MicroRNAs (miRs) are small, noncoding RNA molecules with important regulatory functions whose role in regulating natural killer (NK) cell biology is not well defined. Here, we show that miR-155 is synergistically induced in primary human NK cells after costimulation with IL-12 and IL-18, or with IL-12 and CD16 clustering. Over-expression of miR-155 enhanced induction of IFN-γ by IL-12 and IL-18 or CD16 stimulation, whereas knockdown of miR-155 or its disruption suppressed IFN-γ induction in monokine and/or CD16-stimulated NK cells. These effects on the regulation of NK cell IFN-γ expression were found to be mediated at least in part via miR-155's direct effects on the inositol phosphatase SHIP1. Consistent with this, we observed that modulation of miR-155 overrides IL-12 and IL-18–mediated regulation of SHIP1 expression in NK cells. Collectively, our data indicate that miR-155 expression is regulated by stimuli that strongly induce IFN-γ in NK cells such as IL-12, IL-18, and CD16 activation, and that miR-155 functions as a positive regulator of IFN-γ production in human NK cells, at least in part via down-regulating SHIP1. These findings may have clinical relevance for targeting miR-155 in neoplastic disease.
Introduction
Human natural killer (NK) cells are CD56+CD3− large granular lymphocytes of the innate immune system.1,2 NK cells participate in early responses against infection or malignant transformation. In addition to their potent cytolytic activity, NK cells have an important immunoregulatory function in that they produce cytokines and chemokines when activated. In particular, NK cells produce IFN-γ, a critical cytokine for the clearance of infectious pathogens and tumor surveillance,3 in response to a wide variety of stimuli, including both soluble factors and cellular interactions.4,5 Dendritic cells and monocytes stimulated with bacterial cell wall components release monokines such as IL-12 and IL-18, which synergistically induce rapid and robust production of IFN-γ by NK cells.6
NK cells also express the low-affinity receptor for the Fc fragment of immunoglobulin (Ig)G (FcγRIIIA, CD16), which is the activating receptor required for triggering antibody dependent cellular cytotoxicity (ADCC) as well as the induction of IFN-γ.7 IL-12 monokine stimulation in combination with CD16 activation induces a synergistic induction of IFN-γ in NK cells, but to a lesser extent than does IL-12 and IL-18 costimulation.8 This observation has recently been shown to have implications in the antibody therapy of breast cancer patients. In fact, the antitumor actions of the anti-HER2 monoclonal antibody trastuzumab are enhanced by IL-12 treatment in vivo, and this effect is dependent on NK cell production of IFN-γ.9
The regulation of NK cell IFN-γ production involves positive and negative mediators, such as kinases and phosphatases, as well as transcription factors.10,,,–14 SHIP1 is a hematopoietic cell specific 5′ inositol phosphatase.15 We have previously shown that SHIP1 is expressed differentially in CD56bright and CD56dim NK cell subsets, and is negatively modulated by the costimulation of IL-12 and IL-18.13 SHIP1, by dampening the PI3K pathway, is able to negatively regulate IFN-γ production by monokines and CD16 stimulation in both human and mouse NK cells.13,16
MicroRNAs (miRs) are a highly conserved class of small, noncoding RNAs with important regulatory functions in proliferation, differentiation, signal transduction, immune responses, and carcinogenesis.17 miRs regulate gene expression posttranscriptionally by forming imperfect base pairs with sequences in the 3′ untranslated region (UTR) of genes. In turn, this prevents protein accumulation by repressing translation or by inducing mRNA degradation.18 Recently, a general role of miRs in regulation of NK cell activation, survival, and function has been shown using conditional deletion of Dicer or Dgcr8.19 A specific role of miR-150 in regulating development and maturation of mouse NK cells has also been reported.20 Further, it has been shown that miR-181 promotes human NK cell development by regulating Notch signaling.21 In addition, Fehninger et al have shown that treatment of mouse NK cells with IL-15 increased or decreased the expression of several miRs.22 Among these miRs, miR-223 was down-modulated thereby up-regulating its target gene, granzyme B.22 Among miRs, miR-155 is involved in protective immunity when properly regulated and contributes to malignant transformation when deregulated.23 miR-155 is processed from the non–protein-coding transcript of the BIC gene RNA. Further, it is required for the normal function of B, T, and dendritic cells,24,25 and its expression is increased during B, T, macrophage and dendritic cell activation.23 Transgenic mice with selective overexpression of miR-155 in B cells develop B-cell lymphoma.26 Of interest, miR-155 is overexpressed in NK-cell lymphoma/leukemia and this correlates with low levels of SHIP1 expression and up-regulation of AKT signaling.27 Still, the expression and role of miR-155 in regulating NK cell development and function have yet to be explored. In this report, we characterize the expression of miR-155 in human NK cells, its role in the regulation of NK cell IFN-γ expression, and the mechanism by which this occurs.
Methods
Cells lines, NK cell preparations, and flow cytometry
The human IL-2 dependent NK cell line NK-92 (gift of Dr H. Klingemann, Rush Cancer Center, Chicago, IL) was maintained in culture in RPMI-1640 medium (Invitrogen), supplemented with 20% heat-inactivated FBS (Invitrogen), 2mM l-glutamine and 150 IU/mL rhIL-2 (Hoffman-LaRoche). The amphotropic-packaging cell line 293T was maintained in culture in Dulbecco modified Eagle medium (DMEM; Invitrogen)/10% FBS medium and grown for 16 to 18 hours to 80% confluence before transfection by calcium phosphate-DNA precipitation (Profection System; Promega). Human NK cells were isolated from peripheral blood leukopacks of healthy individuals (American Red Cross) by incubation for 30 minutes with RosetteSep NK cell antibody cocktail (StemCell Technology), followed by Ficoll-Hypaque density gradient centrifugation. NK cell preparations containing > 99% CD56+ NK cells were obtained by positive selection using CD56 MicroBeads and MACS separation columns from Miltenyi (Miltenyi Biotech), as determined by direct immunofluorescence using an anti-CD56 phycoerythrin (PE)–conjugated monoclonal antibody (Ab; Immunotech). IL-18Rα fluorescence-activated cell sorter (FACS) analysis was performed by staining CD56+ NK cells with IL-18Rα PE-conjugated Ab or PE-conjugated nonreactive isotype control Ab (R&D Systems). Samples were acquired using FACSCalibur or LSRII (BD Bioscience) and analyzed with FlowJo Version 7.6.1 (TreeStar). Values for both percent positivity and mean fluorescence intensity (MFI) were obtained and recorded. All work with human materials was approved by the institutional review board of The Ohio State University Comprehensive Cancer Center.
Mice
Wild-type (WT; Bic+/+) and miR-155−/−(Bic−/−) mice (B6, 129S7-Mirn155tm/Brd) were obtained from Mutant Mouse Regional Resource Centers.28 In the spleens of miR-155 KO compared with WT littermates, we observed a similar percentage and total number of NK.1+CD3− NK cells and similar levels of CD94 surface expression (data not shown). All animal work was approved by The Ohio State University Animal Care and Use Committee, and mice were treated in accordance with the institutional guidelines for animal care.
Lentivirus infection of NK-92 cell line and primary human NK cells
Lentiviral vectors encoding miR-155 (Lenti-miR-155), the antisense (miR-Zip155), and empty vectors (Lenti and miR-Zip00) were obtained from SBI System Biosciences. NK-92 cells and primary NK cells were infected followed protocol similar to previously published standards.29 Briefly, infectious supernatant from Lenti, Lenti-miR-155, miR-Zip00, and miR-Zip155 transfected 293T cells were collected after 48 hours and used for 2 or 3 cycles of infections at multiplicity of infection of 10. All vectors contain the gene for green fluorescent protein (GFP). On infection, NK-92 cells and CD56+ NK cells were sorted for GFP expression on a FACSAria II (BD Bioscience). GFP+CD56+ primary NK cells were used for experimentation immediately after sorting. Expression of miR-155 was confirmed by real time RT-PCR (reverse transcription [transcriptase]-polymerase chain reaction) in NK-92 cells.
Cell culture conditions
Before cytokine stimulation, NK-92 cells were cultured in IL-2–free medium containing 10% FBS for 18 or 36 hours. NK-92 and primary NK cells were incubated in medium plus 10% or 20% FBS at 37°C (1 × 106/mL) for the indicated times with the addition of recombinant human or mouse IL-12 (kindly provided by Genetics Institute) or recombinant human or mouse IL-18 (R&D Systems) at the indicated concentration. For experiments using immobilized Abs, wells of flat bottom plate were coated with PBS diluted IgG (100 μg/mL; Sigma-Aldrich) or anti–mouse CD16 (10 μg/mL; R&D Systems) overnight at 4°C.
Western blot analysis
Cells were harvested, washed once with ice-cold PBS and lysed (108 cells/mL RIPA buffer: 0.15M NaCl, 1% NP-40, 0.1% SDS, 50mM Tris, pH 8.0), supplemented with protease and phosphatase inhibitors, 1mM phenylmethylsulfonylfluoride (PMSF), 1mM Na3VO4, 50mM NaF, 10mM β-glycerol-phosphate, 1mM ethylenediaminetetraacetic acid (EDTA), and a protease inhibitor cocktail tablet from Roche Applied Science, as described.30 Alternatively, cells were directly lysed in Laemmli buffer (2 × 105 cells/20 μL). Western blotting was performed according to previously published protocols,30 and Ab-reactive proteins were detected with horseradish peroxidase–labeled sheep anti–rabbit, mouse and/or goat Ig sera and enhanced chemiluminescence (ECL; Amersham). Proteins were analyzed in 4%-15% sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE; Bio-Rad Laboratories) using reducing conditions. To measure the levels of protein expression, the intensity of each band was quantified by densitometry using ImageJ Version 1.43 software (National Institutes of Health; http://rsb.info.nih.gov/ij/docs/index.html), and normalized to the level of ACTIN or GRB2 in each lane. Monoclonal and polyclonal Abs used were: mouse monoclonal anti-SHIP1 and the goat anti-ACTIN from Santa Cruz Biotechnology; and monoclonal anti-GRB2 Ab from Transduction Laboratories.
Real-time RT-PCR
Total RNA was extracted using either RNeasy Mini kits (QIAGEN) or Trizol (Invitrogen). cDNA was generated according to the manufacturer's recommendations (Invitrogen) or TaqMan MicroRNA reverse transcription kit and RT primers specific for miR-155 and RNU44 as control (Applied Biosystem). Real-time RT-PCR reactions were performed as a reaction with primer/probe set specific for the human BIC, SHIP1, miR-155, RNU44 (Applied Biosystem), IFN-γ, and 18S as previously described.12 Water (no template) was used as a negative control. Reactions were performed using an ABI prism 7700 sequence detector (Taqman; PE Applied Biosystems), and data were analyzed with the Sequence Detector Version 1.6 software to establish the PCR cycle at which the fluorescence exceeded a set threshold, CT, for each sample. Data were analyzed according to the comparative CT method using the internal control RNU44 and 18S transcript levels to normalize differences in sample loading and preparation. Results represent the n-fold difference of transcript levels in a particular sample compared with calibrator cDNA samples of unstimulated or control vector infected NK-92 or primary NK cells. Results are expressed as the mean ± SEM of triplicate reaction wells.
IFN-γ ELISA assays
Quantification of human IFN-γ was performed using commercially available mAbs pairs (Endogen). Cell free supernatants were collected after 18 or 24 hours of incubation at 37°C with the indicated stimuli. WT and miR-155−/− purified mouse NK1.1+CD3− NK cells were left untreated or stimulated with monokines and/or immobilized Abs for 18 or 24 hours at 37°C. Cell supernatants were collected and analyzed by enzyme-linked immunosorbent assay (ELISA) using a Kit from R&D Systems. Results are shown as the mean of triplicate wells ± SEM.
Statistics
Data were compared using student 2-tailed t test. A P value < .05 was considered significant.
Results
Expression of miR-155 and BIC in resting and monokine-activated human NK cells
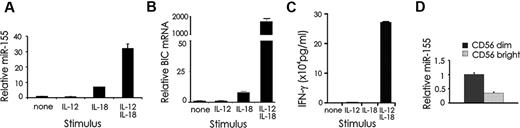
Stimulation with the combination of IL-12 and IL-18 cytokines induces a rapid and high induction of IFN-γ expression in NK cells.6 An Affymetrix profile identified miR-155 as the most highly induced miR by human NK cell costimulation with IL-12 and IL-18 cytokines (data not shown), which was further quantified by performing real-time RT-PCR in 7 different fresh primary human NK samples. After culture in IL-12 plus IL-18 we observed an average induction in miR-155 expression of 27.8-fold (P < .0001; Figure 1A), but only 3.7-fold with IL-18 alone (P < .01, n = 4) and zero with IL-12 alone. In further support of these data, the non–protein-coding miR-155 precursor RNA BIC was induced in a pattern similar to miR-155 (Figure 1B). The kinetics of this induction with IL-12 plus IL-18 were similar to those of IFN-γ release (Figure 1C).6 Furthermore, activated CD56bright NK cells show relatively high production of IFN-γ compared with CD56dim NK cells,31 yet express low levels of miR-155 compared with CD56dim NK cells in both resting (P < .001; Figure 1D), and IL-12 and IL-18 costimulated conditions (not shown).
miR-155 and BIC RNA expression in resting and IL-12 and/or IL-18–stimulated NK cells and NK subsets. CD56+ human NK cells were left unstimulated or stimulated with IL-12 (10 ng/mL), IL-18 (100 ng/mL) or combination of both for 18 hours after which pellets were collected and used to prepare RNA. miR-155 (A) and BIC (B) RNA expression were quantified by real-time RT-PCR. (C) Supernatants were analyzed for IFN-γ production by ELISA. (D) CD56bright and CD56dim NK cells were quantified for miR-155 expression by real-time RT-PCR. This experiment is representative of at least 4 such experiments performed with similar results.
miR-155 and BIC RNA expression in resting and IL-12 and/or IL-18–stimulated NK cells and NK subsets. CD56+ human NK cells were left unstimulated or stimulated with IL-12 (10 ng/mL), IL-18 (100 ng/mL) or combination of both for 18 hours after which pellets were collected and used to prepare RNA. miR-155 (A) and BIC (B) RNA expression were quantified by real-time RT-PCR. (C) Supernatants were analyzed for IFN-γ production by ELISA. (D) CD56bright and CD56dim NK cells were quantified for miR-155 expression by real-time RT-PCR. This experiment is representative of at least 4 such experiments performed with similar results.
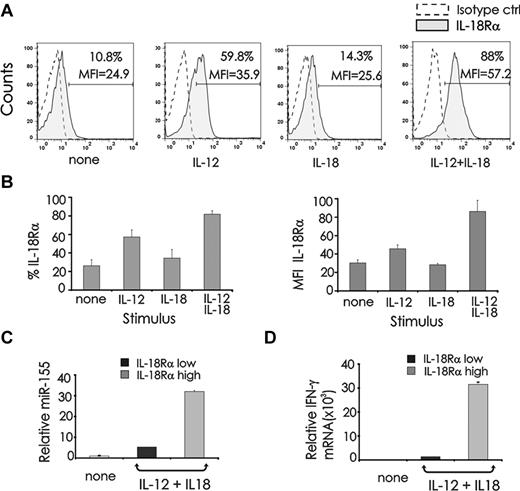
To better understand the basis for this synergistic induction of miR-155 in the presence of IL-18 and IL-12, we assessed IL-18 receptor (R) surface density expression in cytokine stimulated human NK cells. Stimulation of resting NK cells with IL-12 alone significantly induced IL-18Rα, whereas costimulation with IL-12 plus IL-18 induced significantly more IL-18Rα expression as detected by percent of expression and mean fluorescence intensity (Figure 2A-B). Conversely, IL-18Rβ was not visibly induced by IL-12 stimulation alone; however, the combination of IL-12 plus IL-18 led to strong induction (data not shown).
miR-155 expression in IL-18Rαhigh and IL-18Rαlow NK cells stimulated by IL-12 plus IL-18. (A) Freshly isolated CD56+ human NK cells were stimulated with IL-12 (10 ng/mL), IL-18 (100 ng/mL) or their combination for 48 hours after which cells were analyzed by FACS for expression of IL-18Rα. A nonreactive directly conjugated isotype-identical Ab was used as a control. (B) Percent and MFI of IL-18Rα expression from 6 different donors are summarized. (C) NK cells activated for 24 hours with IL-12 plus IL-18 were FACS sorted for IL18Rαhigh and IL18Rαlow expression, and quantified for miR-155 and (D) IFN-γ mRNA expression by real-time RT-PCR. This experiment is representative of at least 4 experiments performed with similar results.
miR-155 expression in IL-18Rαhigh and IL-18Rαlow NK cells stimulated by IL-12 plus IL-18. (A) Freshly isolated CD56+ human NK cells were stimulated with IL-12 (10 ng/mL), IL-18 (100 ng/mL) or their combination for 48 hours after which cells were analyzed by FACS for expression of IL-18Rα. A nonreactive directly conjugated isotype-identical Ab was used as a control. (B) Percent and MFI of IL-18Rα expression from 6 different donors are summarized. (C) NK cells activated for 24 hours with IL-12 plus IL-18 were FACS sorted for IL18Rαhigh and IL18Rαlow expression, and quantified for miR-155 and (D) IFN-γ mRNA expression by real-time RT-PCR. This experiment is representative of at least 4 experiments performed with similar results.
To determine whether the synergistic induction of miR-155 is dependent on the induction of IL-18R, we stimulated primary CD56+ NK cells overnight with the combination of IL-12 and IL-18 and sorted cells into 2 groups: high expression of IL-18Rα and low expression of IL-18Rα. These sorted populations were then analyzed for miR-155 and IFN-γ mRNA (Figure 2C-D). We observed that expression of IL-18Rα directly and significantly correlated with levels of miR-155 (P < .001; Figure 2C) and IFN-γ (Figure 2D).
The role of miR-155 in the regulation of IFN-γ production by IL-12 plus IL-18–stimulated NK cells
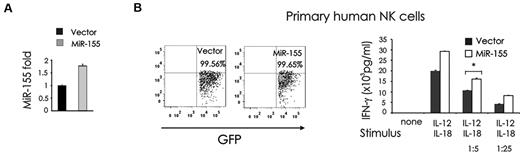
Based on our observation that both miR-155 and IFN-γ are synergistically induced by IL-12 plus IL-18 in human NK cells, we hypothesized that miR-155 could be a positive regulator of IFN-γ production. Thus, we overexpressed miR-155 by infecting the CD3−CD56+ human NK cell line NK-92 and CD3−CD56+ primary human NK cells using a lentiviral vector carrying either the cDNA for GFP alone (Lenti), or GFP plus the miR-155 native stem-loop structure along with 200 to 400 base pairs of upstream and downstream flanking genomic sequence (Lenti-miR-155). Overexpression of miR-155 was confirmed in the GFP+ fraction after NK-92 cell sorting (Figure 3A). Importantly, overexpression of miR-155 led to a significantly greater production of IFN-γ in GFP+ primary human NK cells after costimulation with IL-12 plus IL-18, compared with the mock-infected primary human NK cells (*P < .01; Figure 3B).
miR-155 overexpression enhances IL-12 plus IL-18–induced IFN-γ production in NK cells. (A) NK-92 cells were infected using an empty lentivirus vector (Lenti) containing GFP or a lentivirus vector containing GFP and miR-155 sense (Lenti-miR-155 vector). Cells were then sorted for high GFP expression and analyzed for miR-155 expression by RT-PCR. (B) Primary human CD56+ NK cells were infected using either Lenti or Lenti-miR-155, sorted by FACS for GFP (left), plated in medium and costimulated with IL-12 (10 ng/mL) plus IL-18 (100 ng/mL) or 1/5th or 1/25th the doses of these monokines for 24 hours. Supernatants were then collected and assayed for IFN-γ by ELISA (right). These data are representative of 3 experiments performed with moderate doses, and 2 experiments performed with high and low concentrations of monokines.
miR-155 overexpression enhances IL-12 plus IL-18–induced IFN-γ production in NK cells. (A) NK-92 cells were infected using an empty lentivirus vector (Lenti) containing GFP or a lentivirus vector containing GFP and miR-155 sense (Lenti-miR-155 vector). Cells were then sorted for high GFP expression and analyzed for miR-155 expression by RT-PCR. (B) Primary human CD56+ NK cells were infected using either Lenti or Lenti-miR-155, sorted by FACS for GFP (left), plated in medium and costimulated with IL-12 (10 ng/mL) plus IL-18 (100 ng/mL) or 1/5th or 1/25th the doses of these monokines for 24 hours. Supernatants were then collected and assayed for IFN-γ by ELISA (right). These data are representative of 3 experiments performed with moderate doses, and 2 experiments performed with high and low concentrations of monokines.
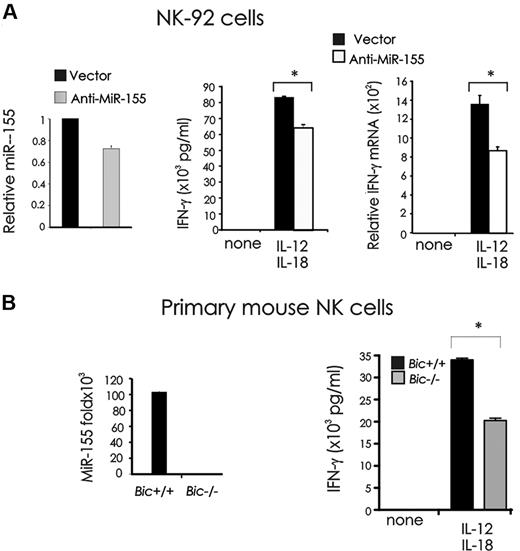
To confirm our observation, we down-modulated miR-155 by infecting NK-92 cells using an empty lentiviral vector called miR-Zip00, or a lentiviral vector encoding antisense miR-155 called miR-Zip155. In the human NK-92 cell line, down modulation of miR-155 significantly lowered IFN-γ production at both protein (*P < .01, n = 3) and mRNA (*P < .001, n = 3) levels after stimulation by IL-12 plus IL-18 compared with mock-infected NK-92 cells (Figure 4A). To address this observation in primary NK cells, we turned to Bic−/− mice. BIC is the precursor mRNA to miR-155, and Bic+/+ and Bic−/− mice were therefore used in this experiment. Primary NK cells from Bic−/− mice also showed significantly less IFN-γ production after stimulation with IL-12 and IL-18, compared with NK cells from Bic+/+ mice (*P < .001, n = 3; Figure 4B).
Effect of miR-155 down-modulation on IFN-γ production in monokine-stimulated NK cells. (A) NK-92 cells were infected with empty vector (miR-Zip00) or a miR-155 antisense encoding vector (miR-Zip155), sorted for GFP+ and analyzed for miR-155 expression by real-time RT-PCR (left). GFP+ miR-Zip00 and miR-Zip155 NK-92 cells were stimulated with IL-12 (10 ng/mL) plus IL-18 (100 ng/mL) for 24 hours and analyzed for IFN-γ production by ELISA (middle) and real-time RT-PCR (right). (B) BIC is the precursor of miR-155. NK cells sorted from spleens of Bic+/+ or Bic−/− mice were analyzed for miR-155 expression by real-time RT-PCR (left), and stimulated with IL-12 (20 ng/mL) and IL-18 (10 ng/mL). Supernatants were then collected and analyzed for IFN-γ by ELISA (right). This experiment is representative of 3 performed with similar results.
Effect of miR-155 down-modulation on IFN-γ production in monokine-stimulated NK cells. (A) NK-92 cells were infected with empty vector (miR-Zip00) or a miR-155 antisense encoding vector (miR-Zip155), sorted for GFP+ and analyzed for miR-155 expression by real-time RT-PCR (left). GFP+ miR-Zip00 and miR-Zip155 NK-92 cells were stimulated with IL-12 (10 ng/mL) plus IL-18 (100 ng/mL) for 24 hours and analyzed for IFN-γ production by ELISA (middle) and real-time RT-PCR (right). (B) BIC is the precursor of miR-155. NK cells sorted from spleens of Bic+/+ or Bic−/− mice were analyzed for miR-155 expression by real-time RT-PCR (left), and stimulated with IL-12 (20 ng/mL) and IL-18 (10 ng/mL). Supernatants were then collected and analyzed for IFN-γ by ELISA (right). This experiment is representative of 3 performed with similar results.
Regulation of SHIP1 expression in resting and IL-12 and/or IL-18 activated primary human NK cells
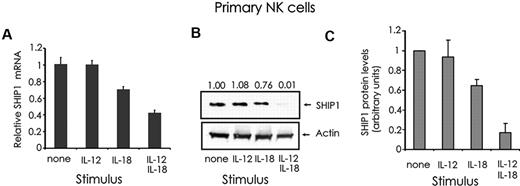
SHIP1 5′ inositol phosphatase is a negative regulator of IFN-γ in both human and mouse NK cells,13,16 and a primary target of miR-155.28,32,33 To address whether there is an inverse correlation between SHIP1 expression and miR-155 expression in NK cells, we first analyzed SHIP1 mRNA and protein expression in primary NK cells stimulated with IL-12 and/or IL-18. SHIP1 was expressed inversely to the pattern seen by miR-155 in that the former was reduced by the stimulation with IL-18 alone and substantially reduced further by the combination of IL-12 plus IL-18 stimulation, but no change was seen with IL-12 alone (Figure 5A-C).
Quantification of SHIP1 expression in monokine-stimulated primary human CD56+ NK cells. (A) CD56+ primary human NK cells were first incubated for 18 hours with medium containing either IL-12 (10 ng/mL) and/or IL-18 (100 ng/mL), and then quantified for SHIP1 (A) mRNA by real-time RT-PCR and (B) protein by Western blot. The relative levels of SHIP1 protein, as determined by densitometry, are indicated above the blot and expressed as densitometric units relative to the control lane (“none”), which is arbitrarily set at 1.00. (C) The graph summarizes the data of densitometric analysis of 3 primary donors. This experiment is representative of at least 3 such experiments performed with similar results.
Quantification of SHIP1 expression in monokine-stimulated primary human CD56+ NK cells. (A) CD56+ primary human NK cells were first incubated for 18 hours with medium containing either IL-12 (10 ng/mL) and/or IL-18 (100 ng/mL), and then quantified for SHIP1 (A) mRNA by real-time RT-PCR and (B) protein by Western blot. The relative levels of SHIP1 protein, as determined by densitometry, are indicated above the blot and expressed as densitometric units relative to the control lane (“none”), which is arbitrarily set at 1.00. (C) The graph summarizes the data of densitometric analysis of 3 primary donors. This experiment is representative of at least 3 such experiments performed with similar results.
Monokine-mediated regulation of SHIP1 expression in human NK cells is mediated via miR-155
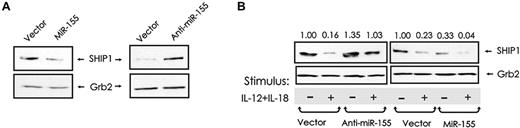
To determine whether miR-155 regulates SHIP1 expression in human NK cells, we first analyzed SHIP1 expression in NK-92 cells overexpressing sense or antisense miR-155, as previously described. SHIP1 expression was down-modulated in NK-92 cells overexpressing miR-155 (Figure 6A left) and up-regulated in cells with reduced expression of miR-155 (Figure 6A right). Next, in the same cell lines, we analyzed the expression of SHIP1 after costimulation with IL-12 plus IL-18. Ectopic expression of miR-155 antisense rescued SHIP1 down-modulation (Figure 6B left panel), whereas ectopic overexpression of miR-155 sense induced additional down-modulation of SHIP1 (Figure 6B right panel). These data provide evidence that miR-155 regulates SHIP1 in both resting and cytokine-activated NK cells and that down-modulation of SHIP1 in NK cells costimulated with IL-12 plus IL-18 is at least partially regulated by and dependent on the expression of miR-155.
SHIP1 is regulated by miR-155 in resting and monokine-stimulated human NK cells. (A) GFP+ NK-92 Lenti and GFP+ NK92 Lenti-miR-155 cells (left) and GFP+ NK-92 miR-Zip00 and NK-92 miR-Zip155 cells (right) were cultured in medium without IL-2 for 24 hours and then quantified for SHIP1 protein expression by Western blot. (B) Comparable NK-92 cells were then cultured in medium containing IL-12 (10 ng/mL) plus IL-18 (100 ng/mL) for 24 hours and then quantified for SHIP1 protein expression by Western blot. The relative levels of SHIP1 protein, as determined by densitometry, are indicated above the blot and expressed as densitometric units relative to the control lane (far left), which is arbitrarily set at 1.00. This experiment is representative of at least 2 such experiments performed with similar results.
SHIP1 is regulated by miR-155 in resting and monokine-stimulated human NK cells. (A) GFP+ NK-92 Lenti and GFP+ NK92 Lenti-miR-155 cells (left) and GFP+ NK-92 miR-Zip00 and NK-92 miR-Zip155 cells (right) were cultured in medium without IL-2 for 24 hours and then quantified for SHIP1 protein expression by Western blot. (B) Comparable NK-92 cells were then cultured in medium containing IL-12 (10 ng/mL) plus IL-18 (100 ng/mL) for 24 hours and then quantified for SHIP1 protein expression by Western blot. The relative levels of SHIP1 protein, as determined by densitometry, are indicated above the blot and expressed as densitometric units relative to the control lane (far left), which is arbitrarily set at 1.00. This experiment is representative of at least 2 such experiments performed with similar results.
miR-155 expression and role in NK cells activated via CD16 and IL-12
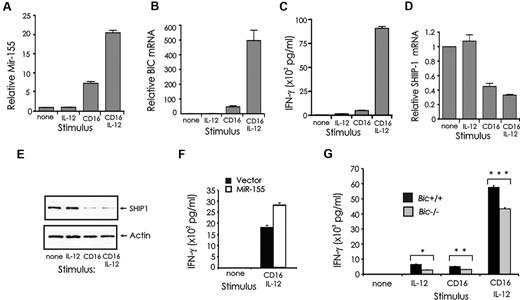
We have previously shown that SHIP1 regulates IFN-γ production induced by FcRγIIIA (CD16) stimulation in both human and mouse NK cells.16 We therefore quantified miR-155 expression in NK cells after activation via CD16 and/or IL-12, because in combination, these signals result in a synergistic induction of IFN-γ production (Figure 1C).8 Both miR-155 and its precursor BIC were induced in primary human NK cells by CD16 stimulation (P < .01, n = 4), whereas a greater induction of miR-155 and BIC resulted from costimulation by CD16 and IL-12 (Figure 7A-B; P < .01, n = 4). CD16 stimulation alone was able to significantly reduce SHIP1 mRNA and protein expression in primary NK cells, whereas costimulation via CD16 and IL-12 resulted in only a modest yet significant further reduction of SHIP1 mRNA (P = .01, n = 4) but not protein (Figure 7D-E).
Expression of miR-155 and SHIP1 in CD16-activated NK cells, and the role of miR-155 in regulating IFN-γ. (A) Primary human NK cells were activated with immobilized human IgG and/or IL-12 (10 ng/mL) for 24 hours and quantified for miR-155 and (B) BIC mRNA expression by real-time RT-PCR. (C) Supernatants were analyzed for IFN-γ production by ELISA. (D) SHIP1 mRNA expression was quantified by real-time RT-PCR and (E) protein expression by Western blot. (F) CD56+ primary human NK cells were infected using Lenti or Lenti–miR-155 vectors, sorted for GFP, and left unstimulated or stimulated with immobilized human IgG and/or IL-12 (10 ng/mL) for IFN-γ secretion. This experiment is representative of 2 experiments performed with identical results. (G) NK cells were isolated from spleens of Bic+/+ and Bic−/− mice, cultured 8 days in medium containing IL-2 (900 IU/mL), stimulated for 24 hours in vitro with immobilized anti-CD16 Ab and/or IL-12 (20 ng/mL) and assayed for IFN-γ.
Expression of miR-155 and SHIP1 in CD16-activated NK cells, and the role of miR-155 in regulating IFN-γ. (A) Primary human NK cells were activated with immobilized human IgG and/or IL-12 (10 ng/mL) for 24 hours and quantified for miR-155 and (B) BIC mRNA expression by real-time RT-PCR. (C) Supernatants were analyzed for IFN-γ production by ELISA. (D) SHIP1 mRNA expression was quantified by real-time RT-PCR and (E) protein expression by Western blot. (F) CD56+ primary human NK cells were infected using Lenti or Lenti–miR-155 vectors, sorted for GFP, and left unstimulated or stimulated with immobilized human IgG and/or IL-12 (10 ng/mL) for IFN-γ secretion. This experiment is representative of 2 experiments performed with identical results. (G) NK cells were isolated from spleens of Bic+/+ and Bic−/− mice, cultured 8 days in medium containing IL-2 (900 IU/mL), stimulated for 24 hours in vitro with immobilized anti-CD16 Ab and/or IL-12 (20 ng/mL) and assayed for IFN-γ.
The role of miR-155 in regulating IFN-γ through CD16 and IL-12 stimulation was analyzed using primary human NK cells overexpressing miR-155 and in primary NK cells from Bic−/− mice. In primary human NK cells overexpressing miR-155, we observed an increase in IFN-γ after costimulation with CD16 and IL-12 compared with mock-infected cells (Figure 7F). In contrast, primary NK cells from Bic−/− mice showed a significant decrease in IFN-γ production after stimulation by CD16 and/or IL-12 compared with WT controls (*P < .01, n = 3; **P < .04, n = 3; ***P < .001, n = 3; Figure 7G).
Discussion
IFN-γ is the prototypic cytokine produced by NK cells. Indeed, its fundamental role during inflammation and tumor immunity has been well established, in that IFN-γ deficiency results in increased susceptibility to infection and/or malignancy.34,35 In contrast, overproduction of IFN-γ can lead to autoimmune disorders.36
MicroRNAs have recently been shown to play a critical role in the immune system as regulators of gene expression.37 The expression of miR-155 in NK cells and its role in regulating NK cell functions has not been investigated. Here we show that miR-155 functions as a regulator of IFN-γ production in activated human NK cells at least in part via the modulation of SHIP1 expression.
Among combinations of cytokines, IL-12 plus IL-18 potently induces NK-cell IFN-γ production, in part because of the synergistic effect of these 2 monokines on this process.6 In this report, we provide clear evidence that like IFN-γ production, the expression of miR-155 and its precursor BIC are synergistically induced in primary human NK cells after stimulation with IL-12 plus IL-18. In fact, IL-18 stimulation alone but not IL-12 alone, modestly induced miR-155 and BIC in human NK cells. Others have previously shown that IL-12 treatment of PBMC induced IL-18R expression in CD56+ NK cells.38 Here we observed that IL-12 specifically and directly induced IL-18Rα expression, but not IL-18Rβ expression (not shown), on purified human NK cells. Further, we found a direct correlation between IL-18Rα, miR-155 and IFN-γ expression in NK cells stimulated by IL-12 plus IL-18. These observations support a model where the synergistic induction of miR-155 by IL-12 and IL-18 depends at least in part on the induction of IL-18Rα by IL-12, and suggest that miR-155 is one of the molecules responsible for the synergistic induction of IFN-γ in NK cells stimulated with IL-12 plus IL-18.
A characteristic of IL-18R signaling is the phosphorylation of IL-1R-associated kinase (IRAK) and the sequential activation of nuclear factor-κB (NF-κB).39,40 NF-κB directly binds to the IFN-γ promoter,39 but its activity is also directly correlated with miR-155 expression.41 Our data suggest that induction of miR-155 by IL-18 in NK cells is probably mediated via NF-κB activation.
Importantly, by performing loss or gain-of-function experiments in primary human and mouse NK cells and in the human NK-92 tumor cell line, we identify miR-155 as a positive regulator of IFN-γ in NK cells.
Notably, despite having multiple targets that include MAF, PU1, SOCS1, and Taff,23 miR-155 appears to be the only known miR that targets the 3′UTR of SHIP1 and regulates its expression.28,32,33 Our previous study identified SHIP1 as a negative regulator of IFN-γ, with a higher level of constitutive expression in resting CD56dim NK cells compared with CD56bright NK cells. One would have therefore predicted that miR-155 expression would be inversely correlated with SHIP1 expression in these resting NK subsets; however, we found the opposite: CD56dim NK cells have a higher level of miR-155 expression compared with CD56bright NK cells. Thus, miR-155 expression does not inversely correlate with the constitutive expression of its target SHIP1 in CD56dim and CD56bright NK cells, suggesting that SHIP1 is probably regulated via miR-155–dependent and –independent mechanisms in these NK subsets as has been previously observed for SHIP1 in other hematopoietic tissues42 and as noted in this section. Further, CD56bright and CD56dim NK cells represent stage 4 and stage 5 of human NK cell development, respectively.43 Whether miR-155 may also target the 3′ UTR of genes important for the transition between stage 4 and stage 5 of NK cell development remains to be investigated.
We showed that SHIP1 is down-modulated in primary human NK cells after costimulation with IL-12 and IL-18,13 and under these conditions we observed an inverse correlation between miR-155 and SHIP1 expression. We prove that miR-155 is down modulating SHIP1 by recapitulating this effect in NK-92 cells ectopically expressing an anti–miR-155, and by amplifying the effect by overexpressing miR-155. These experiments, together with our previous finding that SHIP1 regulates IFN-γ in both human cells and in mouse NK cells in vitro and in vivo,13 identify the inositol phosphatase SHIP1 as one of the intermediaries through which miR-155 exerts its biologic effect on NK cell IFN-γ production.
NK cells mediate ADCC largely through CD16, and this receptor has been the focus of intense research throughout the years. It has been shown by Parihar et al8 that CD16 activation in the presence of IL-12 induces IFN-γ expression in a synergistic fashion in human NK cells. Collaboratively, we also found that SHIP1 was a negative regulator of IFN-γ in both mouse and human NK cells under similar conditions.16 Our findings in this study are in agreement with this, wherein CD16 activation induces miR-155, which is synergistically enhanced by the addition of IL-12. These results suggest that IL-12 primes NK cells for the induction of miR-155 not only via the aforementioned up-regulation of IL-18Rα but also through means that affect other signaling pathways, such as the colocalization of IL-12R and CD16 to cell lipid rafts and synergistic activation of extracellular signal-regulated kinase (ERK).44 In fact, activation of BIC and miR-155 expression by B-cell receptor signaling occurs through the ERK pathway.45
SHIP1 is down-modulated by CD16 activation in NK cells, implicating its involvement in this regulation of IFN-γ. In contrast to the combination of IL-12 and IL-18, we did not observe synergistic down-modulation of SHIP1 protein with IL-12 and CD16 activation, having seen a near-maximal down modulation with CD16 activation alone. Taken with the previous finding that IL-12 synergized with CD16 activation for IFN-γ production,8 and with our current finding that IL-12 significantly enhanced CD16-mediated miR-155 expression, it is probable that (1) activation of CD16 regulates SHIP1 via both miR-155–dependent and –independent mechanisms, and (2) miR-155 enhances IFN-γ production by targeting more than just SHIP1. The latter notion is consistent with the fact that miR-155 has several targets23 and that the regulation of IFN-γ gene expression is complex.14
In human primary NK cells and NK-92 cells ectopically overexpressing miR-155, we did not observe any effect on spontaneous cytotoxicity or ADCC (data not shown). There is still the possibility that long-term culture in IL-2 during infection of primary NK cells and NK-92 cells may compensate for an effect of miR-155 on NK cell cytotoxicity which may have otherwise been observed.
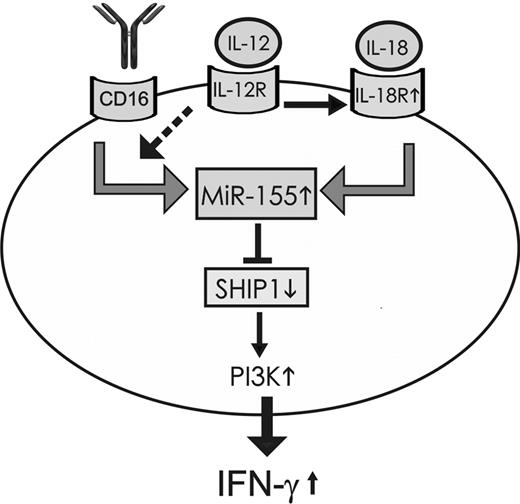
The data from the current study suggest a potential model for the induction of miR-155 and the role of miR-155 in regulating IFN-γ production in NK cells after NK cell activation. The induction of miR-155 expression depends on signaling events induced by triggering the IL-18R or CD16, but not directly via the IL-12 receptor. The combination of IL-12 with either IL-18 or CD16 activation induces miR-155 in a synergistic fashion. The synergistic induction of miR-155 expression after IL-12 and IL-18 costimulation of NK cells depends on the induction of IL-18Rα by IL-12 stimulation whereas the synergistic induction of miR-155 by IL-12 and CD16 stimulation possibly depends on increasing signaling downstream of CD16. In turn miR-155 overexpression targets the 5′ inositol phosphatase SHIP1 with consequent up-regulation of the phosphatidylinositol-3 kinase pathway and subsequent enhanced IFN-γ production (Figure 8).
Schematic representation modeling the induction of miR-155 after NK cell activation and its role in regulating IFN-γ production. miR-155 expression was induced after activation of resting NK cells via IL18 alone, via CD16 activation alone, but not via IL-12 alone; however, the combination of IL-12 with either IL-18 or CD16 activation induced miR-155 in a synergistic fashion. This synergism was dependent on the induction of the IL-18R by IL-12, whereas the synergistic induction of miR-155 noted with CD16 and IL-12 possibly results from enhanced intracellular signaling downstream of CD16 (dashed arrow). miR-155 targets the 5′ inositol phosphatase SHIP1, thereby down-modulating SHIP1 expression, which in turn promotes the prolonged activation of the PI3K pathway and subsequent enhanced production of IFN-γ.
Schematic representation modeling the induction of miR-155 after NK cell activation and its role in regulating IFN-γ production. miR-155 expression was induced after activation of resting NK cells via IL18 alone, via CD16 activation alone, but not via IL-12 alone; however, the combination of IL-12 with either IL-18 or CD16 activation induced miR-155 in a synergistic fashion. This synergism was dependent on the induction of the IL-18R by IL-12, whereas the synergistic induction of miR-155 noted with CD16 and IL-12 possibly results from enhanced intracellular signaling downstream of CD16 (dashed arrow). miR-155 targets the 5′ inositol phosphatase SHIP1, thereby down-modulating SHIP1 expression, which in turn promotes the prolonged activation of the PI3K pathway and subsequent enhanced production of IFN-γ.
In summary, we show that miR-155 plays a central role in NK cell IFN-γ production, whether through monokine signaling or CD16 activation. This is accomplished at least in part by directly regulating the expression of SHIP1, although our data suggest that miR-155 probably targets other components in parallel. By gaining a greater understanding of how miR-155 is induced and how it modulates IFN-γ, we may be able to identify new targets that can control chronic inflammation and/or modulate NK cell antitumor activity.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Susan McClory and Ed Briercheck for helpful discussion.
This work was supported in part by the National Cancer Institute grants CA16058, CA95426, and CA68458 (all M.A.C.).
National Institutes of Health
Authorship
Contribution: R.T. designed the study, performed research work, analyzed data, and wrote the paper; L.C., D.C., and S.J. performed experimental work and analyzed data; C.M. performed cell sorting experiments; L.Y. analyzed data; S.C., J.P.B., S.T., and C.M.C. provided reagents and discussed data; and M.A.C. contributed to the design of the study and to the writing/editing of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rossana Trotta or Michael A. Caligiuri, The Ohio State University Comprehensive Cancer Center, 884 and 886 OSU Biomedical Research Tower, 460 West 12th Ave, Columbus, OH 43210; e-mail: rossana.trotta@osumc.edu or michael.caligiuri@osumc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal