cells from mice receiving SS31 displayed an approximately 20% reduction of ROS and accumulated 2.4 times fewer TKI-resistant cells (Figure 7E in vivo).
To determine whether the addition of SS31 may be beneficial for imatinib treatment, SCID mice bearing GFP+ BCR-ABL1 leukemia were treated with the combination of imatinib and SS31 or SS20. GFP+ cells harvested from BM and spleen of the animals receiving imatinib + SS31 accumulated 4 times fewer TKI-resistant clones than those from mice treated with imatinib + SS20 (Figure 7F left). This observation was confirmed when CD34+ CML-CP cells (TKI-resistant clones not detected at the start of experiment) were cultured with imatinib and SS31 or SS20 in the presence of growth factors (to protect leukemia cells from the toxicity of imatinib). The addition of SS31, but not SS20, inhibited the accumulation of TKI-resistant clones by 4.8-fold (Figure 7F right).
Discussion
We demonstrated that the most primitive CML-CP LSCs, including quiescent cells, and LPCs contain higher levels of ROS and oxidative DNA damage compared with their healthy counterparts. Therefore, we postulated that ROS-induced genomic instability in CML-CP may start as early as in LSCs and continue in LPCs. This is supported by previous observations that TKI-resistant mutations in BCR-ABL1 kinase have been detected in CD34+CD38− and CD34+CD38+ CML-CP cells.31 Leukemia cells are tolerant to DNA damage because of BCR-ABL1–mediated protection from apoptosis and/or modulation of the response to DNA damage.32 Moreover, nuclear localization of FOXO3a, a critical mediator of resistance to physiologic oxidative stress, localized in the nuclei of LSCs in a murine model of CML-CP, suggesting another mechanism of protection from the toxic effects of ROS.33
Genomic instability in LSCs and TKI-resistant LPCs is of particular concern because these cells are not eliminated by TKIs.2 This could be especially important because CML-CP patients at diagnosis have been estimated to have approximately 5 × 107 CD34+ cells displaying innate imatinib resistance. Even patients in complete cytogenetic or major molecular response that are BCR-ABL1− by PCR after imatinib treatment may contain BCR-ABL1+ cells in the CD34+CD38− compartment, most of which belong to the pool of quiescent and/or TKI-refractory LSCs.34 These cells may continue to accumulate genomic instability, especially in patients no longer treated with TKIs.
The quiescent LSCs appear to have higher levels of ROS and oxidative DNA damage than the proliferating population, which consists mostly of LPCs. This observation is consistent with studies indicating that despite metabolic stimulus associated with cell proliferation, cells try to reduce ROS during DNA replication.35 However, the accumulation of ROS-induced oxidative DNA damage in LPCs, especially in those harboring TKI-resistant mutations, should not be underestimated: genomic instability combined with epigenetic alterations may drive them into a more aggressive “secondary stemness,” as has been reported in CML-BP.4,36
Using various experimental strategies, we have shown in the present study that mitochondria are involved in the generation of excessive amounts of ROS in LSCs and LPCs. Despite Warburg's hypothesis that cancer cells display irreversible injury to mitochondria, forcing aerobic glycolysis to replace ATP lost from defective oxidative phosphorylation, numerous lines of evidence demonstrate that mitochondria are functional in many tumors, including CML.37 However, the production of ATP and mitochondrial ·O2− could be down-modulated by adaptation of the cells to hypoxia.38 For example, BCR-ABL1 and other OTKs phosphorylate pyruvate kinase M2 directly on tyrosine 105 to inhibit its enzymatic activity, facilitate aerobic glycolysis, and probably promote leukemia cell growth in hypoxia.13 In agreement with this, we found that the overall generation of cellular ·O2−, including mitochondrial ·O2−, in primitive CD34+ CML-CP cells was diminished in hypoxia. This would suggest that LSCs are more vulnerable to accumulating genomic instability when present in the peripheral blood (normoxia) than when they are in the BM niche (hypoxia). However, the levels of DNA-damaging H2O2 and ·OH appeared to be unaffected by oxygen tension, which is consistent with other reports of paradoxically high levels of H2O2 generated by mitochondria in hypoxia.39 Consistent with this, the accumulation of oxidative DNA damage in CD34+ CML-CP cells was only slightly affected by hypoxia. Therefore, LSCs present in the hypoxic BM niche may also accumulate genetic errors.
We identified MRC-cIII as the major producer of ROS causing oxidative DNA damage in LSCs and LPCs. This effect probably originates from an impaired electron transfer between MRC complexes I-III and II-III (present study), which can facilitate “electron leakage” and the generation of ROS.16 A sustained increase in ROS may in turn depolarize the mitochondrial membrane, leading to reduced ATP production and/or release of proapoptotic cytochrome c. Leukemia cells can compensate for the shortage of mitochondrial ATP by enhancing glucose uptake and facilitating aerobic glycolysis, and proapoptotic cytochrome c is counteracted by BCR-ABL1 antiapoptotic activity.13,40,41 We postulate that the mitochondria present in primitive CML-CP cells display a unique “metabolic-bioenergetic-oxidative” phenotype in which dysfunctional MRC-cIII generates high levels of mitochondrial ROS and low levels of ATP, with the latter effect being compensated for by enhanced glucose uptake and aerobic glycolysis. This hypothesis is supported by the observation that enhanced mitochondrial ROS production has been associated with a failure of electron transport through MRC in imatinib-resistant CML cells.42
ROS-dependent genomic instability in CML-CP is responsible for the generation of TKI-resistant clones and the accumulation of additional chromosomal aberrations, eventually leading to CML-BP clones.7,14 Three independent approaches, ablation of MRC,15 expression of MitCat,30 and addition of mitochondrial ROS-scavenging SS31 polypeptide,17 have shown that genomic instability in BCR-ABL1 leukemia cells is dependent on mitochondrial ROS in vitro and in vivo. The results of the present study, to our knowledge, provide the first direct evidence that ROS generated by MRC-cIII are responsible for the induction of genomic instability in LSCs and primitive LPCs.
PI3K appears to stimulate ROS in BCR-ABL1+ leukemia cells.43 We have shown herein that Rac, which not only interacts with p210 BCR-ABL1 but also forms a positive feedback loop with PI3K,44,45 plays an important role in the generation of ROS in CML. More specifically, a Rac2-GTP–dependent mechanism modifies ΔΨm and electron flow through MRC, resulting in an overproduction of ·O2− by MRC-cIII, which then leads to oxidative DNA damage and genomic instability in LSCs and LPCs. The identification of Rac2, but not Rac1 or Rac3, as a key stimulator of ·O2− production by MRC-cIII may enable the development of more specific novel therapies that stabilize the genome of primitive CML-CP cells, thus preventing the generation of TKI-resistant and/or CML-BP clones (present study), while also helping to eradicate residual LSCs.46,47 For example, Rac2 unique aspartic acid 150 and the C-terminal polybasic domain are both essential for stimulation of ROS production (supplemental Figure 7).
Imatinib failed to inhibit Rac and reduce mitochondrial ROS in CD34+ CML-CP cells in the presence of growth factors, which is consistent with the persistant activation of other signaling molecules such as STAT5, AKT, and MAPK in these conditions.48 In addition, CD34+CD38− CML-CP LSCs can expand in the presence of imatinib and acquire TKI-resistant BCR-ABL1 kinase mutations and additional chromosomal aberrations.49 Accordingly, the Rac2-MRC-cIII pathway may stimulate ROS-induced oxidative DNA damage and genomic instability, not only in imatinib-treated LSCs and/or LPCs harboring the TKI-resistant BCR-ABL1 kinase mutant, but even in LSCs and/or LPCs displaying significant inhibition of BCR-ABL1 kinase.3 Therefore, persistent activation of the Rac2-MRC-cIII–ROS oxidative DNA damage pathway may allow a relatively unabated accumulation of genetic aberrations independently of whether CML-CP patients appear to be responding to imatinib.50
In conclusion, we postulate that the genomic instability responsible for the generation of TKI-resistant and/or CML-BP clones may occur in LSCs and/or LPCs and depends on the Rac2-MRC-cIII axis as a major stimulator of ROS-induced oxidative DNA damage in these cells (supplemental Figure 8). The application of mitochondrial ROS scavengers in patients treated with imatinib may prevent the emergence of mutated clones. These observations may lead to improved treatment strategies that target the mechanisms of genomic instability in therapy-refractory leukemia cells expressing BCR-ABL1 and possibly in other OTKs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Carrie Sims and Yuxia Guan from the University of Pennsylvania School of Medicine for technical support with MRC electron flow assay; Dr Michael Murphy (Mitochondrial Dysfunction MRC Mitochondrial Biology Unit, Cambridge, United Kingdom) for help with obtaining mitochondria-targeted ubiquinone; and Drs Heping Cheng and Xianghua Wang (Peking University, Beijing, China) for assistance with the mt-cpYFP studies.
This work was supported in part by the National Institutes of Health (grants CA123014, CA134458, and CA133646 to T. Skorski; CA134660 to M.S.; T32 HL07574-29 to A.D.; and DK62757 to D.A.W.); by an American Society of Hematology Research Trainee Award (to P.K.K.); by the Polish Ministry of Education and Science (N401 039037 to G.H.); by the Medical University of Warsaw (1M19/NK1W/2009) and the Polish National Science Center (N N402 676540 to T. Stoklosa); by a grant from the Canadian Cancer Research Institute supported by the Canadian Cancer Society (to C.E.); and by grants from Cancer Research UK C1174/A11008, Leukaemia & Lymphoma Research, and the Glasgow Experimental Cancer Medicine Centre (ECMC), which is funded by Cancer Research UK and by the Chief Scientist Office, Scotland (to T.L.H.). Dr Ivan de Curtis (San Raffaele Scientific Institute, Milan, Italy) provided Rac3−/− mice. Dr Jose A. Enriquez (Universidad de Zagaroza, Zagaroza, Spain) provided MRC-cI and MRC-cIII–depleted cell lines. Drs Ilona Seferynska (Institute of Hematology and Blood Transfusion, Warsaw, Poland), Joanna Niesiobedzka-Kretzel (Medical University of Warsaw, Warsaw, Poland), and Deborah White (University of Adelaide, Adelaide, Australia) provided CML samples. Drs Magdalena Ambrozek (Medical Center for Postgraduate Education, Warsaw, Poland) and Artur Slupianek (Temple University, Philadelphia, PA) assisted with the retroviral transfections.
National Institutes of Health
Authorship
Contribution: M.N.-S., P.K.K., R.R., G.H., D.N, S.F., K.C., M.M.R., M.K., T.P., E.G.-M., E.B., C.R., and A.D. performed the experiments; T.L.H., C.J.E., S.C.-R., P.V., A.H., T.P.H., H.v.d.K., W.W.-J., and T. Stoklosa collected, prepared, and supplied the CML cells; T.L.H., C.J.E., P.V., A.H., T.P.H., M.S., T. Stoklosa, and D.A.V. designed and supervised several experiments and/or contributed intellectually to the final version of the manuscript; and T. Skorski conceived the idea, designed the experiments, supervised the project, and wrote the manuscript.
Conflict-of-interest disclosure: T.L.H holds research funding and serves on the advisory board of Novartis. P.V. obtained a research grant and honoraria from Novartis and Bristol-Myers Squibb. A.H. received research support and honoraria from Novartis, Bristol-Myers Squibb, Ariad, and Pfizer. T.P.H. has received grant funding and honoraria from Bristol-Myers Squibb, Novartis, and Ariad. The remaining authors declare no competing financial interests.
Correspondence: Tomasz Skorski, Department of Microbiology and Immunology, School of Medicine, Temple University, 3400 N Broad St, MRB 548, Philadelphia, PA 19140; e-mail: tskorski@temple.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal