Abstract
Glucocorticoid receptor (GR) agonists increase erythropoiesis in vivo and in vitro. To clarify the effect of the dominant negative GRβ isoform (unable to bind STAT-5) on erythropoiesis, erythroblast (EB) expansion cultures of mononuclear cells from 18 healthy (nondiseased) donors (NDs) and 16 patients with polycythemia vera (PV) were studied. GRβ was expressed in all PV EBs but only in EBs from 1 ND. The A3669G polymorphism, which stabilizes GRβ mRNA, had greater frequency in PV (55%; n = 22; P = .0028) and myelofibrosis (35%; n = 20) patients than in NDs (9%; n = 22) or patients with essential thrombocythemia (6%; n = 15). Dexamethasone stimulation of ND cultures increased the number of immature EBs characterized by low GATA1 and β-globin expression, but PV cultures generated great numbers of immature EBs with low levels of GATA1 and β-globin irrespective of dexamethasone stimulation. In ND EBs, STAT-5 was not phosphorylated after dexamethasone and erythropoietin treatment and did not form transcriptionally active complexes with GRα, whereas in PV EBs, STAT-5 was constitutively phosphorylated, but the formation of GR/STAT-5 complexes was prevented by expression of GRβ. These data indicate that GRβ expression and the presence of A3669G likely contribute to development of erythrocytosis in PV and provide a potential target for identification of novel therapeutic agents.
Introduction
Polycythemia vera (PV) is a Philadelphia chromosome–negative myeloproliferative neoplasm (MPN) characterized by increased production of erythroid cells.1 As in other MPNs, PV is associated with a gain-of-function mutation (JAK2V617F) of the JAK2 gene.2-4 JAK2 is the first transduction element of many hematopoietic growth factor receptors, including the receptor for erythropoietin, the primary growth factor that controls erythroid cell production.5 The observation that inhibition of JAK2V617F abrogates erythropoietin-independent growth of erythroid progenitors suggested the hypothesis that in PV, increased erythroid production is caused by constitutive activation of erythropoietin receptor signaling (STAT-5 and/or PI3K/Akt).6-8 Because low and high levels of STAT-5 activation favor maturation and proliferation, respectively, in normal hematopoietic cells,9,10 the presence of the JAK2V617F mutation in PV may increase the intrinsic proliferative potential of erythroid cells by increasing STAT-5 activation.11 This concept is indirectly supported by the observation that hematopoietic progenitor cells from PV patients generate greater numbers of erythroblasts (EBs) in liquid culture than cells from healthy (nondiseased) donors (NDs).7,12
In addition to erythropoietin, erythroid proliferation is also controlled by nuclear receptors such as the glucocorticoid receptor (GR). Evidence for this regulatory role is provided by clinical observations and in vitro studies of cultured EBs. Patients may develop erythrocytosis as the first manifestation of Cushing syndrome.13 In addition, 50% of patients with Diamond-Blackfan anemia,14 a disease caused by inherited mutations that lead to ribosomal dysfunction,15 respond to glucocorticoids. However, little is known about the mechanism by which GR stimulation restores red cell production in Diamond-Blackfan anemia or why only 50% of these patients respond to glucocorticoids. Studies in mice have shown that the presence of glucocorticoids maintains EBs in a self-renewal state, blocking their maturation in response to stem cell factor (SCF) and erythropoietin.16 Loss-of-function studies proved that this effect requires not only erythropoietin receptor and GR but also STAT-5, which suggests that the switch from maturation to self-renewal is downstream of the STAT-5 pathway. The ability of glucocorticoids to maintain erythroid cells in proliferation has been exploited to develop liquid cultures that permit massive amplification of primary murine and human EBs.17-19 Greater numbers (108-1010) of EBs are generated under human erythroid mass amplification (HEMA) conditions, a refinement of this liquid system.17 This greater EB expansion is sustained by GR through both transcriptional (regulation of subsets of erythropoietin- and SCF-sensitive genes18 ) and membrane-associated (inhibition of STAT-5 phosphorylation induced by erythropoietin20 ) activities. However, for unknown reasons, in contrast to the constant numbers of EBs generated in culture from the same donor, great variability (> 1 log) exists in the numbers of EBs generated among donors.17
GR is encoded by a polymorphic gene on human chromosome 5q31-3221,22 (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). This polymorphism produces up to 256 combinations of alternative GR homodimers and heterodimers, which are variably expressed in the human population.23,24 Differences in ligand or DNA binding affinities among isoforms are emerging as the leading cause of heterogeneity in body mass25 and in the response to GR ligands during stress26 and major depression.27 One of these polymorphisms is represented by the A3669G single-nucleotide polymorphism (SNP) in exon 9 of GR, which stabilizes the mRNA and increases the expression of GRβ (half-life of A3669G+ and A3669G− GRβ mRNA is > 6 and < 6 hours, respectively).28 GRβ is generated by alternative splicing of exon 9 and differs from GRα by the 50 carboxy-terminal amino acids, which are replaced by 15 unique amino acids.29,30 These 15 amino acids interfere with protein folding, retaining the receptor within the nucleus and conferring unique biologic functions.31 On ligand activation, GRα migrates to the nucleus both as homodimers and as heterodimeric tetrameric protein complexes with several signaling molecules, including the phosphorylated form of STAT-5.23,24,32 In the nucleus, GRα binds not only glucocorticoid-responsive elements but also elements that are responsive to each type of glucocorticoid complex.23,24,32 GRβ binds poorly to glucocorticoids because its unique structure impairs the ligand-binding domain and induces nuclear retention.30 Therefore, it does not form complexes with other signaling partners, including the phosphorylated form of STAT-5, and by forming heterodimers with GRα, it impairs ligand binding of this isoform and acts as a dominant negative regulator of GR function within the cells.23,24,32 It is not known whether GRβ is expressed in erythroid cells or how the presence of A3669G affects this expression.
Erythroblasts expanded ex vivo from the mononuclear cells (MNCs) of NDs in the presence of dexamethasone express increased proliferation, with delayed maturation similar to that expressed by EBs generated ex vivo by MNCs from PV patients.7,12,17 This similarity prompted us to compare the biologic activity of GR in EBs from NDs and PV patients. First, we confirmed that MNCs from PV patients generated ex vivo greater numbers of EBs than MNCs from NDs. However, EBs from PV patients, which proliferated well without dexamethasone, did not increase in number in response to dexamethasone and were similar in morphology and expression profile to EBs generated ex vivo from NDs in the presence of dexamethasone. The poor response to dexamethasone was likely related to the fact that EBs from all the PV patients tested expressed GRβ in addition to GRα. Thus, in spite of constitutive STAT-5 phosphorylation observed in PV cells, STAT-5 was not bound into transcriptionally active complexes with GRα. Analysis of the frequency of the A3669G SNP in NDs and in MPN patients revealed that this SNP was more frequent in PV patients (55%) and patients with primary myelofibrosis (PMF; 35%) than in NDs (9%) or patients with essential thrombocythemia (ET; 6%). Although the mechanism that activates expression of GRβ in PV EBs is still unknown, these data suggest impaired GR signaling as a unifying mechanism for development of erythrocytosis in syndromes associated with chronic exposure to high levels of glucocorticoids, as in Cushing syndrome and immunosuppressive treatments (glucocorticoid prevention of erythropoietin-induced STAT-5 phosphorylation), PV patients (expression of GRβ and presence of the A3669G SNP), and possibly other idiopathic forms of erythrocytosis.
Methods
Subjects
Peripheral blood was collected from 41 deidentified NDs by the transfusion center of La Sapienza University (Rome, Italy) and the New York Blood Center (New York, NY) according to guidelines established by the local ethics committees for human subject studies. After written informed consent approved by the Mount Sinai School of Medicine was obtained, in accordance with the Declaration of Helsinki, peripheral blood from patients with MPN was collected. Granulocyte DNA was prepared from NDs (n = 22) and from patients with PV (n = 22), ET (n = 15), and PMF (n = 20), and MNCs were cryopreserved for ex vivo culture from 19 PV patients not studied previously. The total number of subjects studied is reported in Table 1. The diagnoses were established according to World Health Organization criteria.33 All MNCs were prepared by standard Ficoll-Hypaque centrifugation (Amersham Pharmacia Biotech). The clinical features of the first 16 patients at the time of blood collection are summarized in supplemental Table 1. The patients were considered homozygous for the JAK2V617F mutation when the granulocyte allele burden was > 50%. Both patients and NDs are sometimes referred to by alphanumeric designations in the figures and in the text (eg, PV732).
Summary of data on the expression (mRNA or protein) of GRβ in erythroblasts expanded ex vivo and on the presence of the A3669G SNP in ND and MPN patients
| . | Ex vivo expanded erythroblasts . | Blood DNA A3669G SNP . | Total subjects, n/N (%) . | |
|---|---|---|---|---|
| mRNA . | Protein . | |||
| ND | 0/10 | 1/9 | 2/22 | 3/41 (7.3) |
| Polycythemia vera | 16/16 | 4/4* | 12/22 | 32/41 (78) |
| Essential thrombocythemia | N/A | N/A | 1/15 | 1/15 (6.6) |
| Primary myelofibrosis | N/A | N/A | 7/20 | 7/20 (35) |
| . | Ex vivo expanded erythroblasts . | Blood DNA A3669G SNP . | Total subjects, n/N (%) . | |
|---|---|---|---|---|
| mRNA . | Protein . | |||
| ND | 0/10 | 1/9 | 2/22 | 3/41 (7.3) |
| Polycythemia vera | 16/16 | 4/4* | 12/22 | 32/41 (78) |
| Essential thrombocythemia | N/A | N/A | 1/15 | 1/15 (6.6) |
| Primary myelofibrosis | N/A | N/A | 7/20 | 7/20 (35) |
N/A indicates not available (not determined).
One of these PV patients was also analyzed for mRNA expression.
Liquid culture of human EBs
MNCs (106 cells/mL) were cultured under HEMA conditions stimulated with SCF (10 ng/mL), erythropoietin (3 U/mL), and IL-3 (1 ng/mL), with or without dexamethasone (10−6M; Sigma-Aldrich), as described previously.17 After 11 and 13 days, cells were collected for further analyses.
Phenotypic analysis
Smears were prepared by cytocentrifugation (Shandon) and either stained with May-Grünwald-Giemsa stain or fixed with paraformaldehyde (4%; Invitrogen), saturated/permeabilized for 30 minutes with NET gel (150mM NaCl, 5mM EDTA, 50mM Tris-HCl pH 7.4, 0.05% NP-40, 0.25% lambda carrageenan gelatin, 0.02% NaN3) and stained for 60 minutes with antibodies specific for GR (sc-8992; Santa Cruz Biotechnology) and then probed with secondary FITC-conjugated antibodies (Invitrogen), as described previously.20 Nuclei were counterstained with DAPI (4,6-diamidino-2-phenylindole; Sigma-Aldrich). As a negative control, samples were incubated with secondary antibody only. Slides were observed with an Axioskop 40 microscope (Carl Zeiss) and images acquired with a CoolSNAP cf digital CCD camera (Photometrics) and analyzed with a MetaMorph Imaging System (Molecular Devices). Differential counts were performed on May-Grünwald-Giemsa–stained cytospin preparations by counting at least 200 cells on randomly selected fields. For flow cytometry, cells were labeled with PE-conjugated CD235a (anti–glycophorin A) and either FITC-conjugated CD36 (an antigen on the thrombospondin receptor34 ) or CD45 (all from BD Biosciences). Dead cells were identified by staining with propidium iodide (5 μg/mL; Sigma-Aldrich). Cell fluorescence was analyzed with a FACSAria (Becton Dickinson).
RNA isolation and gene expression profiling
Total RNA was extracted with TRIzol (Invitrogen Life Technologies) and its concentration purity/integrity established by use of NanoDrop technology (Thermo Scientific). RNA (100 ng) was reverse transcribed with a high-capacity cDNA archive kit (Applied Biosystems). cDNA was diluted with the HotStarTaq master mix kit (QIAGEN). Both regular and quantitative RT-PCR were performed. Regular RT-PCR was used to detect the presence of transcripts for GRα and GRβ. In these experiments, cDNA was amplified for 30 cycles with oligonucleotides that were specific for GRα or GRβ and the housekeeping gene GAPDH, as described previously35 (supplemental Figure 1), with a vapo.protect thermocycler (Eppendorf AG). Amplified products were fractioned by electrophoresis on agarose gels (1%), stained with ethidium bromide, and visualized with ultraviolet light. The specificity of the amplification was confirmed by sequencing. Quantitative RT-PCR determinations were performed as described previously.36,37 cDNA was dissolved in TaqMan universal PCR master mix (KAPA Biosystems) and each sample amplified in triplicate with TaqMan gene expression assays for the different gene targets with the ABI PRISM 7300 HT sequence detection system (Applied Biosystems). The variation in threshold cycle observed in replicate independent amplifications of the same sample was < 5%. In these experiments, RNase P was amplified concurrently as a housekeeping control gene (Applied Biosystems). Expression levels were calculated with the algorithm ΔCt = CtX−CtRNaseP, in which Ct was the average threshold cycle and X was the gene being analyzed, and are presented as 2−ΔCt.
Western blot and immunoprecipitation analyses
Whole-cell extracts were prepared as described previously,20 and proteins (30 μg) were separated on SDS-PAGE; transferred to nitrocellulose membranes that were first incubated with anti-GRβ (PA3-514; Affinity Bioreagents), anti-GRα, anti–glucocorticoid-induced leucine zipper protein (GILZ FL-134: sc-33780), or anti-actin (sc-1616) antibodies (Santa Cruz Biotechnology); and then incubated with appropriate HRP-coupled secondary antibodies (Calbiochem). Whole-cell lysates from mouse 293T cells transfected with the GILZ gene were used as control. Immune complexes were detected with an enhanced chemiluminescence kit (Amersham). For immunoprecipitation studies, cell extracts (30-50 μg) were incubated with either the anti–STAT-5 or anti-GRα antibody overnight at 4°C with rotation and then with Ultralink immobilized protein A/G sepharose (Pierce Biotech) for 2 hours at room temperature. Immune complexes were dissociated by boiling the beads for 5 minutes in loading buffer, separated on SDS-PAGE, transferred to nitrocellulose membranes, and analyzed by Western blot (WB) with either anti-STAT5pTyr antibody (1:1000, 9351S; Cell Signaling), anti-GRα, or anti-GRβ antibodies.
Nuclear extract preparation and EMSA analyses for STAT-5
Nuclear extracts were prepared with the nuclear extraction kit (Thermo Scientific) and incubated (4 μg per reaction with the biotin-labeled STAT-5 probe of an EMSA kit; Panomics, Affymetrix) as described by the manufacturer. The specificity of the reaction was assessed by competition with unlabeled probe. After the reaction, the samples were resolved on 6% nondenaturing PAGE at 4°C in 90mM Tris-borate–2mM EDTA buffer at 120 V. Transfer was performed by electroblotting on presoaked Biodyne B nylon membrane with 0.5× Tris-borate-EDTA for 30 minutes at 300 mA. Oligonucleotides were then fixed on the membrane by UV cross-linking for 1 minute, and the membrane was incubated in blocking buffer and then with HRP-coupled secondary antibodies (15 minutes each). EMSA was revealed with Hyperfilm ECL.
JAK2V617F mutation and A3669G GR SNP determinations
DNA was isolated from blood granulocytes or from cultured cells by standard techniques. JAK2V617F mutant allele burden was measured by quantitative RT-PCR, as described previously,38 with the ABI Prism 7300 analyzer (Applied Biosystems). The JAK2V617F allele burden was calculated by comparison with standard curves obtained with DNA from a PV patient with 100% allele burden and an ND (positive and negative controls, respectively). PCR/single-strand conformation polymorphism analysis of the A3669G SNP in exon 9 of GR was performed as described previously.28 Amplified products were purified with the QIAquick kit (QIAGEN Sciences) and sequenced by the core facility of Mount Sinai School of Medicine.
Statistical Analysis
Statistical analysis was performed with GraphPad Prism Version 4.0 software for Windows (GraphPad Software). To compensate for differences in numbers of observations among experimental groups, statistical significance was calculated with the Mann-Whitney test (2-tailed) and was defined as P < .05. Expression levels of GATA1, GATA2, NF-E2, WT1, and β-globin were analyzed by nonparametric tests that included the independent Kruskal-Wallis test, Dunn multiple comparison post hoc test, and paired Wilcoxon signed rank test. JAK2V617F expression was compared by 1-way ANOVA and Bonferroni multiple comparisons post hoc test. The frequency of the A3669G SNP in the different populations was compared by the Fisher exact test.
Results
MNCs from PV patients do not respond to dexamethasone ex vivo
The number of hematopoietic cells generated by MNCs from NDs and PV patients in liquid culture with or without dexamethasone was compared (Figure 1). ND MNCs, in the absence of dexamethasone, generated both EBs (55%-57% CD235+ cells) and nonerythroid cells (37%-39% CD45+ cells; supplemental Figure 3; supplemental Table 2). Approximately 50% of the EBs were mature, as shown by morphology and FACS profiling for CD36/CD235a expression (Figure 1B-C; Table 2). The fact that increased CD235a expression was correlated with increased maturation was confirmed by the expression profile of CD36+ EBs prospectively isolated by sorting into classes by CD235a expression (Figure 2A). Expression of GATA1 and β-globin increased, whereas that of GATA2 decreased in EBs expressing progressively more CD235a, which confirms that CD235a expression correlates with maturation (Figure 2B). As expected,17 with dexamethasone, ND MNCs generated 2-5× greater cell numbers (fold increase over time 0: 1 vs 3-15, depending on the donor; Figure 1A; Migliaccio et al17,20 ), the majority (> 85%) of which were CD45− EBs (supplemental Figure 3; supplemental Table 2). Approximately 50% of the EBs expressed low levels of CD235a (Table 2). In addition, ND EBs generated in the presence of dexamethasone expressed levels of GATA1 (2−ΔCt: 10-1 vs 2 × 100) and β-globin (2−ΔCt: 102 vs 103) that were approximately 10-fold lower than those expressed by CD235ahigh EBs generated without dexamethasone (Figure 2B,D). These EBs were therefore primarily immature. On erythropoietin exposure for 48-96 hours, these cells matured, as revealed by increased CD235a expression and partial loss of CD36 expression (Figure 2C; supplemental Figure 4) and gene expression profiling (GATA1, β-globin, and GATA2 expression became similar to that of nondexamethasone-treated CD235ahigh EBs; Figure 2B,D).
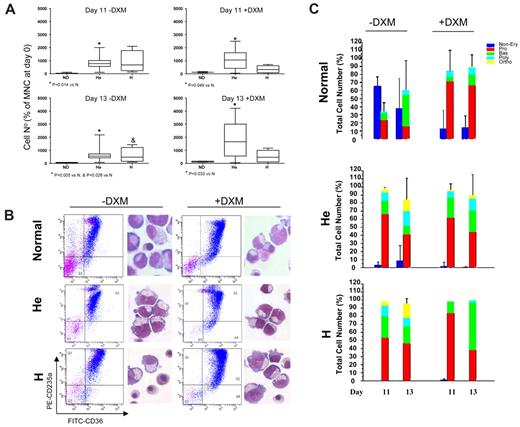
Dexamethasone increases the numbers of EBs generated in cultures of NDs but not of PV patients. Numbers (at days 11 and 13; A) and representative maturation profiles and morphology (at day 13; B) of EBs generated in liquid cultures stimulated with growth factors with or without dexamethasone (DXM) by MNCs from NDs8 or from PV patients homozygous (H; n = 3) or heterozygous (He; n = 13) for the JAK2V617F mutation. Magnification ×40. Forward-scatter/side-scatter analyses and propidium iodide staining were comparable (∼ 10%-15% propidium iodide–positive cells) in all cultures analyzed and are not shown. P values are provided when differences between ND and PV were statistically significant. Statistical analyses of the FACS profiles are presented in Table 2. Differential counts of cultured cells (by morphology) are presented in panel C. The different populations are color coded: blue for non-EBs; red for Pro-EBs; and green, blue, and yellow for basophilic (Bas), polychromatic (Poly), and orthochromatic (Ortho) EBs, respectively.
Dexamethasone increases the numbers of EBs generated in cultures of NDs but not of PV patients. Numbers (at days 11 and 13; A) and representative maturation profiles and morphology (at day 13; B) of EBs generated in liquid cultures stimulated with growth factors with or without dexamethasone (DXM) by MNCs from NDs8 or from PV patients homozygous (H; n = 3) or heterozygous (He; n = 13) for the JAK2V617F mutation. Magnification ×40. Forward-scatter/side-scatter analyses and propidium iodide staining were comparable (∼ 10%-15% propidium iodide–positive cells) in all cultures analyzed and are not shown. P values are provided when differences between ND and PV were statistically significant. Statistical analyses of the FACS profiles are presented in Table 2. Differential counts of cultured cells (by morphology) are presented in panel C. The different populations are color coded: blue for non-EBs; red for Pro-EBs; and green, blue, and yellow for basophilic (Bas), polychromatic (Poly), and orthochromatic (Ortho) EBs, respectively.
Progression of erythroid maturation, defined by levels of CD235a expression, in cultures of MNCs from NDs and PV patients
| . | CD235alow . | CD235amed . | CD235ahigh . | |||
|---|---|---|---|---|---|---|
| −DXM . | +DXM . | −DXM . | +DXM . | −DXM . | +DXM . | |
| Normal (5 donors) | ||||||
| Day 11 | 21.4 ± 3.8* | 50.0 ± 24.9 | 25.9 ± 6.4 | 12.4 ± 13.0 | 49.3 ± 4.8* | 20.7 ± 25.0 |
| Day 13 | 20.5 ± 1.6* | 49.0 ± 22.0 | 25.5 ± 9.2 | 12.1 ± 15.0 | 51.9 ± 10.4* | 18.5 ± 26.5 |
| Heterodimers (10 donors) | ||||||
| Day 11 | 6.8 ± 3.1† | 26.3 ± 7.2 | 29.7 ± 8.7† | 28.1 ± 7.6 | 63.0 ± 10.5† | 44.0 ± 13.6 |
| Day 13 | 13.5 ± 11.7† | 21.2 ± 5.6 | 37.1 ± 13.4† | 30.0 ± 11.4 | 48.0 ± 20.1† | 48.3 ± 15.5 |
| Homodimers (3 donors) | ||||||
| Day 11 | 8.5 ± 2.6† | 29.3 ± 17.1 | 27.2 ± 4.6 | 26.8 ± 4.5 | 63.4 ± 5.3† | 42.8 ± 21.3 |
| Day 13 | 19.2 ± 10.9† | 26.5 ± 15.6 | 32.7 ± 12.7 | 27.1 ± 9.7 | 44.0 ± 13.4 | 45.5 ± 25.4 |
| . | CD235alow . | CD235amed . | CD235ahigh . | |||
|---|---|---|---|---|---|---|
| −DXM . | +DXM . | −DXM . | +DXM . | −DXM . | +DXM . | |
| Normal (5 donors) | ||||||
| Day 11 | 21.4 ± 3.8* | 50.0 ± 24.9 | 25.9 ± 6.4 | 12.4 ± 13.0 | 49.3 ± 4.8* | 20.7 ± 25.0 |
| Day 13 | 20.5 ± 1.6* | 49.0 ± 22.0 | 25.5 ± 9.2 | 12.1 ± 15.0 | 51.9 ± 10.4* | 18.5 ± 26.5 |
| Heterodimers (10 donors) | ||||||
| Day 11 | 6.8 ± 3.1† | 26.3 ± 7.2 | 29.7 ± 8.7† | 28.1 ± 7.6 | 63.0 ± 10.5† | 44.0 ± 13.6 |
| Day 13 | 13.5 ± 11.7† | 21.2 ± 5.6 | 37.1 ± 13.4† | 30.0 ± 11.4 | 48.0 ± 20.1† | 48.3 ± 15.5 |
| Homodimers (3 donors) | ||||||
| Day 11 | 8.5 ± 2.6† | 29.3 ± 17.1 | 27.2 ± 4.6 | 26.8 ± 4.5 | 63.4 ± 5.3† | 42.8 ± 21.3 |
| Day 13 | 19.2 ± 10.9† | 26.5 ± 15.6 | 32.7 ± 12.7 | 27.1 ± 9.7 | 44.0 ± 13.4 | 45.5 ± 25.4 |
The percentages of EBs expressing high, medium, and low levels of CD235a were calculated on the basis of cells included in the CD235a+ gate. Values statistically different (P < .05) from parallel cultures without dexamethasone (*) or from those observed in the corresponding group from ND (†) are indicated. Representative FACS analyses are presented in Figure 1. The gates defining CD235alow, CD235amed, and CD235ahigh cells are the same as in the top panel of Figure 1A.
DXM indicates dexamethasone.
EBs generated ex vivo by MNCs of PV patients with or without dexamethasone express levels of GATA1, NF-E2, and WT1 similar to those expressed by EBs generated by MNCs from NDs with dexamethasone. Maturation profiles (by FACS analysis for CD36/CD235a expression; A,C) and gene expression profile (GATA1, GATA2, and β-globin; B,D) of EBs obtained from NDs in the presence of growth factors (GFs) without (A-B) or with (C-D) dexamethasone (DXM). Because of the great contamination from non-EBs (> 50%; Figure 1C and Table 1), EBs generated in the absence of DXM were purified by sorting into classes of progressively more mature populations on the basis of CD235a expression (CD235alow, CD235amedium, and CD235ahigh), and gene expression by individual populations was compared. Erythroblasts obtained in the presence of GFs plus DXM were analyzed before and after induction of maturation with erythropoietin (EPO) for 48 hours. Expression levels are expressed as 2−ΔCt and are presented as mean ± SD of at least 3 separate experiments. (E) Levels of GATA1, GATA2, NF-E2, WT1, and β-globin expressed by EBs generated by heterozygous (top panels) and homozygous (middle panels) PV patients and by NDs (bottom panels) with and without DXM. Because of the heterogeneity of their cell composition, samples from NDs cultured without DXM were not included in the analyses. Expression levels are presented as 2−ΔCt and as mean ± SD of different experiments. If differences were significant, P values are provided. Erythroblasts obtained from heterozygous PV patients with DXM expressed levels of GATA2 and β-globin significantly different from those expressed by EBs obtained from NDs with DXM and are indicated in red. The numbers of experiments included in each group are indicated by n.
EBs generated ex vivo by MNCs of PV patients with or without dexamethasone express levels of GATA1, NF-E2, and WT1 similar to those expressed by EBs generated by MNCs from NDs with dexamethasone. Maturation profiles (by FACS analysis for CD36/CD235a expression; A,C) and gene expression profile (GATA1, GATA2, and β-globin; B,D) of EBs obtained from NDs in the presence of growth factors (GFs) without (A-B) or with (C-D) dexamethasone (DXM). Because of the great contamination from non-EBs (> 50%; Figure 1C and Table 1), EBs generated in the absence of DXM were purified by sorting into classes of progressively more mature populations on the basis of CD235a expression (CD235alow, CD235amedium, and CD235ahigh), and gene expression by individual populations was compared. Erythroblasts obtained in the presence of GFs plus DXM were analyzed before and after induction of maturation with erythropoietin (EPO) for 48 hours. Expression levels are expressed as 2−ΔCt and are presented as mean ± SD of at least 3 separate experiments. (E) Levels of GATA1, GATA2, NF-E2, WT1, and β-globin expressed by EBs generated by heterozygous (top panels) and homozygous (middle panels) PV patients and by NDs (bottom panels) with and without DXM. Because of the heterogeneity of their cell composition, samples from NDs cultured without DXM were not included in the analyses. Expression levels are presented as 2−ΔCt and as mean ± SD of different experiments. If differences were significant, P values are provided. Erythroblasts obtained from heterozygous PV patients with DXM expressed levels of GATA2 and β-globin significantly different from those expressed by EBs obtained from NDs with DXM and are indicated in red. The numbers of experiments included in each group are indicated by n.
PV MNCs generated greater numbers (500 to 2000-fold increase over time 0) of EBs that contained rare nonerythroid CD45+ cells (supplemental Figure 3; supplemental Table 2) both with and without dexamethasone (Figure 1A). The majority of these EBs were immature as assessed by morphology (Figure 1B-C), but > 40%-60% of them expressed high levels of CD235a (Figure 1B; Table 2), an indication that in PV, EB-generated ex vivo CD235a expression may not correlate with maturation. As reported previously,39 the relative level of JAK2V617F expressed by these cultured EBs (18%-50%) was lower than that expressed by blood granulocytes (22%-80%; supplemental Figure 5).
Gene expression analyses confirmed the immature nature of PV EBs generated ex vivo both in the absence and in the presence of dexamethasone (Figure 2E). PV EBs also expressed low levels of GATA1 similar to those expressed by ND EBs generated with dexamethasone (2−ΔCt: 0.28 ± 0.46 in heterozygous and 1.44 ± 1.49 in homozygous PV vs 0.10 ± 0.06 in ND). The levels of β-globin expressed by EBs from heterozygous PV patients were significantly greater than those expressed by dexamethasone-stimulated ND EBs (2−ΔCt: 5.44 ± 5.70 vs 0.444 ± 0.39, P = .0078; Figure 2E), but they remained lower than those expressed by CD235ahigh ND EBs generated without dexamethasone (2−ΔCt: ∼ 103; Figure 2B). The delayed maturation of ex vivo expanded PV EBs was further demonstrated by the fact that cells generated both in the presence and in the absence of dexamethasone failed to mature (Figure 1B-C) and increased in numbers in the presence of erythropoietin alone for 96 hours (supplemental Figure 4). In addition, dexamethasone-stimulated and -unstimulated EBs from PV patients also expressed levels of NF-E2 (2−ΔCt: 0.73 ± 0.51 and 8.21 ± 9.76 vs 0.35 ± 0.17) and WT1 (2−ΔCt: 0.0030 ± 0.0021 and 0.0021 ± 0.0016 vs 0.0028 ± 0.0025) that were similarly higher than those expressed by ND EBs generated with dexamethasone (Figure 2E). These results suggest that increased NF-E2 and WT1 expression is associated with great EB expansion not only in PV patients but also in NDs stimulated with dexamethasone. By contrast, GATA2 expression was uniquely high in EBs from heterozygous PV patients (2−ΔCt: 0.055 ± 0.03 vs 0.025 ± 0.019, P = .0147), which may be linked to the extremely high proliferative potential of these EBs ex vivo (Figure 1A).
In conclusion, PV MNCs generated greater numbers of immature EBs in culture than ND MNCs either with or without dexamethasone.
Constitutive activation of STAT5 in EBs generated ex vivo from PV patients
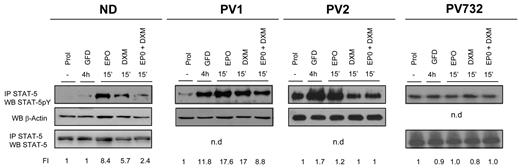
Dexamethasone exerts its biologic action by binding to a receptor, GRα, that is normally bound to the microfilaments of the cytoplasm.23,24 Binding of dexamethasone to this receptor induces the formation of homodimers that may also bind other signaling molecules, such as AP-1 and STAT-5, forming active complexes that translocate to the nucleus, where they regulate expression of gene subsets by binding to consensus sequences specific for each complex.23,32 When simultaneously stimulated with erythropoietin and dexamethasone, ND EBs fail to phosphorylate STAT-5 (Figure 3),20 which suggests that a block of STAT-5 phosphorylation and formation of STAT-5/GR complexes allows EBs to continue to proliferate in the presence of dexamethasone. By contrast, EBs expanded ex vivo from PV patients express higher levels of STAT-5 phosphorylation than NDs in response to growth factor deprivation (Figure 3). Further stimulation of the cells with erythropoietin modestly increased (fold increase [FI] = 17.6 vs 11.8, PV1) or had no effect (FI = 1.0-1.2 vs 0.9-1.7, PV732 and PV2) on STAT-5 phosphorylation. The addition of dexamethasone, alone or in combination with erythropoietin, either had no effect (FI = 0.8-1 vs 0.9-1.7, PV732 and PV2) or had little effect (FI = 17-8.8 vs 11.8, PV1) on STAT-5 phosphorylation.
Constitutive phosphorylation and nuclear translocation of STAT-5 in EBs generated ex vivo from PV patients. Levels of STAT-5 phosphorylation in cell extracts of EBs obtained in HEMA culture (Prol) from 1 ND and 3 PV patients and in cultures deprived of growth factor for 4 hours (GFD) and then treated with erythropoietin (EPO; 3 U/mL) and dexamethasone (DXM; 10−6M), alone or in combination, as indicated. STAT-5 phosphorylation was analyzed by WB of cell extracts immunoprecipitated (IP) with anti–STAT-5 antibody. The cell lysates were then analyzed by WB for β-actin and/or STAT-5 as quantitative control. The intensity of the signal was quantified by densitometry and expressed as a ratio (FI indicates fold increase) with the signal from cells in Prol. Similar data on STAT-5 phosphorylation of EBs from 10 additional NDs were reported previously.20
Constitutive phosphorylation and nuclear translocation of STAT-5 in EBs generated ex vivo from PV patients. Levels of STAT-5 phosphorylation in cell extracts of EBs obtained in HEMA culture (Prol) from 1 ND and 3 PV patients and in cultures deprived of growth factor for 4 hours (GFD) and then treated with erythropoietin (EPO; 3 U/mL) and dexamethasone (DXM; 10−6M), alone or in combination, as indicated. STAT-5 phosphorylation was analyzed by WB of cell extracts immunoprecipitated (IP) with anti–STAT-5 antibody. The cell lysates were then analyzed by WB for β-actin and/or STAT-5 as quantitative control. The intensity of the signal was quantified by densitometry and expressed as a ratio (FI indicates fold increase) with the signal from cells in Prol. Similar data on STAT-5 phosphorylation of EBs from 10 additional NDs were reported previously.20
Erythroblasts generated ex vivo from PV patients express the dominant negative GRβ isoform
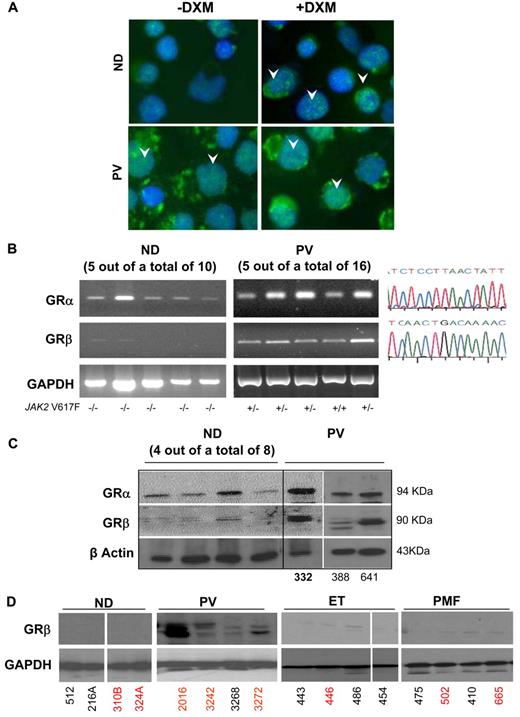
The inducible versus constitutive phosphorylation of STAT-5 in EBs generated ex vivo from NDs and PV patients led us to compare the nuclear localization of GR in the 2 cell types (Figure 4A). In ND EBs, treatment with dexamethasone induced nuclear localization with a punctate appearance of GR similar to that expressed by cells that ectopically expressed green fluorescent protein–labeled GRα.40 By contrast, nuclear localization of GR was diffuse in EBs from PV patients and increased only slightly when the cells were stimulated with dexamethasone, resembling that of cell lines that express a green fluorescent protein–labeled GRβ isoform.40 These similarities led us to determine the presence of GRα and GRβ mRNA in ex vivo expanded EBs from NDs10 and PV patients16 (Figure 4B). By RT-PCR, EBs from all 16 PV patients tested expressed both GRα and GRβ mRNA, whereas those from 10 NDs expressed GRα mRNA only. In addition, the levels of GRβ expressed by EBs from 4 PV patients and 4 NDs were compared by quantitative RT-PCR. Erythroblasts from PV patients expressed levels of GRβ 3-fold greater than those expressed by EBs expanded from NDs (2−ΔCt = 117.3 ± 11.5 vs 34.8 ± 8.4, respectively, P < .001).
Constitutive nuclear localization of GR in EBs generated ex vivo from PV MNCs is associated with expression of the dominant negative GRβ isoform. (A) Immunostaining for GR of EBs obtained from 1 representative ND and PV patient with and without dexamethasone (DXM). Arrowheads indicate representative nuclear localization of GR. In EBs from ND, nuclear staining for GR was observed after DXM stimulation and had the punctuated appearance expected for GRα. By contrast, in EBs from PV, GR staining of the nucleus had a diffuse DXM-independent pattern characteristic of GRβ.40 Magnification ×40. (B) RT-PCR analyses for expression of GRα and GRβ of EBs expanded ex vivo from 5 representative ND and PV patients. The homozygous (+/+) or heterozygous (+/−) allele status of the JAK2V617F mutation of the PV patients is indicated on the bottom. Similar results were obtained with additional NDs (n = 5) and PV patients (n = 11; not shown). In all cases, the identity of the band was confirmed by sequencing (on the right). GAPDH was amplified as control. (C) WB analyses with GRα- and GRβ-specific antibodies of EBs generated from 8 additional NDs (not analyzed at the mRNA level) and from 3 PV patients (the first patient was analyzed at the mRNA level in the second lane in panel B). β-actin was analyzed as a loading control. The proteins recognized by the GRα- and GRβ-specific antibodies migrated with an apparent molecular weight of 94 and 90 kDa, respectively. (D) WB analyses for GRβ of MNCs from NDs and from PV, ET, and PMF patients (4 each). Data are representative of those observed in a total of 10 subjects per group. In panels C and D, ND and MPN patients are identified by the same unique alphanumeric codes used in Figure 6. SNP-negative and SNP-positive subjects are indicated in black and red fonts, respectively. The SNP status of PV332 (in bold) is not known.
Constitutive nuclear localization of GR in EBs generated ex vivo from PV MNCs is associated with expression of the dominant negative GRβ isoform. (A) Immunostaining for GR of EBs obtained from 1 representative ND and PV patient with and without dexamethasone (DXM). Arrowheads indicate representative nuclear localization of GR. In EBs from ND, nuclear staining for GR was observed after DXM stimulation and had the punctuated appearance expected for GRα. By contrast, in EBs from PV, GR staining of the nucleus had a diffuse DXM-independent pattern characteristic of GRβ.40 Magnification ×40. (B) RT-PCR analyses for expression of GRα and GRβ of EBs expanded ex vivo from 5 representative ND and PV patients. The homozygous (+/+) or heterozygous (+/−) allele status of the JAK2V617F mutation of the PV patients is indicated on the bottom. Similar results were obtained with additional NDs (n = 5) and PV patients (n = 11; not shown). In all cases, the identity of the band was confirmed by sequencing (on the right). GAPDH was amplified as control. (C) WB analyses with GRα- and GRβ-specific antibodies of EBs generated from 8 additional NDs (not analyzed at the mRNA level) and from 3 PV patients (the first patient was analyzed at the mRNA level in the second lane in panel B). β-actin was analyzed as a loading control. The proteins recognized by the GRα- and GRβ-specific antibodies migrated with an apparent molecular weight of 94 and 90 kDa, respectively. (D) WB analyses for GRβ of MNCs from NDs and from PV, ET, and PMF patients (4 each). Data are representative of those observed in a total of 10 subjects per group. In panels C and D, ND and MPN patients are identified by the same unique alphanumeric codes used in Figure 6. SNP-negative and SNP-positive subjects are indicated in black and red fonts, respectively. The SNP status of PV332 (in bold) is not known.
To clarify whether GRβ mRNA was translated into protein, lysates of EBs from 8 additional NDs and those from 3 PV patients (1 of whom was previously analyzed at the mRNA level) were analyzed by WB with GRα- and GRβ-specific antibodies (Figure 4C). Erythroblasts from only 1 of the 8 NDs analyzed expressed low levels of GRβ, whereas EBs from all PV patients analyzed expressed robust levels of both GRα and GRβ. WB analyses for GRβ of MNCs from the blood of NDs and of PV, ET, and PMF patients showed that GRβ was expressed only by MNCs (likely T cells and monocytes) generated in vivo by PMF patients (Figure 4D).
Reduced formation of active STAT-5/GRα complexes in the nucleus of ND EBs stimulated by dexamethasone and erythropoietin in combination and in the nucleus of PV EBs
We have previously shown that stimulation with dexamethasone and erythropoietin in combination prevents formation of STAT-5/GRα complexes in ND EBs.20 In the present study, we performed additional immunoprecipitation to compare the formation of STAT-5/GRα and GRα/GRβ complexes in EBs from NDs and PV patients that were deprived of growth factor for 4 hours and then stimulated with either erythropoietin or dexamethasone alone and in combination (Figure 5A). WB of immunoprecipitation with STAT-5 antibodies of ND EBs stimulated with erythropoietin or dexamethasone alone, but not in combination, contained proteins detected by WB with anti–STAT-5pY and anti-GRα antibodies (Figure 5A). As expected, WBs of immunoprecipitation with anti-GRα antibodies of these cells contained STAT-5 but not GRβ. By contrast, WB of immunoprecipitation of PV EBs with anti–STAT-5 was positive for STAT-5pY regardless of the stimulation but did not contain proteins recognized by the anti-GRα antibody. Conversely, WB of immunoprecipitation of these cells with anti-GRα reacted with anti-GRβ but not with anti–STAT-5 (Figure 5A). These results suggest that in ND EBs stimulated with either erythropoietin or dexamethasone alone, STAT-5 is phosphorylated and forms complexes with GRα but that stimulation of these cells with erythropoietin and dexamethasone together fails to induce STAT-5 phosphorylation and formation of STAT-5/GRα complexes. By contrast, in PV EBs, GRα forms a complex with GRβ that does not associate with STAT-5 in spite of its constitutive phosphorylation status.
Both ND EBs exposed to excess glucocorticoids (by costimulation with erythropoietin and glucocorticoids) and PV EBs expressing the dominant negative GRβ isoform exhibit impaired functional GR/STAT-5 nuclear interaction. Erythroblasts were generated at day 10 of HEMA (Prol) and analyzed either as such or after growth factor deprivation for 4 hours (GFD) followed by treatment with erythropoietin (EPO; 3 U/mL) and dexamethasone (DXM; 10−6M), alone or in combination. (A) Erythroblasts obtained from 1 ND and 1 PV patient were immunoprecipitated (IP) with either STAT-5– or GRα-specific antibodies and the immunoprecipitations were analyzed by WB with either anti–STAT-5 or anti-GRα, as loading control, or with anti–STAT-5pY and anti-GRβ antibody. Similar results were obtained in 3 additional experiments, each with a separate donor. (B) EMSA with STAT-5–specific labeled probes of nuclear extract of EBs from 1 ND and 1 PV patient with (+) and without (−) DXM for 24 hours. Lane 1 is labeled probe only; lane 2, labeled probe with ND EBs without DXM; lane 3, labeled probe with ND EBs treated with DXM; lane 4, labeled probe with PV EBs treated with DXM; and lane 5, labeled probes with PV EBs that competed with excessive unlabeled probe. The position of the STAT-5–bound and –free probe is indicated by arrows. Similar results were obtained in 3 additional experiments. (C) WB analyses for GILZ and GAPDH (as loading control) expression of EBs generated at day 10 of HEMA (Prol) from ND or PV as indicated and analyzed either as such (Prol) or after growth factor deprivation for 4 hours (GFD) followed by treatment with erythropoietin (EPO; 3 U/mL) and DXM (10−6M), alone or in combination. Similar results were obtained in 3 additional experiments, each with a separate donor. Murine 293T cells overexpressing GILZ were analyzed as positive control. (D) Proposed model for development of erythrocytosis because of inhibition of GR/STAT-5 interactions by exposure to excess glucocorticoids and expression of the dominant negative GRβ isoform. In patients chronically stimulated with glucocorticoids who express GRα only, costimulation with erythropoietin and dexamethasone impairs regulation of GR/STAT-5–responsive genes by inhibiting STAT-5 phosphorylation and formation of GR/STAT-5 tetrameric complexes (A) and reducing the DNA-binding activity of STAT-5 (B). In PV, STAT-5 is constitutively phosphorylated by JAK2V617F (A) but cannot form complexes with GRα because this protein is largely retained in the nucleus as GRβ heterodimer (B). Therefore, in PV EBs, the DNA-binding activity is reduced (B), and expression of GR/STAT-5–responsive genes, such as GILZ, is also defective (C).
Both ND EBs exposed to excess glucocorticoids (by costimulation with erythropoietin and glucocorticoids) and PV EBs expressing the dominant negative GRβ isoform exhibit impaired functional GR/STAT-5 nuclear interaction. Erythroblasts were generated at day 10 of HEMA (Prol) and analyzed either as such or after growth factor deprivation for 4 hours (GFD) followed by treatment with erythropoietin (EPO; 3 U/mL) and dexamethasone (DXM; 10−6M), alone or in combination. (A) Erythroblasts obtained from 1 ND and 1 PV patient were immunoprecipitated (IP) with either STAT-5– or GRα-specific antibodies and the immunoprecipitations were analyzed by WB with either anti–STAT-5 or anti-GRα, as loading control, or with anti–STAT-5pY and anti-GRβ antibody. Similar results were obtained in 3 additional experiments, each with a separate donor. (B) EMSA with STAT-5–specific labeled probes of nuclear extract of EBs from 1 ND and 1 PV patient with (+) and without (−) DXM for 24 hours. Lane 1 is labeled probe only; lane 2, labeled probe with ND EBs without DXM; lane 3, labeled probe with ND EBs treated with DXM; lane 4, labeled probe with PV EBs treated with DXM; and lane 5, labeled probes with PV EBs that competed with excessive unlabeled probe. The position of the STAT-5–bound and –free probe is indicated by arrows. Similar results were obtained in 3 additional experiments. (C) WB analyses for GILZ and GAPDH (as loading control) expression of EBs generated at day 10 of HEMA (Prol) from ND or PV as indicated and analyzed either as such (Prol) or after growth factor deprivation for 4 hours (GFD) followed by treatment with erythropoietin (EPO; 3 U/mL) and DXM (10−6M), alone or in combination. Similar results were obtained in 3 additional experiments, each with a separate donor. Murine 293T cells overexpressing GILZ were analyzed as positive control. (D) Proposed model for development of erythrocytosis because of inhibition of GR/STAT-5 interactions by exposure to excess glucocorticoids and expression of the dominant negative GRβ isoform. In patients chronically stimulated with glucocorticoids who express GRα only, costimulation with erythropoietin and dexamethasone impairs regulation of GR/STAT-5–responsive genes by inhibiting STAT-5 phosphorylation and formation of GR/STAT-5 tetrameric complexes (A) and reducing the DNA-binding activity of STAT-5 (B). In PV, STAT-5 is constitutively phosphorylated by JAK2V617F (A) but cannot form complexes with GRα because this protein is largely retained in the nucleus as GRβ heterodimer (B). Therefore, in PV EBs, the DNA-binding activity is reduced (B), and expression of GR/STAT-5–responsive genes, such as GILZ, is also defective (C).
The STAT-5 DNA binding activity in nuclear extract from ND EBs (stimulated with and without dexamethasone) and PV EBs is compared in Figure 5B. Nuclear extracts from ND EBs and from PV EBs stimulated with dexamethasone were 10-fold less efficient in binding STAT-5–specific oligonucleotides than nuclear extracts from untreated ND EBs. These results confirm that the nuclei of both dexamethasone-stimulated ND and PV EBs have reduced STAT-5 DNA-binding activity.
As further proof of impaired GRα transcriptional activity in PV EBs, we measured the levels of expression of the glucocorticoid-responsive GILZ gene by WB (Figure 5C). GILZ was readily detectable by WB in ND EBs but not in PV EBs. The GILZ content of ND EBs was slightly increased by stimulations that increased STAT-5 phosphorylation (the same cells were analyzed in Figure 5A,C) but was not affected by short-term (15 minutes) stimulation with combined erythropoietin and dexamethasone, which suppressed STAT-5 phosphorylation. We believe that this failure is related to the fact that the half-life of GILZ is longer than 15 minutes. These data provide further support for the hypothesis that GRα is inactive in PV EBs.
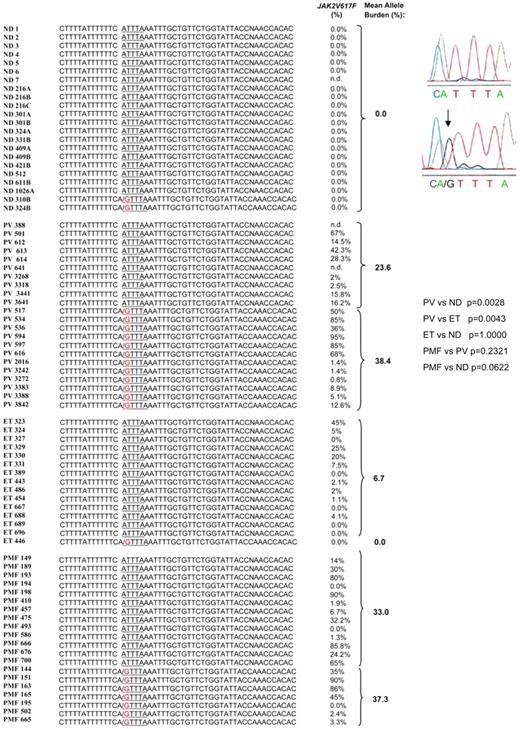
Frequency of A3669G in exon 9β of GR in ND and patients with MPN
The A3669G SNP in exon 9 of GR was detected in the heterozygous state in 55% of 22 PV patients and 35% of 20 PMF patients tested but in only 1 (6%) of 15 ET patients and 2 (9%) of 22 NDs tested (Figure 6). The frequency of the SNP in the nondiseased population (9%) was consistent with that (1%) reported by other studies,28 whereas the frequency observed in PV patients was slightly greater than that previously reported for autoimmune disorders (rheumatoid arthritis [36%] and systemic lupus erythematosus [42%]) and for subjects predisposed to central adiposity (27.6%).25,28
Presence of the A3669G polymorphism in NDs and in patients with PV, ET, and PMF. Individual subjects are indicated by unique alphanumeric codes. The presence of the GTTTA SNP was determined by PCR genotyping and confirmed by sequencing. Representative sequences are presented on the right. P values for the frequencies of the polymorphism in different groups were calculated with Fisher exact test and are presented on the right. The JAK2V617F status of the patients is reported for comparison. There was no difference in the mean JAK2V617F allele burden between patients with and without the polymorphism (P = .85 and .66 for PV and PMF patients, respectively, by Wilcoxon rank sum test).
Presence of the A3669G polymorphism in NDs and in patients with PV, ET, and PMF. Individual subjects are indicated by unique alphanumeric codes. The presence of the GTTTA SNP was determined by PCR genotyping and confirmed by sequencing. Representative sequences are presented on the right. P values for the frequencies of the polymorphism in different groups were calculated with Fisher exact test and are presented on the right. The JAK2V617F status of the patients is reported for comparison. There was no difference in the mean JAK2V617F allele burden between patients with and without the polymorphism (P = .85 and .66 for PV and PMF patients, respectively, by Wilcoxon rank sum test).
In conclusion, the increased prevalence of A3669G in PV patients suggests that improved mRNA stability may favor expression of GRβ mRNA (and protein) in EBs derived from these patients.
Discussion
We have previously shown that simultaneous stimulation of ND EBs with erythropoietin and dexamethasone leads to inhibition of STAT-5 phosphorylation and is correlated with the level of EB expansion obtained ex vivo from different NDs under HEMA conditions.17,41 Here, we provide data indicating that the expression profile of ND EBs expanded in the presence of dexamethasone is similar to that of PV EBs, with low levels of GATA1 (the transcription factor required for EB maturation)42 and β-globin expression and high levels of GATA2, NF-E2, and WT1 expression. High levels of GATA2 expression have been associated previously with EB proliferation.43 NF-E2 has also been reported to be overexpressed in neutrophils and other hematopoietic cells from PV patients,44 and its ectopic expression in normal CD34+ cells favors EB proliferation ex vivo.45 WT1, a gene overexpressed in leukemia, was found by differential microarray analyses to be the gene most up-regulated in CD34+ cells from PV patients.46 The observation that EBs generated ex vivo from NDs in the presence of dexamethasone express levels of NF-E2 and WT1 similar to those expressed by PV EBs suggests that overexpression of these 2 genes is not unique to PV but is also associated with massive expansion of ND EBs ex vivo in the presence of dexamethasone.
The observations that (1) ND EBs failed to phosphorylate STAT-5 when simultaneously challenged with dexamethasone and erythropoietin, (2) EBs from all 19 PV patients analyzed expressed the dominant negative GRβ isoform, (3) expression of GRβ impaired formation of STAT-5/GRα complexes and expression of the GRα-specific GILZ gene in PV EBs, and (4) EBs expanded ex vivo from one patient with idiopathic erythrocytosis expressed only GRβ mRNA (not shown) suggest a unifying mechanism for increased EB expansion in PV and other forms of erythrocytosis based on inhibition of STAT-5/GRα complex formation and consequent deregulation of NF-E2 and WT1 (and GATA2) expression (Figure 5D). According to this model, inhibition of STAT-5/GRα interactions by chronic stimulation with glucocorticoids favors expansion of ND EBs and may contribute to development of JAK2-negative forms of erythrocytosis such as Cushing syndrome, whereas decreased STAT-5/GRα interactions caused by expression of GRβ contribute to erythrocytosis in PV. This model predicts that erythrocytosis mediated by glucocorticoids may be eliminated by blocking GR activation and restoring STAT-5 phosphorylation, whereas erythrocytosis in PV may persist as long as GRβ is expressed. This hypothesis is supported by the observation that ND EBs matured when exposed to erythropoietin only, whereas PV EBs did not mature when exposed to erythropoietin alone (supplemental Figure 5).
GRβ mRNA and protein were detectable in the EBs from all 19 patients with PV analyzed. By contrast, GRβ mRNA was not detectable by RT-PCR in EBs from 10 NDs, and GRβ protein was expressed at low levels in EBs from only 1 of 9 NDs analyzed (Figure 4; Table 1). By quantitative RT-PCR, EBs from 4 PV patients were found to express levels of GRβ mRNA 3-fold greater than those expressed by EBs from 4 NDs. These results suggest the existence of mechanisms that activate alternative splicing and stabilize GRβ mRNA in EBs from PV patients. In airway epithelial cell lines, dexamethasone treatment activates alternative splicing, which leads to GRβ expression and is associated with glucocorticoid resistance.47 However, GRβ mRNA was not detectable by sensitive RT-PCR in EBs from NDs stimulated with dexamethasone, SCF, or erythropoietin, which indicates that in these cells, the alternative splicing is not inducible by HEMA components. We suggest that the presence of JAK2V617F may be responsible for activation of alternative GR splicing in MPN EBs. Once the alternative splicing is activated, the presence of the A3669G polymorphism, by stabilizing GRβ mRNA, may favor its translation, generating amounts of protein adequate for GRα neutralization. In agreement with this hypothesis, the A3669G polymorphism was detected in the heterozygous state in 12 of 22 PV patients investigated; however, 2 of the A3669G-negative PV patients expressed robust levels of GRβ protein. Exon 9 of the GR gene contains 3 additional putative polyadenylation sites, and > 250 GR mutations have been described in the human population that encode GRα isoforms with different dexamethasone or DNA-binding activities.23,24 Presently, in A3669G− PV, we do not know whether GRβ mRNA is stabilized by polymorphisms in one of these additional polyadenylation sites or whether the levels of GRβ protein translated from this less stable mRNA are sufficient to neutralize less active GRα isoforms. Our observations add to the ongoing discussion on the mechanisms by which a single mutation, JAK2V617F, may cause disease phenotypes as different as PV, ET, or PMF.11,48 The consensus is that although the JAK2V617F mutation may increase proliferation of hematopoietic cells in general, the lineage affected (erythroid [PV] or megakaryocytic with increased [ET] or decreased [PMF] maturation) is determined by host genetic modifiers still to be identified.11,48 The observation that the A3669G polymorphism is detectable at low frequency in patients with ET (Figure 6; Table 1) suggests that polymorphisms such as A3669G that lead to GRβ expression in EBs may represent genetic modifiers that determine development of PV rather than ET. Further studies on the regulation of GR expression in EBs with larger numbers of NDs and MPN patients, as well as sequencing of the entire GR locus, are necessary to fully characterize the relationship between the GR polymorphism and development of PV and to translate the present study into a clinically relevant bioassay to predict GR activity in PV patients.
JAK2V617F constitutively activates both the PI3K/AKT/FOXO and STAT-5 pathways.7,8 The observation that STAT-5 is likely to be at least partially inactivated by GRβ in these patients suggests that erythrocytosis may result from lack of balance rather than absolute increases in activation levels of the 2 pathways. As such, targeted therapies for PV should not attempt to decrease STAT-5 activity49 but should instead increase STAT-5 activity and/or decrease PI3K/AKT/FOXO activity, ie, restore the balance between the 2 pathways. Several investigators have reported that JAK2 inhibitors effectively treat polycythemia in JAK2V617F transgenic mice49 ; however, the beneficial effects of these inhibitors in PV patients have been modest50 and thus far have suggested that combinatorial therapies may be required in these diseases.51,52 We suggest that the poor correspondence between results obtained with these inhibitors in transgenic mice and in PV patients occurs because mice either do not express GRβ53 or express a slightly different GRβ isoform generated through an alternative splicing that differs from that which is active in humans.54 Double human GRβ/JAK2V617F transgenic mice may represent better models for preclinical drug evaluation. Hypothesis-driven research has played a pivotal role in the identification of JAK2 mutations in MPN,11 but recent progress in the field, such as the identification of a JAK2 SNP that predisposes to development of JAK2V617F in MPN, has been accomplished through powerful, but expensive, genome-wide association methodologies.55 The results of the present study highlight the important role that hypothesis-driven experiments may still play in the identification of the combinatorial therapies much needed for these diseases.
In conclusion, the data presented in the present study indicate that expression of GRβ and the A3669G polymorphism is frequently associated with PV and suggest that GR may represent a diagnostic tool to predict development of erythrocytosis, as well as a potential therapeutic target for PV.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Drs Maria Elena Fabucci, Emilia Stellacci, and Angela Di Baldassarre are acknowledged for technical support. Amy Rodriquez, RN, is thanked for obtaining written informed consent and for collecting the blood from the MPN patients.
This work was supported by a grant from the National Cancer Institute (P01-CA108671), a grant from the National Center for Research Resources (U54-RR026134), the NYSTAR foundation (C-06066), and institutional funds from the Istituto Superiore Sanità, Rome, Italy. Human recombinant SCF was provided by Amgen (Thousand Oaks, CA; MTA no. 19982634-005). E.M. was supported by a Rotary Foundation Ambassadorial Scholarship.
National Institutes of Health
Authorship
Contribution: L.V., E.M., E.A., and B.G. performed experiments and analyzed data; A.M.V., D.R., and R.H. provided patients and revised the manuscript; W.Z. performed JAK2 allele burden determinations; J.G. analyzed the data; and A.B., G.M., C.W., and A.R.M. designed research, analyzed data, and wrote the manuscript. All authors have read the manuscript, concur with its content, and state that its content has not been submitted elsewhere.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anna Rita Migliaccio, Tisch Cancer Center, Mount Sinai School of Medicine, One Gustave L. Levy Pl, Box 1079, New York, NY 10029; e-mail: annarita.migliaccio@mssm.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal