HIV infection results in a decrease in circulating CD4+ T-cell and naive T-cell numbers. If such losses were associated with an erosion of T-cell receptor (TCR) repertoire diversity in the peripheral T-cell pool, this might exacerbate the state of persistent immunodeficiency. Existing methods for the analysis of the TCR repertoire have demonstrated skewed distributions of TCR genes in HIV-infected subjects but cannot directly measure TCR diversity. Here we used AmpliCot, a quantitative assay based on DNA hybridization kinetics, to measure TCR diversity in a cross-sectional comparison of 19 HIV-infected persons to 18 HIV-uninfected controls. HIV-infected persons had a 10-fold decrease in total TCR repertoire diversity in 1.5 mL of blood compared with uninfected controls, with decreased diversity correlating most closely with a lower CD4+ T-cell percentage. Nonetheless, the TCR repertoire diversity of sort-purified T-cell subpopulations in HIV-infected and HIV-uninfected subjects was comparable. These observations suggest that the TCR repertoire diversity changes in whole blood during HIV disease progression are primarily the result of changes in the number and proportion of T-cell subpopulations and that most HIV-infected persons may retain a sufficiently diverse TCR repertoire to permit immune reconstitution with antiretroviral therapy alone, without thymopoiesis.
Introduction
Because CD4+ T cells are progressively lost in most HIV-infected persons, the absolute CD4+ T-cell count has proven useful for staging the degree of immunosuppression and for predicting the risk of opportunistic infections and cancers in these patients.1,–3 Hidden within the absolute count, however, is a range of features that are critical for the function of the immune system, including the relative proportion of thymically derived naive T cells and peripherally expanded memory/effector subpopulations as well as the repertoire of T-cell receptors found within each of these subpopulations. Clearly, substantial changes in the composition of the T-cell compartment occur during disease progression, with loss of naive T cells and expansion of memory/effector cells.4 Less well understood is the impact of HIV infection on the TCR repertoire of the total CD4+ and CD8+ T-cell compartments and of their constituent subpopulations.
Previous studies have found that HIV disease alters the normal distribution of TCRs in the repertoire.5 Moreover, analysis of patients who had recurrent opportunistic infections, even after effective antiretroviral therapy, demonstrated that these patients had lost antigen-specific responses, despite having high CD4+ T-cell counts.6 Data such as these raised the possibility that the immunodeficiency of HIV disease might be the result, at least in part, of loss of TCR repertoire diversity, resulting in patients lacking the optimal TCR specificities for recognizing and responding to pathogens.5,6 There was concern that TCR specificities might be permanently lost, especially in patients whose HIV had not been treated until late-stage AIDS. To address this concern, efforts were made to monitor thymic activity in HIV-infected patients7,–9 and to boost thymic function (eg, with growth hormone or IL-7)10,,–13 so that naive T cells with a diverse TCR repertoire might be made anew.
However, evidence supporting the hypothesis that HIV causes a reduction in TCR diversity is limited. Most investigations of the TCR repertoire in HIV disease have measured either Vβ gene usage (determined by flow cytometry or quantitative PCR) or the length distribution of VC rearrangements (analyzed with spectratyping/immunoscope).5,14,–16 These studies have found a skewed distribution of circulating TCR clones in HIV-infected patients, with certain clones expanded relative to others. Unfortunately, these methods are both qualitative and insensitive and cannot distinguish between 2 possible mechanisms of skewing: the expansion of selected clones versus the loss of others.
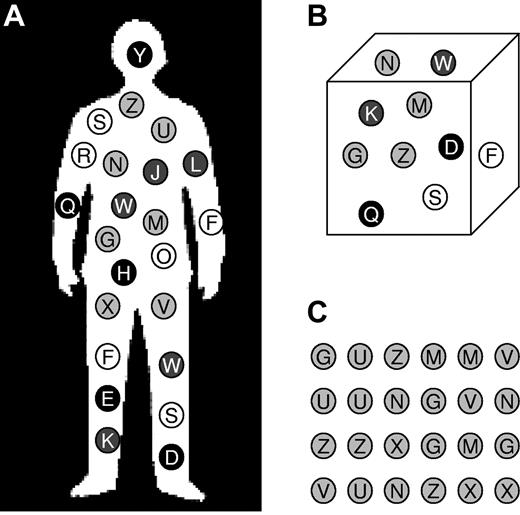
Here we apply the quantitative AmpliCot method17,–19 to a cross-sectional study of HIV-infected and -uninfected subjects to clarify the impact of HIV disease on the TCR repertoire. One challenge with quantitative measurements of TCR diversity is that they are dependent on the nature of the sample used. Three potential approaches are schematized in Figure 1. The “whole body repertoire” (Figure 1A) is, in theory, the frame of reference that defines whether clones are truly permanently lost. In practice, the repertoire of the estimated 1011 T cells in the body is impossible to measure.20 Even if it were possible to enumerate all TCR clones, rare clones may not be available at the required anatomic sites for immune responses or immune reconstitution. Figure 1B depicts what we will call the “whole-blood TCR repertoire,” or the number of unique TCRs found in a fixed volume of whole blood (essentially, the operational definition of the repertoire most often used by clinical researchers). It reflects the absolute abundance of different T-cell subpopulations in the blood and may define the current state of the immune system's ability to recognize a pathogen. Finally, we measured the “subpopulation TCR repertoire,” or the number of unique TCRs found in a fixed number of cells from defined T-cell subpopulations (Figure 1C). This more detailed examination may define the potential diversity that could be used to reconstitute the immune system if cells of these subpopulations were expanded.
Frames of reference for TCR diversity measurements. (A-C) In this schematic, circles represent T cells, their shading represents their phenotype (eg, naive CD4+ T cells), and their letter represents their TCR clonotype. (A) Total body TCR diversity (ie, unique TCR sequences in the entire body). (B) TCR diversity per fixed volume of blood or tissue (eg, unique TCR sequences per 1.5 mL of whole blood). (C) TCR diversity per fixed number of cells, often of a defined subpopulation (eg, number of unique TCR sequences found in a sample of 1.2 million naive CD4+ cells).
Frames of reference for TCR diversity measurements. (A-C) In this schematic, circles represent T cells, their shading represents their phenotype (eg, naive CD4+ T cells), and their letter represents their TCR clonotype. (A) Total body TCR diversity (ie, unique TCR sequences in the entire body). (B) TCR diversity per fixed volume of blood or tissue (eg, unique TCR sequences per 1.5 mL of whole blood). (C) TCR diversity per fixed number of cells, often of a defined subpopulation (eg, number of unique TCR sequences found in a sample of 1.2 million naive CD4+ cells).
We find that there is a significant decrease in the whole-blood TCR diversity in untreated HIV-infected subjects as opposed to well-matched HIV-uninfected subjects and argue that these defects may contribute to the immune deficits associated with untreated HIV disease. In contrast, the TCR repertoires of constituent naive and memory/effector subpopulations, when analyzed on a per-cell basis, are largely indistinguishable between infected and uninfected subjects, suggesting that that the decrease in whole-blood TCR diversity is largely the result of changes in the relative representation of different T-cell subpopulations. Consequently, it should be possible to reconstitute a T-cell compartment with a diverse TCR repertoire in most HIV-infected subjects, even in the absence of thymic function.
Methods
Subjects
HIV-infected subjects, recruited from the SCOPE cohort, were infected for over 6 months and had not received antiretroviral drugs in the 6 months before enrollment. HIV-uninfected subjects were recruited from the community to match the demographics and disease exposures of the HIV-infected participants and were also enrolled into the SCOPE cohort. Subjects were excluded from the study if they were born outside the United States, Canada, or Western Europe; had known chronic HBV infection, or present or past HCV infection; had taken systemic corticosteroids or immunosuppressive drugs; had a history of disseminated malignancy, organ or bone marrow transplant, pregnancy, or hemophilia; or had used intravenous drugs within the previous 6 months. Subjects had their participation deferred for infections requiring systemic antibiotics, immunizations, or blood transfusions in the past month, fever, cough diarrhea, or travel to Asia, Africa, or Latin America in the past week. Subjects provided demographic information and information on routine exposures via questionnaire. All human subject research was conducted under protocols approved by the University of California, San Francisco Committee on Human Research, and all subjects gave written informed consent in accordance with the Declaration of Helsinki.
Blood samples
HIV-infected and HIV-uninfected subjects donated 40 mL and 100 mL blood, respectively. Samples were processed within 6 hours of donation. Leukocytes from 1.5 mL of blood were isolated by hypotonic lysis, followed by lysis of the leukocyte pellet in buffer RLT (QiAmp RNA kit, QIAGEN). The lysate was homogenized using shredding columns and then stored at −80°C before batch RNA isolation according to the manufacturer's instructions.
Plasma was isolated by centrifugation of blood (400g for 10 minutes and 800g for 10 minutes). Mononuclear cells were obtained by Ficoll preparation of the remaining blood and were stained for the markers CD3, CD4, CD8β, CD45RA, CCR7, and CD57 and sorted for the phenotypes (see Table 3). To measure the percentage of cells of each phenotype using particular Vβ families, required for calculations of absolute TCR diversity, 2 aliquots of cells were additionally stained with either Vβ1, Vβ3, Vβ4; or Vβ2, Vβ17, Vβ22.
Cell samples were sorted with a FACSAria (BD Biosciences) into chilled 1.5-mL collection tubes containing 250 μL of PBS plus 2% FCS. Cell yields were determined from cytometer event counts, after verification using a hemocytometer. Viability and purity were assessed by reanalysis of propidium iodide-stained sorted cells. Viability was greater than 95%, and purity (defined in terms of contamination with other T-cell subpopulations) was greater than 98%. Cells were centrifuged at 4°C for 10 minutes at 840g and then lysed in buffer RLT.
AmpliCot and quantitative PCR
Total RNA was reverse transcribed and amplified for measuring VJ family sequence diversity, as described.17,19 V primers for the most abundant Vβ gene families (V9, V20, and V28) were used, corresponding to the Vβ1, Vβ2, and Vβ3 antibodies used for flow cytometry, respectively.21 J primers used were J2S1, J2S2, and J2S3. Measurements of up to 9 VJ families were averaged to estimate total sample diversity. For whole-blood samples, the diversity of the sample is expressed as the sum of the Cot1/2 values for these VJ families. The Cot1/2 value is the concentration by time product required for half of the sample to reanneal under stringent conditions and is proportional to the sequence diversity of the sample. For whole-blood samples, the wide variation of clone frequency makes it difficult to translate Cot1/2 values into an absolute number of sequences. For sorted T-cell subpopulations, which have more uniformity in their clone frequency, the absolute number of sequences of a given VJ family in the sample was calculated using quantitative standards as described.18 This value was then used to estimate the total number of sequences in the sample by multiplication with measured parameters of VJ family usage as described previously.17,19 The error resulting from selection of individual VJ families was assessed by measuring the reproducibility of measurements of the same samples using different VJ families (supplemental Figure 3, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). This estimate of sequences per sample was more accurate for highly diverse cell types, such as CD4+ naive T cells, than for low diversity cell subpopulations, such as CD8+ central memory T cells. More details of the AmpliCot method are available at http://amplicot.ucsf.edu.
Other measurements
Viral loads were measured by the SFGH Clinical Laboratory using the bDNA Version 3 Versant assay (Siemens). Complete blood counts were performed by the SFGH Clinical Laboratory. Transcription Mediated Amplification for hepatitis B and C viruses was performed by Dr Leslie Tobler of the Blood Systems Research Institute. Serologic tests for HBV IgG, HCV IgG, Helicobacter pylori IgG, EBV VCA IgG, and CMV IgG used Bioquant EIA kits following the manufacturer's instructions.
Results
Subjects
We recruited HIV-infected subjects with a range of CD4+ T-cell counts to reflect the spectrum of disease progression. Study participants were naive to antiretroviral medications, except for 2 persons who had been off treatment for at least 6 months. Because our goal was an internally valid study (to be followed by later generalizable studies) and because much remains to be learned about the factors that determine TCR repertoire diversity, strict enrollment criteria were used. As a matched comparator group, HIV-uninfected subjects were selected to have prespecified characteristics that might impact on the TCR repertoire. In addition to age and CMV serostatus, factors shown to affect repertoire diversity,22,,,–26 subject groups were also similar for sex, race, and prior exposure to HCV or HBV (Table 1). H pylori seropositivity, a proxy for unmeasured socioeconomic variables affecting infectious disease exposure history,27 was similar in both groups (Table 1). Because the analysis of TCR repertoire diversity was carried out before the coinfection status could be ascertained, data were also gathered from subjects who did not meet all of the prespecified characteristics. Data from these additional subjects, however, did not differ noticeably from that of the predefined subjects (see, for instance, open circles in Figure 2A) and are therefore included to increase the total number of data points. Little is known about the effects of acute exposures on the TCR repertoire; accordingly, a conservative list of exclusion criteria was applied, and (as detailed in this section) potential subjects were excluded if they had a recent history of an acute infection, immunization, or travel.
Study subjects
| . | Prespecified comparison groups . | All subjects . | ||
|---|---|---|---|---|
| HIV+ . | HIV− . | HIV+ . | HIV− . | |
| n | 15 | 9 | 19 | 18 |
| Age, y | 46 (35-58) | 46 (29-65) | 46 (35-58) | 50 (29-68) |
| Sex, no. (%) | ||||
| Male | 15 (100) | 9 (100) | 19 (100) | 18 (100) |
| Female | 0 | 0 | 0 | 0 |
| Race, no. (%) | ||||
| Black | 5 (33) | 4 (44) | 9 (50) | 6 (33) |
| Native American | 1 (7) | 2 (22) | 1 (5) | 3 (17) |
| Asian | 1 (7) | 1 (11) | 1 (5) | 1 (6) |
| White | 8 (53) | 2 (22) | 8 (42) | 8 (44) |
| Hispanic | 3 (22) | 0 (0) | 3 (16) | 2 (11) |
| MSM (ever) | 14 (93) | 7 (78) | 17 (89) | 15 (83) |
| IVDU (ever) | 1 (7) | 0 (0) | 1 (5) | 0 (0) |
| Pet at home | 3 (20) | 3 (33) | 3 (16) | 5 (28) |
| Child under age 10 at home | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| CD4 count/μL (median, range) | 260 (54-388) | 1203 (749-1990) | 244 (18-392) | 1060 (326-1990) |
| HIV VL/mL (median, range) | 44 000 (135-382 000) | NA | 46 000 (135-382 000) | NA |
| History of ART, no. (%) | 2 (13) | NA | 2 (11) | NA |
| Serologies, no. (%) | ||||
| EBV VCA IgG+ | 8 (53) | 4 (44) | 10 (53) | 6 (33) |
| H pylori IgG+ | 6 (40) | 5 (56) | 8 (42) | 11 (61) |
| CMV IgG+ | 15 (100) | 9 (100) | 19 (100) | 12 (68)* |
| HCV IgG+ with negative HCV viral load | 0 (0) | 0 (0) | 1 (5) | 2 (11) |
| Nucleic acid tests, no. (%) | ||||
| HCV | 0 (0) | 0 (0) | 0 (0) | 1 (6) |
| HBV | 0 (0) | 0 (0) | 3 (16) | 1 (6) |
| . | Prespecified comparison groups . | All subjects . | ||
|---|---|---|---|---|
| HIV+ . | HIV− . | HIV+ . | HIV− . | |
| n | 15 | 9 | 19 | 18 |
| Age, y | 46 (35-58) | 46 (29-65) | 46 (35-58) | 50 (29-68) |
| Sex, no. (%) | ||||
| Male | 15 (100) | 9 (100) | 19 (100) | 18 (100) |
| Female | 0 | 0 | 0 | 0 |
| Race, no. (%) | ||||
| Black | 5 (33) | 4 (44) | 9 (50) | 6 (33) |
| Native American | 1 (7) | 2 (22) | 1 (5) | 3 (17) |
| Asian | 1 (7) | 1 (11) | 1 (5) | 1 (6) |
| White | 8 (53) | 2 (22) | 8 (42) | 8 (44) |
| Hispanic | 3 (22) | 0 (0) | 3 (16) | 2 (11) |
| MSM (ever) | 14 (93) | 7 (78) | 17 (89) | 15 (83) |
| IVDU (ever) | 1 (7) | 0 (0) | 1 (5) | 0 (0) |
| Pet at home | 3 (20) | 3 (33) | 3 (16) | 5 (28) |
| Child under age 10 at home | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| CD4 count/μL (median, range) | 260 (54-388) | 1203 (749-1990) | 244 (18-392) | 1060 (326-1990) |
| HIV VL/mL (median, range) | 44 000 (135-382 000) | NA | 46 000 (135-382 000) | NA |
| History of ART, no. (%) | 2 (13) | NA | 2 (11) | NA |
| Serologies, no. (%) | ||||
| EBV VCA IgG+ | 8 (53) | 4 (44) | 10 (53) | 6 (33) |
| H pylori IgG+ | 6 (40) | 5 (56) | 8 (42) | 11 (61) |
| CMV IgG+ | 15 (100) | 9 (100) | 19 (100) | 12 (68)* |
| HCV IgG+ with negative HCV viral load | 0 (0) | 0 (0) | 1 (5) | 2 (11) |
| Nucleic acid tests, no. (%) | ||||
| HCV | 0 (0) | 0 (0) | 0 (0) | 1 (6) |
| HBV | 0 (0) | 0 (0) | 3 (16) | 1 (6) |
MSM indicates men who have sex with men; IVDU, intravenous drug use; and NA, not applicable.
P < .05.
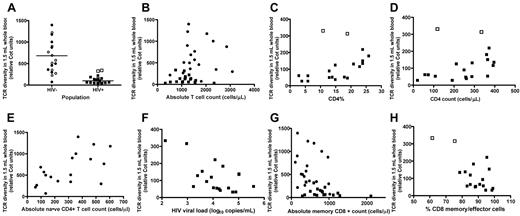
Impact of HIV infection on TCR repertoire diversity in a fixed volume of whole blood. In this figure, as well as those that follow, circles represent HIV− subjects; and squares, HIV+ subjects. (A) Whole-blood TCR repertoire diversity compared for HIV-infected and -uninfected subjects. Diversity is expressed in relative Cot units.17 Solid symbols represent subjects meeting prespecified criteria for comparison; and open symbols, subjects that did not meet these prespecified criteria. Diversity values for HIV-infected subjects were significantly lower (P = .0005; 2-tailed Mann-Whitney test for prespecified comparison groups). (B) Absolute T-cell count did not correlate with whole-blood TCR repertoire diversity (Spearman r = −0.07; P = .79 for HIV−; Spearman r = −0.26; P = .27 for HIV+; Spearman r = 0.25; P = .14 for all subjects). (C) CD4+ T-cell percentage and whole-blood TCR repertoire diversity measured for HIV-infected subjects. Solid symbols represent subjects with viral loads > 1000 copies/mL; and open symbols, subjects with viral loads < 1000 copies/mL. CD4% and sum TCR repertoire diversity were significantly correlated for all HIV-infected subjects (Spearman r = 0.55; P = .01); the strength of correlation increases if the 2 subjects with viral loads < 1000 copies/mL are excluded (Spearman r = 0.72; P = .001). (D) CD4+ T-cell absolute counts and whole-blood TCR repertoire diversity measured for HIV-infected subjects. Solid symbols represent subjects with viral loads > 1000 copies/mL; and open circles, subjects with viral loads < 1000 copies/mL. Absolute CD4 count and whole-blood TCR repertoire diversity were significantly correlated (Spearman r = 0.46; P = .048), but the strength of this correlation improved if the 2 subjects with viral loads < 1000 copies/mL are excluded (Spearman r = 0.61; P = .01). (E) Naive CD4+ T-cell absolute counts and whole-blood TCR repertoire diversity measured for HIV-uninfected subjects (Spearman r = 0.61; P = .008). (F) HIV viral load and blood TCR diversity for HIV-infected subjects. The correlation was not significant (Spearman r = −0.42, P = .07). (G) Absolute memory CD8+ cell count and whole-blood TCR diversity. Memory cells were defined as in Table 3 (Spearman r = −0.21, P = .41 for HIV−; Spearman r = −0.27, P = .29 for HIV+). (H) Percentage CD8 effector/memory cells of all CD8 cells and whole-blood TCR diversity in HIV+ subjects. Open symbols represent subjects with viral loads < 1000 copies/mL. Memory and effector cells were defined as in Table 3 (Spearman r = −0.52, P = .03).
Impact of HIV infection on TCR repertoire diversity in a fixed volume of whole blood. In this figure, as well as those that follow, circles represent HIV− subjects; and squares, HIV+ subjects. (A) Whole-blood TCR repertoire diversity compared for HIV-infected and -uninfected subjects. Diversity is expressed in relative Cot units.17 Solid symbols represent subjects meeting prespecified criteria for comparison; and open symbols, subjects that did not meet these prespecified criteria. Diversity values for HIV-infected subjects were significantly lower (P = .0005; 2-tailed Mann-Whitney test for prespecified comparison groups). (B) Absolute T-cell count did not correlate with whole-blood TCR repertoire diversity (Spearman r = −0.07; P = .79 for HIV−; Spearman r = −0.26; P = .27 for HIV+; Spearman r = 0.25; P = .14 for all subjects). (C) CD4+ T-cell percentage and whole-blood TCR repertoire diversity measured for HIV-infected subjects. Solid symbols represent subjects with viral loads > 1000 copies/mL; and open symbols, subjects with viral loads < 1000 copies/mL. CD4% and sum TCR repertoire diversity were significantly correlated for all HIV-infected subjects (Spearman r = 0.55; P = .01); the strength of correlation increases if the 2 subjects with viral loads < 1000 copies/mL are excluded (Spearman r = 0.72; P = .001). (D) CD4+ T-cell absolute counts and whole-blood TCR repertoire diversity measured for HIV-infected subjects. Solid symbols represent subjects with viral loads > 1000 copies/mL; and open circles, subjects with viral loads < 1000 copies/mL. Absolute CD4 count and whole-blood TCR repertoire diversity were significantly correlated (Spearman r = 0.46; P = .048), but the strength of this correlation improved if the 2 subjects with viral loads < 1000 copies/mL are excluded (Spearman r = 0.61; P = .01). (E) Naive CD4+ T-cell absolute counts and whole-blood TCR repertoire diversity measured for HIV-uninfected subjects (Spearman r = 0.61; P = .008). (F) HIV viral load and blood TCR diversity for HIV-infected subjects. The correlation was not significant (Spearman r = −0.42, P = .07). (G) Absolute memory CD8+ cell count and whole-blood TCR diversity. Memory cells were defined as in Table 3 (Spearman r = −0.21, P = .41 for HIV−; Spearman r = −0.27, P = .29 for HIV+). (H) Percentage CD8 effector/memory cells of all CD8 cells and whole-blood TCR diversity in HIV+ subjects. Open symbols represent subjects with viral loads < 1000 copies/mL. Memory and effector cells were defined as in Table 3 (Spearman r = −0.52, P = .03).
HIV disease associated with decreased whole-blood TCR repertoire diversity
Previous studies using methods, such as immunoscope/spectratyping, have demonstrated that HIV disease is associated with a skewing of the T-cell receptor repertoire (ie, an increased representation of some clones relative to others).5,14 Although these data have been used to suggest that TCR specificities are lost from the repertoire during the course of HIV disease, they could simply reflect the preferential expansion of selected T-cell clones in HIV-infected persons. AmpliCot, an assay taking advantage of DNA hybridization kinetics and amenable to the evaluation of very large numbers of diverse sequences,17,–19 enabled the quantitative assessment of TCR repertoire diversity of human blood or cell samples. AmpliCot has been validated, using both oligonucleotide libraries and titrated naive T cells, to make reproducible distinctions between samples with 2-fold differences in diversity.17,–19 Interassay reproducibility for replicate samples collected in this study is shown in the supplemental Appendix. For samples containing single types of cells with relatively similar frequencies of TCR genes, it is possible to use standards to estimate the absolute number of unique TCR rearrangements in the sample. By contrast, because the whole-blood repertoire contains a mixture of lymphocyte subtypes with an uneven distribution of TCR rearrangements, absolute counts are both technically difficult and less meaningful. For whole blood, we have accordingly expressed our diversity measurements in relative Cot units, which are proportional to the diversity of TCR rearrangements in a sample.
When whole blood was studied in this fashion, HIV disease was associated with a 10-fold reduction in TCR repertoire diversity of a 1.5-mL blood sample (Figure 2A). No other coinfections or exposures were significantly associated with whole-blood T-cell diversity in the HIV-uninfected subjects (Table 2), but the small number of subjects limited our power to detect such associations. In particular, although large clonal expansions have been found in elderly persons with CMV infection,22,,,–26 we did not detect significantly decreased diversity in CMV-infected subjects with AmpliCot. Perhaps our subjects were not old enough to demonstrate this phenomenon. A less likely possibility is that AmpliCot, compared with the methods used in previous studies, may have more sensitivity for residual diversity in the presence of large clonal expansions. Although subjects had wide variability in the number of T cells contained in the 1.5-mL blood sample, there was no significant correlation between absolute T-cell count and whole-blood T-cell diversity (Figure 2B).
Patient variables not significantly associated with whole-blood TCR diversity among 18 HIV− study subjects
| Variable . | No. positive . | Statistical test . | P . |
|---|---|---|---|
| CMV IgG+ | 12 | Mann-Whitney | .89 |
| EBV IgG+ | 6 | Mann-Whitney | .28 |
| H pylori IgG+ | 11 | Mann-Whitney | .86 |
| HBV+ (NAT) or HCV+ (antibody or NAT) | 4 | Mann-Whitney | .87 |
| Pet at home | 4 | Mann-Whitney | .22 |
| Age | NA | Linear regression | .08 |
| Variable . | No. positive . | Statistical test . | P . |
|---|---|---|---|
| CMV IgG+ | 12 | Mann-Whitney | .89 |
| EBV IgG+ | 6 | Mann-Whitney | .28 |
| H pylori IgG+ | 11 | Mann-Whitney | .86 |
| HBV+ (NAT) or HCV+ (antibody or NAT) | 4 | Mann-Whitney | .87 |
| Pet at home | 4 | Mann-Whitney | .22 |
| Age | NA | Linear regression | .08 |
NA indicates not applicable.
We examined the relationship between T-cell subpopulation counts and percentages and whole-blood TCR repertoire diversity. In HIV-infected subjects, the strongest correlation was between CD4+ T-cell percentages and whole-blood TCR repertoire diversity (P = .007, Figure 2C). CD4+ T-cell absolute counts were also significantly correlated with whole-blood TCR repertoire diversity in the HIV-infected subjects if those with low viral loads (< 1000 RNA copies/mL) were excluded (P = .01, Figure 2D). Focusing on naive CD4+ T cells, as opposed to all CD4+ T cells, did not increase the strength of these correlations in HIV-infected subjects. By contrast, in HIV-uninfected subjects there was no significant correlation between CD4+ T-cell percentages and counts and whole-blood TCR repertoire diversity. However, in the uninfected subjects, there were significant correlations between naive CD4+ T-cell counts and percentages and whole-blood TCR diversity (P = .006, Figure 2E) as well as between naive CD4+ T-cell percentages and whole-blood TCR diversity (P = .007).
The 2 patients with the lowest plasma HIV RNA levels (off therapy) had the highest whole-blood TCR repertoire diversity measurements (even overlapping with those observed in HIV-uninfected subjects; Figure 2C,E), but an inverse correlation trend between viral load and whole-blood TCR repertoire diversity was not significant for the HIV-infected group as a whole (Figure 2F). Because AmpliCot measurements, like all TCR repertoire diversity measurements, can be affected by large numbers of expanded clones, we were concerned that the whole-blood TCR diversity changes seen in HIV-infected subjects were solely an artifact of memory CD8+ T-cell expansions seen in the context of HIV disease. Arguing against this possibility, there was no significant correlation between absolute memory CD8+ T-cell counts and whole-blood TCR diversity in the HIV-infected or HIV-uninfected subjects (Figure 2G). There was, however, a significant correlation between the percentage of CD8+ cells that were of memory/effector phenotypes and the whole-blood TCR diversity in the HIV-infected subjects, largely driven by the 2 HIV+ outliers with low viral loads (Figure 2H).
TCR repertoire diversity preserved within discrete T-cell subpopulations
Loss of whole-blood TCR repertoire diversity in HIV-infected subjects could be the result of lower numbers of T cells per unit volume, perturbations in the proportions of different T-cell subpopulations, and/or loss of TCR repertoire diversity within discrete T-cell subpopulations. To address the latter possibility, various subpopulations of CD4+ and CD8+ T cells were sort-purified (using the phenotypes shown in Table 3) and subjected to TCR repertoire analysis with AmpliCot. Because it is difficult to normalize diversity measurements to cell number, we selected samples containing equal numbers of cells of a given type for diversity comparisons. This led to the exclusion of some HIV-infected subjects whose very low numbers of certain subpopulations (particularly naive CD4+ T cells) led to inadequate yields of sorted cells. An analysis plotting input cell number versus measured diversity that contained all patient samples, regardless of size, showed no evidence of a diversity difference between the HIV-infected and -uninfected subjects (supplemental Figure 1).
Sorted T-cell subpopulations
| Phenotype . | Defining markers . | Cell number goal . |
|---|---|---|
| Naive CD4+ | CD3+CD4+CD45RA+CCR7+CD57− | 1.2 × 106 |
| Central memory CD4+ | CD3+CD4+CD45RA−CCR7+CD57− | 1.2 × 106 |
| Effector memory CD4+ | CD3+CD4+CD45RA−CCR7−CD57− | 1.2 × 106 |
| Naive CD8+ | CD3+CD8β+CD45RA+CCR7+CD57− | 1.2 × 106 |
| Central memory CD8+ | CD3+CD8β +CD45RA−CCR7+CD57− | 0.4 × 106 |
| Effector memory CD8+ | CD3+CD8β +CD45RA−CCR7−CD57− | 0.4 × 106 |
| Phenotype . | Defining markers . | Cell number goal . |
|---|---|---|
| Naive CD4+ | CD3+CD4+CD45RA+CCR7+CD57− | 1.2 × 106 |
| Central memory CD4+ | CD3+CD4+CD45RA−CCR7+CD57− | 1.2 × 106 |
| Effector memory CD4+ | CD3+CD4+CD45RA−CCR7−CD57− | 1.2 × 106 |
| Naive CD8+ | CD3+CD8β+CD45RA+CCR7+CD57− | 1.2 × 106 |
| Central memory CD8+ | CD3+CD8β +CD45RA−CCR7+CD57− | 0.4 × 106 |
| Effector memory CD8+ | CD3+CD8β +CD45RA−CCR7−CD57− | 0.4 × 106 |
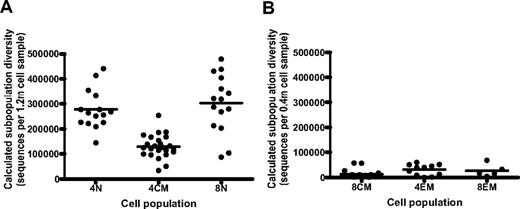
In healthy donors, and as expected, the greatest degree of diversity in the TCR repertoire was found to reside within naive CD4+ and CD8+ T cells, and in central memory CD4+ T cells (Figure 3A), whereas much lower levels of TCR repertoire diversity were found in effector memory CD4+ and CD8+ T cells and central memory CD8+ T cells (Figure 3B). The relatively high diversity of central memory CD4+ T cells is corroborated by the results of sequencing studies.28,29 The number of unique TCRs found within the naive T-cell samples was less than the number of cells examined, a discrepancy likely because of bottlenecks resulting from the splitting of samples during the RT-PCR protocol (which limited the diversity in each Vβ PCR reaction).
Subpopulation diversity of purified T cells from HIV-uninfected subjects. (A) Diversity (TCR sequences measured per 1.2 million cells) of naive CD4+ T cells (4N; CD3+CD4+CCR7+CD45RA+CD57−), central memory CD4+ T cells (4CM; CD3+CD4+CCR7+CD45RA−CD57−), and naive CD8+ T cells (8N; CD3+CD4+CCR7+CD45RA+CD57−) sort-purified from HIV-uninfected subjects. (B) Absolute diversity (TCR sequences measured per 0.4 million cells) of central memory CD8+ T cells (8CM; CD3+CD8+CCR7+CD45RA−CD57−), effector memory CD4+ T cells (4EM; CD3+CD4+CCR7−CD45RA+), and effector memory CD8+ T cells (8EM; CD3+CD4+CCR7−CD45RA+) sorted from HIV-uninfected subjects. One-way ANOVA for the 6 populations was significant (Kruskal-Wallis test, P < .0001). The 4N and 8N populations did not have a significant difference in sequences per sample (Mann-Whitney test, P = .32), but the 4CM population had significantly less diversity than 4N and 8N and significantly more diversity than 8CM (all comparisons with 2-tailed Mann-Whitney tests, P < .0001).
Subpopulation diversity of purified T cells from HIV-uninfected subjects. (A) Diversity (TCR sequences measured per 1.2 million cells) of naive CD4+ T cells (4N; CD3+CD4+CCR7+CD45RA+CD57−), central memory CD4+ T cells (4CM; CD3+CD4+CCR7+CD45RA−CD57−), and naive CD8+ T cells (8N; CD3+CD4+CCR7+CD45RA+CD57−) sort-purified from HIV-uninfected subjects. (B) Absolute diversity (TCR sequences measured per 0.4 million cells) of central memory CD8+ T cells (8CM; CD3+CD8+CCR7+CD45RA−CD57−), effector memory CD4+ T cells (4EM; CD3+CD4+CCR7−CD45RA+), and effector memory CD8+ T cells (8EM; CD3+CD4+CCR7−CD45RA+) sorted from HIV-uninfected subjects. One-way ANOVA for the 6 populations was significant (Kruskal-Wallis test, P < .0001). The 4N and 8N populations did not have a significant difference in sequences per sample (Mann-Whitney test, P = .32), but the 4CM population had significantly less diversity than 4N and 8N and significantly more diversity than 8CM (all comparisons with 2-tailed Mann-Whitney tests, P < .0001).
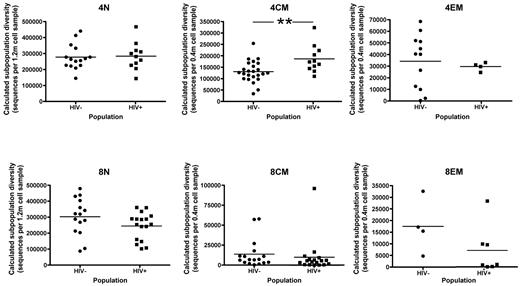
The diversity of purified T-cell subpopulations from HIV-infected and -uninfected subjects is compared in Figure 4. Surprisingly, when equal numbers of cells were analyzed, HIV-infected subjects did not appear to have significant changes in the diversity of their CD4+ naive T cells in the sample sizes examined. Indeed, all T-cell subpopulations examined had equal numbers of TCR sequences on a per-cell basis in HIV-infected and -uninfected subjects, with one exception: central memory CD4+ T cells from HIV-infected subjects harbored a significantly more diverse TCR repertoire (P = .007). The relatively high diversity of CD4+ central memory T cells in HIV-infected subjects may explain why whole-blood TCR diversity was no more strongly correlated with counts of naive CD4+ T cells than with total CD4+ T cells in these subjects. This result suggests that HIV disease is associated with higher rates of naive to central memory cell differentiation and/or changes in the turnover rate of central memory CD4+ T cells.30,31 Several features unique to HIV disease might contribute to these changes in the CD4+ central memory repertoire, including direct responses to HIV antigens, increased immune activation, and responses to unmeasured coinfections that are more prevalent in HIV-infected subjects.
Subpopulation diversity of sort-purified T cells in HIV-uninfected versus -infected subjects. Data are shown for those HIV-uninfected and -infected subjects (regardless of prespecified comparison group) from whom the target number of cells (1.2 million cells or 0.4 million cells) could be collected. Abbreviations for cell subpopulations and the markers used to define them for flow cytometric purification are the same as in Figure 3. All diversity comparisons were nonsignificant, except that the TCR repertoire of the central memory CD4+ T-cell subpopulation from HIV-infected subjects was significantly more diverse than that found in HIV-uninfected subjects (2-tailed Mann-Whitney test, P = .007).
Subpopulation diversity of sort-purified T cells in HIV-uninfected versus -infected subjects. Data are shown for those HIV-uninfected and -infected subjects (regardless of prespecified comparison group) from whom the target number of cells (1.2 million cells or 0.4 million cells) could be collected. Abbreviations for cell subpopulations and the markers used to define them for flow cytometric purification are the same as in Figure 3. All diversity comparisons were nonsignificant, except that the TCR repertoire of the central memory CD4+ T-cell subpopulation from HIV-infected subjects was significantly more diverse than that found in HIV-uninfected subjects (2-tailed Mann-Whitney test, P = .007).
Changes in whole-blood TCR repertoire diversity in HIV disease largely explained by changes in the numbers of T-cell subpopulations
To integrate the aforementioned measurements of TCR repertoire diversity in whole-blood and in sort-purified T-cell subpopulations, we charted a representation of the component subpopulations in a 1.5 mL whole-blood sample for the subjects in the prespecified comparison groups. Two representative charts are shown in Figure 5, and charts for the remaining subjects are provided as supplemental data. The rectangles represent the number of cells of each subpopulation in 1.5 mL of blood (x-axis) and the calculated number of unique TCR sequences in that number of cells from that subpopulation (y-axis; see figure legend for details). The rectangles are summed to reflect the total number of cells in the blood sample on the x-axis and the estimated diversity in the blood sample on the y-axis. For a given T-cell subpopulation, cells from HIV-infected and HIV-uninfected subjects have similar TCR repertoire diversity when equal cell numbers are measured. However, the decreased number of cells from high diversity subpopulations in the peripheral blood (ie, naive CD4+ and CD8+ T cells and, to a lesser extent, central memory CD4+ T cells) results in a significant overall decrease in sum TCR repertoire diversity in whole-blood samples from HIV-infected subjects.
TCR repertoire diversity maps in HIV-infected and -uninfected subjects. (A-B) Plots represent the T cells present in 1.5 mL of blood, with each color block representing a different subpopulation. The x-axis represents the numbers of cells; and y-axis, TCR repertoire diversity (number of unique sequences found in the cell sample). We calculated this using several simplifying assumptions. First, we assumed that all receptors were present at equal frequency within a given T-cell subpopulation. Based on the results of our diversity measurements of highly diverse naive samples, we estimated that the maximal measurable diversity in a sample of n cells was n/2.5. Finally, we assumed that, if the number of a specific subpopulation of cells in a 1.5-mL blood volume was close to the diversity of that population, the actual diversity seen in 1.5 mL would be reduced because of sampling, according to the Poisson distribution. We therefore plotted the diversity of each subpopulation as the smallest of the 3 values: n/2.5; d; or [(n/2.5)+d)/3], where n = number of cells of that subpopulation in a 1.5-mL blood volume and d = maximum measured sequence diversity of that subpopulation in a sample of any size. Sequences within subpopulations are depicted as nonoverlapping; sequencing experiments suggest that this is largely the case with clone nucleotide sequences32,–34 but was not directly verified in our study. Dashed lines indicate total cell number and total TCR repertoire diversity within the 1.5-mL blood sample. The 2 plots are for representative subjects from the prespecified comparison group who are either HIV-uninfected (A) or HIV-infected (B). Plots for all subjects in the prespecified comparison groups are available as supplemental data. All plots use identical scales for cell number and TCR diversity. (C) Correlation between subpopulation and sum blood measurements. For all subjects in the prespecified comparison group, the measured sum TCR repertoire diversity in whole blood (expressed in relative Cot units, as in Figure 2) is shown on the x-axis. Subpopulation TCR repertoire diversity values (sequences expected per 1.5 mL whole blood), plotted on the y-axis, were calculated as the sum of each of the absolute values of the T-cell subpopulations that could be evaluated. The r2 value was 0.72 (P < .0001).
TCR repertoire diversity maps in HIV-infected and -uninfected subjects. (A-B) Plots represent the T cells present in 1.5 mL of blood, with each color block representing a different subpopulation. The x-axis represents the numbers of cells; and y-axis, TCR repertoire diversity (number of unique sequences found in the cell sample). We calculated this using several simplifying assumptions. First, we assumed that all receptors were present at equal frequency within a given T-cell subpopulation. Based on the results of our diversity measurements of highly diverse naive samples, we estimated that the maximal measurable diversity in a sample of n cells was n/2.5. Finally, we assumed that, if the number of a specific subpopulation of cells in a 1.5-mL blood volume was close to the diversity of that population, the actual diversity seen in 1.5 mL would be reduced because of sampling, according to the Poisson distribution. We therefore plotted the diversity of each subpopulation as the smallest of the 3 values: n/2.5; d; or [(n/2.5)+d)/3], where n = number of cells of that subpopulation in a 1.5-mL blood volume and d = maximum measured sequence diversity of that subpopulation in a sample of any size. Sequences within subpopulations are depicted as nonoverlapping; sequencing experiments suggest that this is largely the case with clone nucleotide sequences32,–34 but was not directly verified in our study. Dashed lines indicate total cell number and total TCR repertoire diversity within the 1.5-mL blood sample. The 2 plots are for representative subjects from the prespecified comparison group who are either HIV-uninfected (A) or HIV-infected (B). Plots for all subjects in the prespecified comparison groups are available as supplemental data. All plots use identical scales for cell number and TCR diversity. (C) Correlation between subpopulation and sum blood measurements. For all subjects in the prespecified comparison group, the measured sum TCR repertoire diversity in whole blood (expressed in relative Cot units, as in Figure 2) is shown on the x-axis. Subpopulation TCR repertoire diversity values (sequences expected per 1.5 mL whole blood), plotted on the y-axis, were calculated as the sum of each of the absolute values of the T-cell subpopulations that could be evaluated. The r2 value was 0.72 (P < .0001).
We also used these data to test whether various distributions of T-cell subpopulations might have affected the accuracy of our whole-blood TCR diversity measurements. Figure 5C shows a plot of the total TCR diversity in 1.5 mL of blood estimated from the summation of cell subpopulations versus the measured Cot values from 1.5 mL of whole blood from these subjects. Although these 2 measurements have a linear relationship, the best-fit line does not go through the origin, indicating moderate error in absolute terms. This discrepancy is probably the result of the effect of large clonal expansions, reducing the Cot values of whole-blood AmpliCot measurements, but may also reflect overestimates in our modeling of the diversity of the subpopulations of T cells found in these samples.
Discussion
Association of HIV disease with reduced T-cell receptor repertoire diversity per volume of blood
Here we have used a novel quantitative assay and a cross-sectional study design to demonstrate that progressive HIV disease is associated with decreased whole-blood TCR diversity. The decreased diversity of available TCRs per volume of blood may impair pathogen recognition of pathogens, thereby contributing to the immune dysfunction associated with HIV infection. Despite the strong associations found, our cross-sectional study design makes it impossible to establish a causal relationship between HIV infection and TCR diversity loss. Because in part of this concern, we were careful in selecting our control population, so that we could minimize unmeasured confounding variables. We therefore only included subjects who were CMV seropositive and who lacked evidence of hepatitis B or C exposure. We also matched subjects based on demographics and excluded persons with recent infections and other conditions potentially affecting the immune system. Two lines of evidence that the observed diversity differences are probably attributable to HIV infection are that no other measured coinfection was significantly associated with TCR diversity and the relationship between HIV infection and reduced diversity was consistently seen, whether or not the study analysis was restricted to the prespecified groups of participants. Because we only enrolled subjects without active opportunistic infections or cancers, it is possible that our HIV-infected subjects may have been biased toward those who are healthy, which would decrease the observed effects of HIV infection on immune repertoire diversity.
Clinical significance of TCR diversity
In HIV-infected subjects, we found that blood TCR diversity correlated with CD4+ T-cell counts and percentages, which are important albeit imperfect predictors of the risk of opportunistic infections. Is TCR diversity independent of T-cell numbers clinically significant? Certainly, HLA diversity is important for pathogen resistance on a population level.35 In principle, at least some TCR diversity (per fixed volume of blood or tissue) is necessary for basic antigen recognition, but how much is necessary is not known. Current data are equivocal about the importance of TCR diversity, although recent studies of aging in animal models suggest that skewing of the TCR repertoire predicts vulnerability to infection.36,37 Although we do not yet know the clinical significance of the 10-fold decrease in whole-blood TCR diversity associated with HIV infection, deficits of this magnitude exceed the physiologic reserve of most organ systems. Future studies will determine whether TCR diversity is a clinically relevant predictor of a patient's risk of opportunistic infections or cancer, potentially complementing counts of T-cell subpopulations.
Although the CD4+ T-cell count is a well-validated and useful marker, it does not provide perfect stratification of patient risks of opportunistic infections and cancers.1,–3 TCR diversity could potentially provide additional prognostic information to a CD4+ T-cell count, perhaps explaining cases of opportunistic infections that occur in patients with high CD4+ T-cell counts.6 Although CD4+ T-cell counts are associated with the loss of immune function during HIV disease progression, the relationship between CD4+ T-cell count and reconstitution of immune function after treatment has begun is less well substantiated.38,–40 Part of the benefit of antiretroviral therapy might be mediated through improvements in T-cell diversity (eg, through increases in the CD8+ naive:memory ratio), not just CD4+ T-cell counts. More evidence that CD4+ T-cell counts alone may not fully measure immunocompetence is the finding that IL-2, which increases CD4+ T-cell counts without increasing repertoire diversity, provides no clinical benefit to HIV-infected patients.40
The 2 HIV-infected subjects with low plasma HIV RNA levels also had higher whole-blood TCR diversity. Because HIV-specific T cells represent a small fraction of total T cells,41 this association is probably not a simple consequence of the low abundance of HIV antigen in these patients but may be a result of HIV immunopathogenesis. This result, if corroborated, might provide an explanation for 2 clinical phenomena. First, in cohort studies, viral load and CD4 counts are independent factors that predict progression to AIDS.42 High viral loads might not simply lead to AIDS by depletion of CD4+ T-cell number, but also by reducing T-cell diversity per volume of blood. Second, patients on antiretroviral therapy have improved clinical outcomes, even if their CD4+ T-cell counts are not restored. Longitudinal studies of persons beginning antiretroviral therapy will best address the question of whether suppression of viremia can result in rapid increases in TCR diversity. Clonal expansions have been observed to decrease once ART is started16 ; and if these cells are replaced by more diverse T cells, TCR diversity per volume of blood may therefore increase.
Preserved diversity of T-cell subpopulations and the potential for immune reconstitution
Our analysis of T-cell subpopulations did not detect significant changes in TCR diversity between HIV-uninfected and HIV-infected subjects, except for CD4+ central memory T cells, which surprisingly were significantly more diverse in persons with HIV. We therefore did not find evidence that HIV infection was associated with permanent losses of TCR diversity in T-cell subpopulations. Consonant with this finding, it is unusual for patients to contract opportunistic infections after T-cell reconstitution with antiretroviral therapy, and prophylaxis regimens can be safely discontinued. The preservation of a significant reservoir of TCR repertoire diversity in the naive cell populations might permit a TCR repertoire to be successfully reconstituted without de novo thymic activity. IL-7 is a promising agent that might reconstitute the immune system in this way in patients who do not achieve reconstitution with antiretroviral therapy alone. Human trial data suggest it may increase TCR diversity per blood/tissue volume by selectively stimulating proliferation of high-diversity cell populations rather than by stimulating thymopoiesis.43
Sampling considerations
We do not yet know what sample size for repertoire diversity measurements (ie, the number of cells or the volume of blood or tissue) is most clinically relevant. On the one hand, the body must certainly contain more than a single cell of a given specificity for it to be useful in either immunocompetence or potential immune reconstitution, suggesting that a significantly smaller unit of examination would be appropriate. The fact that individual memory or effector clones are represented by thousands of cells in the body is further evidence that the unit of examination should be smaller than the whole body. On the other hand, if the unit of examination is too small, clinically significant differences in TCR diversity might be masked.
Although the optimal sample sizes of blood and cells remain to be determined, we must underscore that our results are a function of the sample sizes we collected. If we had sampled larger volumes of whole blood, we might have found an even larger diversity difference between HIV-infected and HIV-uninfected subjects. Similarly, our samples of naive cells represented only a tiny fraction of those found in the body, and our data suggest that our sample size was limiting for these cells (as opposed to memory-effector subpopulations). Had we been able to examine larger samples of cells, we might have detected a greater difference in diversity between, for example, naive CD4+ T cells and central memory CD4+ T cells. Moreover, the 1.2 million naive CD4+ T-cell samples we collected represent the cells found in an average of 5 mL of blood in the HIV-uninfected subjects but an average of 27 mL of blood in the HIV-infected subjects. If we had been able to measure the total body naive repertoires of our subjects, we might have found a significant reduction in diversity in our HIV-infected subjects because they have fewer total body naive cells. However, the importance of the total body naive repertoire for immune reconstitution is unknown.
Our study has several limitations, including its cross-sectional nature. We only sampled blood, and the TCR diversity of the lymphoid tissues that contain the bulk of T cells was not directly measured, although the extent to which these repertoires differ is unclear.44 The TCR-β chain repertoire is probably related to, but is not identical to, the functional TCR repertoire: our measurements did not capture TCR-α diversity, binding specificity, or cellular function beyond that suggested by phenotypic markers. Our method cannot discern whether a repertoire of a given diversity may be depleted or enriched for important clones.45 Finally, AmpliCot measurements, although rapid and economical, are prone to error because of unequal frequencies of TCRs in the sample, particularly if they are not evenly distributed across all VJ families. As more TCR sequencing data become available, it may be possible to model these frequency distributions to reduce this source of error. Further discussion of technical limitations of our procedure is included in the supplemental Appendix.
In conclusion, although HIV infection is associated with a quantitative reduction in TCR repertoire diversity per volume of peripheral blood, diversity seems to be preserved on a per-cell basis for individual subpopulations. Thus, although significant efforts have been made to increase the diversity of the TCR repertoire of HIV-infected patients (eg, through stimulation of thymic function), our results suggest that this may not be necessary for the restoration of a diverse repertoire per volume of whole blood. Whereas this study was necessarily small because it required well-defined subject groups for internal validity and involved large-volume cell sorting, future studies of whole-blood diversity in larger numbers of subjects will test the generalizability of our findings and permit multivariate analysis of variables, such as age, sex, and coinfections. Longitudinal studies of patients beginning antiretroviral therapy will help elucidate the possible relationship between viral load and blood TCR diversity. Our work provides a clinically relevant example of the use of AmpliCot for the direct measurement of repertoire diversity. Our method for measuring TCR diversity may also have application in low-resource settings because it could permit the measurement of immunologic status and viral loads with the same nucleic acid amplification technology.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank study volunteers for their participation; Marcia Smith and Joy Madamba for help with recruitment, questionnaires and sample collection; Leslie Tobler for nucleic acid testing of samples; Saunak Sen for statistical consultation; and members of the J.M.M. laboratory for discussions and comments on the manuscript.
This work was supported by the National Institutes of Health (grant K23 AI 073100, P.D.B.; and R37 AI40312 and UO1 AI43864, J.M.M.) and the California HIV/AIDS Research Program (an IDEA Grant, P.D.B.). J.M.M. is a recipient of the National Institutes of Health Director's Pioneer Award Program, part of the National Institutes of Health Roadmap for Medical Research (grant DPI OD00329). This SCOPE cohort was supported in part by the National Institute of Allergy and Infectious Diseases (RO1 AI087145, K24AI069994), the University of California San Francisco Center for AIDS Research (PO AI27763), the University of California San Francisco Clinical and Translational Sciences Institute (UL1 RR024131), the Cleveland Immunopathogenesis Consortium (AI 76174), and Center for AIDS Research Network of Integrated Systems (R24 AI067039).
National Institutes of Health
Authorship
Contribution: P.D.B. and J.M.M. designed the experiments; R.H., S.D., and J.M. managed the SCOPE study; P.D.B., D.S., and R.H. recruited study subjects; P.D.B. performed flow analysis and sorting; P.D.B., J.J.Y., and Q.Z. performed AmpliCot and quantitative PCR analyses; P.D.B. and Q.Z. performed ELISAs; M.B. performed viral NAT assays; and P.D.B. and J.M.M. wrote the manuscript with assistance from all coauthors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paul D. Baum, Division of Experimental Medicine, University of California San Francisco, Box 1234, San Francisco, CA 94143-1234; e-mail: paul.baum@ucsf.edu.

![Figure 5. TCR repertoire diversity maps in HIV-infected and -uninfected subjects. (A-B) Plots represent the T cells present in 1.5 mL of blood, with each color block representing a different subpopulation. The x-axis represents the numbers of cells; and y-axis, TCR repertoire diversity (number of unique sequences found in the cell sample). We calculated this using several simplifying assumptions. First, we assumed that all receptors were present at equal frequency within a given T-cell subpopulation. Based on the results of our diversity measurements of highly diverse naive samples, we estimated that the maximal measurable diversity in a sample of n cells was n/2.5. Finally, we assumed that, if the number of a specific subpopulation of cells in a 1.5-mL blood volume was close to the diversity of that population, the actual diversity seen in 1.5 mL would be reduced because of sampling, according to the Poisson distribution. We therefore plotted the diversity of each subpopulation as the smallest of the 3 values: n/2.5; d; or [(n/2.5)+d)/3], where n = number of cells of that subpopulation in a 1.5-mL blood volume and d = maximum measured sequence diversity of that subpopulation in a sample of any size. Sequences within subpopulations are depicted as nonoverlapping; sequencing experiments suggest that this is largely the case with clone nucleotide sequences32–34 but was not directly verified in our study. Dashed lines indicate total cell number and total TCR repertoire diversity within the 1.5-mL blood sample. The 2 plots are for representative subjects from the prespecified comparison group who are either HIV-uninfected (A) or HIV-infected (B). Plots for all subjects in the prespecified comparison groups are available as supplemental data. All plots use identical scales for cell number and TCR diversity. (C) Correlation between subpopulation and sum blood measurements. For all subjects in the prespecified comparison group, the measured sum TCR repertoire diversity in whole blood (expressed in relative Cot units, as in Figure 2) is shown on the x-axis. Subpopulation TCR repertoire diversity values (sequences expected per 1.5 mL whole blood), plotted on the y-axis, were calculated as the sum of each of the absolute values of the T-cell subpopulations that could be evaluated. The r2 value was 0.72 (P < .0001).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/15/10.1182_blood-2011-11-395384/5/m_zh89991289260005.jpeg?Expires=1769109133&Signature=HI8o6s4kcNlWX5sI6LkHennmoJyZYCCG0Ij3RS1UTCN0ReuNYrmXbpRFHwsCYICw8AI8ClI1aC-O7-u5e1sxyDPS63mST6sXB-pa8gBZfOz5URqp~6FBgFI23qkD-IFdkKHF5r3fVQFPsZVYJGXn-Af6B0CyOjWwayhgI0P-15HgS8y5PM9rv40Zb0d~7trnueRsbBg-KMoB2MCGiiK7r4xxjgfW7D5TLuXZTB-VseJiTrbv5pXEoTgHmq8PioS6egOqQ~5HteXDoYKnMhyKQhROhBCNKTozs91B7ke6Wz2LOIaAyXnCA0UfTqvVbD7N1wc7Vclo8JHWpfOcVKqNYw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal