Abstract
Intrachromosomal amplification of chromosome 21 (iAMP21) defines a distinct subgroup of childhood B-cell precursor acute lymphoblastic leukemia (BCP-ALL) that has a dismal outcome when treated with standard therapy. For improved diagnosis and risk stratification, the initiating genetic events need to be elucidated. To investigate the genetic basis of BCP-ALL, genomes of 94 iAMP21 patients were interrogated by arrays, FISH, and multiplex ligation-dependent probe amplification. Most copy number alterations targeted chromosome 21, reinforcing the complexity of this chromosome. The common region of amplification on chromosome 21 was refined to a 5.1-mb region that included RUNX1, miR-802, and genes mapping to the Down syndrome critical region. Recurrent abnormalities affecting genes in key pathways were identified: IKZF1 (22%), CDKN2A/B (17%), PAX5 (8%), ETV6 (19%), and RB1 (37%). Investigation of clonal architecture provided evidence that these abnormalities, and P2RY8-CRLF2, were secondary to chromosome 21 rearrangements. Patient outcome was uniformly poor with standard therapy irrespective of the presence or absence of these changes. This study has provided evidence that chromosome 21 instability is the only anomaly among those so far investigated that is common to all iAMP21 patients, and therefore the initiating event is likely to be found among the complex structural rearrangements of this abnormal chromosome.
Introduction
Chromosomal changes are important in the diagnosis and prediction of outcome in acute lymphoblastic leukemia (ALL). Several clinically relevant abnormalities are used in risk stratification for treatment within the context of clinical trials.1 One such abnormality is intrachromosomal amplification of chromosome 21 (iAMP2), which occurs at an incidence of ∼ 2% in older children with B-cell precursor ALL (BCP-ALL).2-4 iAMP21 was originally identified as a consequence of routine screening for the presence of the ETV6-RUNX1 (TEL-AML1) fusion by FISH5 and is defined by multiple copies of the RUNX1 gene. In interphase nuclei, patients exhibit 3 or more additional RUNX1 FISH signals, which are duplicated along the arms of an abnormal chromosome 21.6 iAMP21 has been shown to be linked to a dismal outcome when patients are treated with standard therapy, because it is associated with an increased risk of both early and late relapses.7,8 Currently, FISH with probes directed to the RUNX1 gene provides the only reliable detection method for iAMP21, and therefore improved diagnostic approaches are required for accurate risk stratification of these patients.
Using cytogenetics, FISH, and bacterial artificial chromosome array-based comparative genomic hybridization (aCGH), we previously revealed the complex and heterogeneous nature of the chromosome 21 structure in patients with iAMP21.2,6,9 A 6.6-mb common region of amplification (CRA) on chromosome 21 containing RUNX1 and a 3.3-mb common region of deletion (CRD) at the telomere were identified in 100% and 77% of patients, respectively.6,9 We also showed that microarray-based global gene-expression profiling of 8 iAMP21 patients did not detect differential expression of any biologically relevant genes on chromosome 21, or specifically within the CRA or CRD, compared with other ALL subtypes.9 Although these findings assisted in the genomic characterization of iAMP21, the relevant genetic mechanisms leading to the development of ALL in these patients remained unknown.
We have recently reported a genomic alteration involving deletion within pseudoautosomal region 1 of the sex chromosomes10 that leads to a P2RY8-CRLF2 fusion, resulting in overexpression of the type 1 cytokine receptor CRLF2.10-12 This abnormality has been found at a high incidence of 38% in iAMP21 patients.10 High-resolution array-based whole-genome studies have identified recurrent deletions in childhood BCP-ALL, which disrupt several genes with important roles in B-cell differentiation and cell-cycle regulation. These deletions contribute directly to the pathogenesis of BCP-ALL.13-15 Particularly interesting was the observation that focal deletions involving the IKZF1 (Ikaros) gene were associated with a poor outcome in high-risk childhood BCP-ALL.16 However, the incidence and involvement of IKZF1, as well as the other recurrent abnormalities, have not been fully investigated within individual ALL subtypes, including iAMP21.
In the present study, we have expanded our previous investigations to include a more detailed analysis of the iAMP21 genome in a cohort of 94 patients, and demonstrate that chromosomal instability of chromosome 21 is the likely primary genetic event.
Methods
Patients
All patients had a diagnosis of BCP-ALL confirmed by morphology and immunophenotyping. Informed consent was obtained in accordance with the Declaration of Helsinki, and approval was granted by the institutional review boards of all participating institutions. Diagnostic cytogenetics were performed in regional cytogenetics laboratories and data were collected centrally by the Leukemia Research Cytogenetics Group, as described previously.17 A total of 94 patients were investigated, 78 at diagnosis and 15 at both diagnosis and relapse (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
FISH and MLPA
Patients with iAMP21 were identified using the dual-color probe kit LSI TEL/AML1 ES (Abbott Diagnostics), which is designed to detect the translocation t(12;21)(p13;q22) or specific probes targeting the RUNX1 gene, as described previously.2,6 A cutoff value of 3 or more additional RUNX1 signals was used to define iAMP21. In those cases with a normal karyotype or failed cytogenetic result with no metaphases available to confirm the presence of the abnormal chromosome 21, FISH using a probe specific for the sub-telomeric region of chromosome 21 (TelVysion21q; Abbott Diagnostic) was used to confirm that the extra RUNX1 signals had not arisen from the gain of intact copies of chromosome 21, as might be seen in high-hyperdiploid karyotypes. FISH was also used to exclude the presence of the established chromosomal abnormalities BCR-ABL1, ETV6-RUNX1, and MLL rearrangements.5 The copy number of chromosome X was determined using specific centromere probes (Cytocell). Deletions of CDKN2A, IKZF1, PAX5, and the pseudoautosomal region 1 resulting in P2RY8-CRLF2 were determined by FISH. All FISH probes were grown, hybridized, and analyzed as described previously.18 Deletions within these genes and others were also detected using the SALSA multiplex ligation-dependent probe amplification (MLPA) kit P335-A1 (MRC Holland), as described previously.19 Deletions of exons 1-4 of ETV6 were also detected using the LSI TEL/AML1 ES probe. Evidence of clonal heterogeneity was investigated using FISH probes targeting RUNX1 to identify those cells with the abnormal chromosome 21 characteristic of iAMP21, together with IKZF1, PAX5, or P2RY8-CRLF2. The FISH probes used in this study are listed in supplemental Table 2.
aCGH
DNA was extracted from diagnostic bone marrow samples of 17 of the 94 diagnostic iAMP21 patients and from 1 relapse sample using the DNeasy blood and tissue kit (QIAGEN). aCGH was performed with a Human Genome CGH Microarray kit 185A (Agilent Technologies) according to the manufacturer's instructions. Data were processed and analyzed as described previously,10 and further analyses were performed using custom software: SNPView2 v2.6 (Ilixa) and Partek Genomics Suite v6.5 (Partek). Data are available for download from Gene Expression Omnibus (GSE26192).
RUNX1 and JAK mutational analysis
Primers designed to cover the RUNX1 coding exons (exons 2-7) and the JAK gene mutational hot spots JAK1 (exons 13-14), JAK2 (exons 14, 18, 19), and JAK3 (exon 18) were used to amplify genomic DNA sequences from 14 diagnostic samples and 1 relapse sample (supplemental Table 2). The amplicons were screened for mutations by denaturing HPLC using the WAVE system (Transgenomic). Amplicons with potential mutations were validated by direct Sanger sequencing using BigDye Terminator sequencing chemistry (Applied Biosystems).
miR-802 expression analysis
Total RNA was isolated using the TRIzol method (Invitrogen) from patients with: iAMP21 (n = 2), gain of chromosome 21 (n = 6), ETV6-RUNX1 fusion (n = 5), unclassified karyotypes with no known established abnormalities (n = 6), or a normal karyotype (n = 1). miRNA expression levels were assessed by quantitative real-time PCR using Taqman miRNA assays (Applied Biosystems) and the 7900HT Real-Time PCR instrument (Applied Biosystems). Each sample was analyzed in triplicate in 3 independent replicates. The comparative Ct (ΔΔCt) method was used to assess the expression level of mir-802 relative to 2 endogenous controls, RNU6B and RNU43, and each test sample was normalized to the mirVana miRNA Reference Panel Version 9.1 software (Applied Biosystems).
Statistical analysis
Event-free survival was measured as the time from diagnosis to the first adverse event (death or relapse). An event was described as early if it occurred while the patient was on treatment (2 years for girls and 3 years for boys) or 6 months after the end of treatment. A late event refers to any time after this 6-month period. Event-free survival was estimated using the Kaplan-Meier method, as described previously.20 Hazard ratios comparing outcome between subgroups were estimated using univariate Cox models. All tests were conducted at the 1% significance level. All analyses were done using Intercooled Stata Version 11.0 software for Windows.
Results
Demographic, clinical, and cytogenetic details of patients with iAMP21
This study included 94 BCP-ALL patients defined as iAMP21 using our specified criteria of 3 or more additional RUNX1 signals (supplemental Table 1). The cohort consisted of 44 females and 50 males with a median age of 9.5 years (range 2-30 years). Their median white blood cell count was 4.6 × 109/L (range 0.5-226 × 109/L). Cytogenetic analysis was attempted on all 94 diagnostic samples. In 17 patients (18%), analysis failed, whereas 8 (8.5%) had a normal karyotype, including 1 with a constitutional translocation (8987). Apart from 1 patient with 52 chromosomes, all abnormal karyotypes were near-diploid (45-48 chromosomes) with visible abnormalities of chromosome 21 or loss of 1 copy of chromosome 21 plus an unidentified marker chromosome. The abnormal chromosome 21 was described, according to its morphology, as duplicated (dup) or ring (r), etc, and the karyotypes were written in accordance with the International System for Human Cytogenetic Nomenclature.21 In 12 patients, the abnormal chromosome 21 was the sole visible cytogenetic abnormality, whereas 15 patients had a simple karyotype comprising 1 abnormality in addition to the abnormal chromosome 21. The remaining 42 patients showed karyotypes with 3 or more abnormalities. The most frequently observed additional abnormality was gain of an X chromosome in 13 patients.
Genomic analysis of chromosome 21
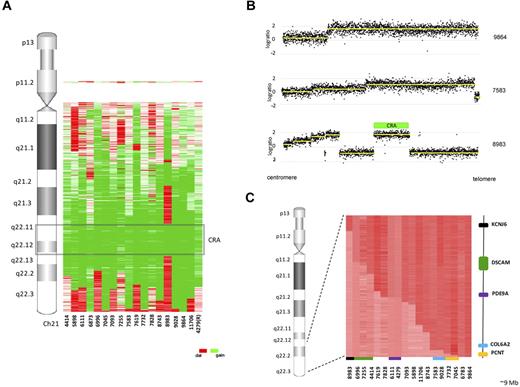
To identify a common abnormality that may be the initiating event giving rise to iAMP21, we initially focused our investigations on chromosome 21. High-resolution aCGH analysis of 17 iAMP21 diagnostic samples and 1 relapse sample identified a mean of 4.8 (range 2-10) copy number abnormalities (CNAs) involving chromosome 21 per patient (supplemental Table 3). Further detailed analysis of the probes mapping to chromosome 21 indicated a high level of complexity indicative of chromosomal instability of this chromosome (Figure 1A). The aCGH profiles of 2 patients showed amplification in a single step with no evidence of a telomeric deletion, whereas the majority showed a stepwise increase in copy number from the centromere toward the telomere, followed by a sharp decrease at the telomere (Figure 1B). In 4 patients, the profile was highly complex, with 9 or 10 abnormalities mapping to chromosome 21. The CRA, as defined by patient 8983, was refined from 6.6 mb to 5.1 mb spanning from 32 813 553 to 37 941 425 bp (Figure 1A-B). This region included 47 known protein-coding genes, including RUNX1 and genes located within the Down syndrome critical region (DSCR), 20 pseudogenes, 18 noncoding RNAs, and a single miRNA, miR-802 (supplemental Table 4). Expression levels of miR-802, 1 of 5 miRNAs mapping to chromosome 21, were determined in 2 iAMP21 patients and compared with 18 ALL patients from different cytogenetic subtypes. Despite an obvious increase in copy number in the iAMP21 patients, this did not translate into a relative increase in expression in iAMP21 patients compared with the other subtypes. In fact, the expression of miR-802 was low among all patients tested, although no statistical analysis could be reliably performed due to the low numbers (supplemental Figure 1).
Summary of chromosome 21 abnormalities in 18 iAMP21 patients. (A) Heat map of chromosome 21 abnormalities detailing the regions of deletion (red), gain (green), and normal copy number (white) relative to genomic location. The CRA, as defined by sample 8983, is highlighted and R denotes that 4279 is a relapse sample. (B) Examples of chromosome 21 profiles from 3 iAMP21 patients clearly demonstrating the complexity and diversity of abnormalities targeting this chromosome. Sample 9864 has a single region of amplification and no telomeric deletion, sample 7583 has a characteristic stepwise profile with a telomeric deletion, and sample 8983 demonstrates a complex stepwise profile with interspersed regions of deletion. The CRA was defined by sample 8983, which is highlighted by a green bar. (C) Heat map showing the telomeric break points (indicated by a change from dark to light red) on chromosome 21 for the 18 patients profiled by aCGH. The region spans ∼ 9 mb, and the most centromeric and telomeric genes with break points, their relative location to the heat map, and the corresponding samples are shown. KCNJ6 is shown in black and PCNT in orange. Recurrent break points were identified in 4 genes: PDE9A (purple) and COL6A2 (blue) had identical break points, but the break points in DSCAM (green) and PCNT (orange) occurred at different genomic positions.
Summary of chromosome 21 abnormalities in 18 iAMP21 patients. (A) Heat map of chromosome 21 abnormalities detailing the regions of deletion (red), gain (green), and normal copy number (white) relative to genomic location. The CRA, as defined by sample 8983, is highlighted and R denotes that 4279 is a relapse sample. (B) Examples of chromosome 21 profiles from 3 iAMP21 patients clearly demonstrating the complexity and diversity of abnormalities targeting this chromosome. Sample 9864 has a single region of amplification and no telomeric deletion, sample 7583 has a characteristic stepwise profile with a telomeric deletion, and sample 8983 demonstrates a complex stepwise profile with interspersed regions of deletion. The CRA was defined by sample 8983, which is highlighted by a green bar. (C) Heat map showing the telomeric break points (indicated by a change from dark to light red) on chromosome 21 for the 18 patients profiled by aCGH. The region spans ∼ 9 mb, and the most centromeric and telomeric genes with break points, their relative location to the heat map, and the corresponding samples are shown. KCNJ6 is shown in black and PCNT in orange. Recurrent break points were identified in 4 genes: PDE9A (purple) and COL6A2 (blue) had identical break points, but the break points in DSCAM (green) and PCNT (orange) occurred at different genomic positions.
A decrease in copy number at the telomere of chromosome 21 occurred in 88% (16 of 18) of iAMP21 patients, resulting in a region of deletion or normal copy number at the telomere, with the break point locations spanning a 9-mb region from within the KCNJ6 gene at position 37 941 425 bp to the telomere (Figure 1C). The region common to the 16 patients (termed CRD) was defined by patient 7045, with the smallest telomeric deletion of 283.8 bp. The break point mapped to a 13.75-kb region (46 646 724-46 660 477 bp) and occurred within the PCNT gene (supplemental Table 3). Identical break points were identified within 2 genes in 2 patients each, PDE9A (4279 and 6111) and COL6A2 (7583 and 9028). Break points were also identified in the DSCAM and PCNT genes in 3 (4414, 6996, and 7255) and 2 (7045 and 7732) patients, respectively, although at different genomic positions within both genes.
No evidence of RUNX1 mutations in patients with iAMP21
Because the RUNX1 gene was always found to be present in the CRA of patients with iAMP21 and abnormalities of this gene are common in leukemia, we searched for mutations in the coding exons 2-7 of RUNX1. No mutations were found in 15 patients (14 diagnostic and 1 relapse sample).
Investigation of recurrent events throughout the genome of patients with iAMP21
A search for other recurrent events elsewhere within the genome of patients with iAMP21 was carried out on the 18 patients analyzed by aCGH (Figure 2). In addition to the 4.8 CNAs involving chromosome 21, a mean of 8.7 (range 1-18) CNAs were identified per patient (excluding common copy number variations such as those involving the antigen receptor loci; supplemental Table 3). Overall, deletions were more frequent than gains, with 64 regions involving focal deletions (< 1 mb) among the autosomes compared and only 11 regions of focal gain. The gain of chromosome X was the only whole-chromosome gain observed by aCGH, in agreement with the findings from the cytogenetic analysis. Recurrent deletions involving genes known to regulate B-cell differentiation and the cell cycle were detected, including deletions of CDKN2A/B (9p21.3; n = 4), IKZF1 (7p13; n = 4), and PAX5 (n = 2). The IKZF1 and PAX5 deletions were heterozygous, whereas CDKN2A deletions were more complex, with some patients having subclones with either heterozygous or homozygous deletions that were confirmed by FISH and/or MLPA (supplemental Figure 2). Deletions were also detected by aCGH in other genes previously associated with leukemia, RB1 (n = 4) and ETV6 (n = 4), but also in regions containing poorly characterized genes, such as TP53TG3 on chromosome 16 (n = 5).
Global abnormalities identified in the iAMP21 genome as detected by aCGH. Idiograms indicating CNAs detected in 18 iAMP21 patients. Each vertical line represents a genomic abnormality identified in a single patient, and the length indicates the extent and position of each CNA. Deletions are shown in red and gains in green.
Global abnormalities identified in the iAMP21 genome as detected by aCGH. Idiograms indicating CNAs detected in 18 iAMP21 patients. Each vertical line represents a genomic abnormality identified in a single patient, and the length indicates the extent and position of each CNA. Deletions are shown in red and gains in green.
Because our global analysis identified recurrent abnormalities affecting genes in key pathways, we screened available samples from the full cohort of 94 iAMP21 patients using a combination of FISH and MLPA to determine their frequency. As shown in Table 1, RB1 deletions were present at the highest incidence of 37%, whereas PAX5 deletions occurred at the lowest incidence of 8%. We also determined that the incidence of P2RY8-CRLF2 by FISH and MLPA was 22%. Gain of chromosome X was confirmed in 24% of patients; 6 also had P2RY8-CRLF2. These abnormalities were not found to be mutually exclusive (supplemental Table 1).
Incidence of recurrent abnormalities detected by FISH and/or MLPA in diagnostic samples of iAMP21 patients
| . | n . | % . | No. with survival data . | No. of relapses . | Early relapses . | Late relapses . | 5-y EFS . | EFS HR (95%CI) . | P . | |
|---|---|---|---|---|---|---|---|---|---|---|
| Total patients | 94 | 100 | 33 | 26 | 9 | 17 | 23% (10%, 38%) | — | — | |
| IKZF1 | N | 56 | 78 | 16 | 12 | 5 | 7 | 35% (13%,58%) | 1.3 (0.41,4.18) | .66 |
| D | 16 | 22 | 5 | 4 | 1 | 3 | 20% (1%,58%) | |||
| CDKN2A | N | 63 | 83 | 20 | 15 | 5 | 10 | — | — | — |
| D | 13 | 17 | 0 | 0 | 0 | 0 | — | — | — | |
| PAX5 | N | 56 | 92 | 15 | 12 | 4 | 8 | — | — | — |
| D | 5 | 8 | 3 | 2 | 1 | 1 | — | — | ||
| ETV6 | N | 66 | 81 | 8 | 6 | 4 | 2 | — | — | — |
| D | 15 | 19 | 0 | 0 | 0 | 0 | — | — | — | |
| RB1 | N | 22 | 62 | 6 | 5 | 3 | 2 | — | — | — |
| D | 13 | 37 | 2 | 1 | 1 | 0 | — | — | — | |
| +X | A | 63 | 76 | 27 | 22 | 7 | 15 | 20% (8%,37%) | 1.22 (0.42,3.56) | .72 |
| P | 20 | 24 | 5 | 4 | 2 | 2 | 20% (1%,58%) | |||
| P2RY8-CRLF2 | A | 61 | 78 | 17 | 14 | 6 | 8 | 27% (9%,49%) | 0.52 (0.15,1.81) | .3 |
| P | 17 | 22 | 5 | 3 | 1 | 2 | 40% (5%,75%) | |||
| . | n . | % . | No. with survival data . | No. of relapses . | Early relapses . | Late relapses . | 5-y EFS . | EFS HR (95%CI) . | P . | |
|---|---|---|---|---|---|---|---|---|---|---|
| Total patients | 94 | 100 | 33 | 26 | 9 | 17 | 23% (10%, 38%) | — | — | |
| IKZF1 | N | 56 | 78 | 16 | 12 | 5 | 7 | 35% (13%,58%) | 1.3 (0.41,4.18) | .66 |
| D | 16 | 22 | 5 | 4 | 1 | 3 | 20% (1%,58%) | |||
| CDKN2A | N | 63 | 83 | 20 | 15 | 5 | 10 | — | — | — |
| D | 13 | 17 | 0 | 0 | 0 | 0 | — | — | — | |
| PAX5 | N | 56 | 92 | 15 | 12 | 4 | 8 | — | — | — |
| D | 5 | 8 | 3 | 2 | 1 | 1 | — | — | ||
| ETV6 | N | 66 | 81 | 8 | 6 | 4 | 2 | — | — | — |
| D | 15 | 19 | 0 | 0 | 0 | 0 | — | — | — | |
| RB1 | N | 22 | 62 | 6 | 5 | 3 | 2 | — | — | — |
| D | 13 | 37 | 2 | 1 | 1 | 0 | — | — | — | |
| +X | A | 63 | 76 | 27 | 22 | 7 | 15 | 20% (8%,37%) | 1.22 (0.42,3.56) | .72 |
| P | 20 | 24 | 5 | 4 | 2 | 2 | 20% (1%,58%) | |||
| P2RY8-CRLF2 | A | 61 | 78 | 17 | 14 | 6 | 8 | 27% (9%,49%) | 0.52 (0.15,1.81) | .3 |
| P | 17 | 22 | 5 | 3 | 1 | 2 | 40% (5%,75%) | |||
EFS indicates event-free survival; HR, hazard ratio; 95%CI, 95% confidence interval; N, no evidence of deletion within gene; D, deletion within gene; A, abnormality is absent; P, abnormality is present; and —, too few samples for statistical analysis.
No evidence of mutations in the JAK gene family members in patients with iAMP21
Clonal heterogeneity of associated abnormalities indicates that iAMP21 is a primary genetic event
The significance of the abnormalities associated with iAMP21 was further explored by comparing the diagnostic and relapse paired samples from 9 patients (supplemental Table 1). The same FISH probes and MLPA kit used to determine the incidence of deletions of IKZF1, PAX5, CDKN2A and the presence of P2RY8-CRLF2 at diagnosis were used to screen the matched relapse samples. Evidence of de novo deletions of these genes at relapse was detected in 2 patients: patient 4780 gained a deletion of IKZF1, whereas patient 10 542 gained deletions of both CDKN2A/B and PAX5 due to the formation of an isochromosome 9 and P2RY8-CRLF2. This isochromosome, i(9)(q10), is a known recurrent abnormality in ALL that provides a mechanism of gene deletion for this patient and 2 others in the series (3382 and 19 578), with corresponding deletions of PAX5 and CDKN2A. Patients 7255 and 4561 had deletions of IKZF1 and P2RY8-CRLF2 fusion, respectively, at diagnosis that were retained at relapse. In patient 4316, the P2RY8-CRLF2 fusion and the PAX5 deletion seen at diagnosis were not observed at relapse; this finding was possibly due to a low blast count in the relapse sample. None of these abnormalities was present in the remaining patients tested.
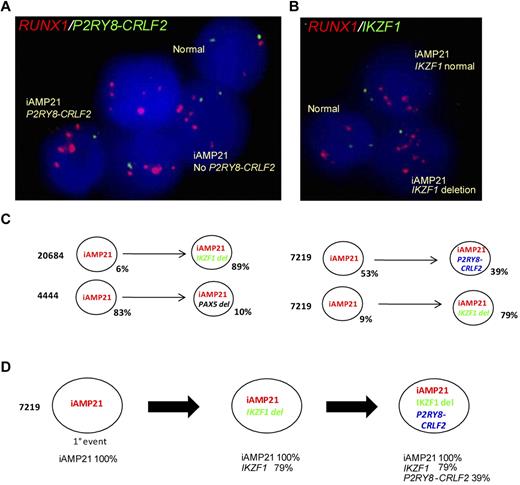
Clonal architecture was investigated among 19 patients who had 1 of the abnormalities specified in the preceding paragraph in association with iAMP21. A dual-color FISH approach was designed to detect amplification of RUNX1 as an indication of the presence of iAMP21 and a second abnormality simultaneously (supplemental Table 1). Examples of the populations detected using dual probes to detect RUNX1/iAMP21 and the presence of P2RY8-CRLF2 and IKZF1, respectively, are shown in Figure 3A-B. Of the patients tested, 3 showed evidence of mixed populations with and without the second abnormality (Figure 3C). In a single sample, 7219, it was possible to predict the temporal order in which the genetic events occurred by studying the clonal architecture as determined by FISH. From sample 7219, we postulated that iAMP21 was the primary event, with IKZF1 deletions and P2RY8-CRLF2 occurring later (Figure 3C). Although accounting for only small numbers of patients, these results indicate clonal heterogeneity within the iAMP21 samples, suggesting that these abnormalities occur as secondary events to iAMP21.
FISH evidence that P2RY8-CRLF2, IKZF1, and PAX5 are secondary events to iAMP21 and a putative model showing the clonal architecture of sample 7219. (A) Five interphase nuclei from patient 7219 hybridized with a probe targeting RUNX1 (red), which detects the multiple copies of this gene that defines iAMP21, and a probe targeting CSF2RA and IL3RA, centromeric to CRLF2, which when deleted indicates the presence of P2RY8-CRLF2 (green). The example shows evidence of clonal heterogeneity; populations of iAMP21-positive cells both with and without P2RY8-CRLF2 are indicated. (B) Three interphase nuclei from patient 7219 hybridized using the same RUNX1 probe (red) and a probe for IKZF1 (green) showing iAMP21-positive cells both with and without the IKZF1 deletion. (C) Examples showing data indicating subclonal architecture in 3 patients: 20684, 7219, and 4444. In patients 7219 and 20684, 9% and 6% of cells, respectively, had amplification of RUNX1 with no deletion of IKZF1, whereas 79% and 89%, respectively, showed an IKZF1 deletion. In patient 4444, 83% of cells had amplification of RUNX1 with no PAX5 deletion, whereas 10% of cells showed the deletion. The major clone (53%) detected in patient 7219 showed iAMP21 without P2RY8-CRLF2, whereas 39% showed the P2RY8-CRLF2 fusion. These observations provide evidence that these events are secondary to iAMP21. (C) Putative model predicting the temporal order of events in the diagnostic sample from patient 7219; apparent linear architecture with 3 populations indicating that iAMP21 is the primary event, followed by deletion of IKZF1 and the presence of P2RY8-CRLF2, is shown.
FISH evidence that P2RY8-CRLF2, IKZF1, and PAX5 are secondary events to iAMP21 and a putative model showing the clonal architecture of sample 7219. (A) Five interphase nuclei from patient 7219 hybridized with a probe targeting RUNX1 (red), which detects the multiple copies of this gene that defines iAMP21, and a probe targeting CSF2RA and IL3RA, centromeric to CRLF2, which when deleted indicates the presence of P2RY8-CRLF2 (green). The example shows evidence of clonal heterogeneity; populations of iAMP21-positive cells both with and without P2RY8-CRLF2 are indicated. (B) Three interphase nuclei from patient 7219 hybridized using the same RUNX1 probe (red) and a probe for IKZF1 (green) showing iAMP21-positive cells both with and without the IKZF1 deletion. (C) Examples showing data indicating subclonal architecture in 3 patients: 20684, 7219, and 4444. In patients 7219 and 20684, 9% and 6% of cells, respectively, had amplification of RUNX1 with no deletion of IKZF1, whereas 79% and 89%, respectively, showed an IKZF1 deletion. In patient 4444, 83% of cells had amplification of RUNX1 with no PAX5 deletion, whereas 10% of cells showed the deletion. The major clone (53%) detected in patient 7219 showed iAMP21 without P2RY8-CRLF2, whereas 39% showed the P2RY8-CRLF2 fusion. These observations provide evidence that these events are secondary to iAMP21. (C) Putative model predicting the temporal order of events in the diagnostic sample from patient 7219; apparent linear architecture with 3 populations indicating that iAMP21 is the primary event, followed by deletion of IKZF1 and the presence of P2RY8-CRLF2, is shown.
No association between abnormalities and time to relapse
In the childhood ALL treatment trial ALL97, we noted not only a high incidence of relapse among iAMP21 patients, but also that these events occurred early (while the patients were on treatment and the first 6 months after) and late (6 months or more after the end of treatment).20 Therefore, we examined the time to relapse within the cohort of 33 patients with follow-up data from this trial in relation to the associated abnormalities (Table 1). There was no difference in time to relapse related to the presence of these abnormalities.
Discussion
Rearrangements targeting chromosome 21 are the hallmark of childhood BCP-ALL with iAMP21, and detailed analysis of chromosome 21 has demonstrated that they are highly complex and heterogeneous in structure. More than a quarter of the copy number alterations detected by aCGH targeted chromosome 21 and included telomeric break points in 88% of patients. The majority of patients showed a stepwise amplification of their chromosome 21 aCGH profile, which was frequently interspersed by additional regions of genomic gain and loss to various levels of complexity, culminating in a decrease in copy number at the telomere. In this study, we refined the CRA to 5.1 mb, including the RUNX1 gene, miR-802, and genes mapping to the DSCR.24,25 Although RUNX1 is a well-known leukemia-related gene, it does not appear to be the target of iAMP21 because: (1) no break points were detected within the gene by aCGH; (2) no RUNX1 mutations were detected, as has also been shown by others26,27 ; and (3) RUNX1 did not show significant overexpression compared with other BCP-ALL subtypes in our previous transcriptome analysis.9
In the present study, we showed that miR-802 expression was low in patients with iAMP21, with no relative difference in expression seen compared with different BCP-ALL cytogenetic subtypes. This agrees with our previous findings of no differential expression of genes within the CRA by gene-expression profiling.9 The ratios of changes corresponding to genes within the CRA and CRD comparing iAMP21 patients with other ALL subtypes, those with ETV6-RUNX1 fusion (rearrangement and, in some cases, gain of whole or part of chromosome 21), high hyperdiploidy (1 or more additional copies of chromosome 21), and unclassified patients (with no established chromosomal abnormalities) from the present study are summarized in supplemental Table 4. Although patient numbers were low, it also appears that miR-802 was not the target.
The overlap between the CRA and the DSCR at 21q22.3 is intriguing because 2 other abnormalities were also detected at high incidence in iAMP21 patients that mirror findings in DS-ALL: gain of the X chromosome and the presence of P2RY8-CRLF2 in 24% and 22% of patients, respectively. The presence of both abnormalities in association with iAMP21 was confirmed in this study and has been reported previously.2,6,10 Somatically acquired activating mutations in JAK2 have been identified in DS-ALL at a high incidence of 18%-28%23,28,29 ; however, we found no mutations in the hot spots of JAK1, JAK2, or JAK3 in patients with iAMP21. In view of the genetic changes in common with DS-ALL, other than JAK mutations, the eventual understanding of the underlying mechanism may emerge from more detailed analysis of similarities and differences between iAMP21 and DS-ALL.
In addition to the CRA, we have more accurately defined the CRD, which was described previously by FISH6 and by bacterial artificial chromosome aCGH9 to be a 283-kb region. Although this deleted region encompasses 3 coding genes, PRMT2, DIP2A, and S100B, which are recognized in several different diseases and significant pathways,30-32 telomeric deletions were not detected in all patients with iAMP21 and are therefore unlikely to be the target of this abnormality. Nevertheless, the higher-resolution aCGH profiles in this study support our previous proposal that iAMP21 arose from a series of breakage-fusion-bridge (BFB) cycles.6,33 Detailed mapping of the CRD break points, the most likely site of the first BFB event, showed that they were variable, with only 2 genes, PDE9A and COL6A2, having identical, recurrent break points in 2 cases each. Recently, the inverted repeat structure or the “fold-back inversions” characteristic of BFB has been reported in pancreatic cancer.34 Therefore, it is interesting to speculate whether the telomeric deletion and the sequence architecture surrounding it represent the initiating event in iAMP21.
The number of CNAs throughout the genome, excluding chromosome 21, was comparable to other ALL subtypes.16,35 Global analysis by genomic arrays identified recurrent abnormalities affecting genes in key pathways, and the frequencies were determined in a larger cohort by FISH and/or MLPA: IKZF1 (22%), CDKN2A/B (17%), PAX5 (8%), ETV6 (19%), and RB1 (37%). The incidences of IKZF1, CDKN2A/BI, and PAX5 are comparable to other BCP-ALL subtypes,15 and therefore are unlikely to be the initiating events in iAMP21 patients. ETV6 and RB1 abnormalities have previously been associated with ALL,13,15,33,36 and, more specifically, focal deletions of ETV6 were described previously in iAMP21 patients.36 In the present study, we showed that RB1 deletions occurred at an incidence of 37%, compared with ∼ 9% in BCP-ALL overall,13-15,35 highlighting a strong association with iAMP21.
There is increasing awareness of clonal heterogeneity among genetic changes in ALL.37 We studied the genetic architecture at the subclonal level in relation to iAMP21 and saw heterogeneity in these patients similar to that observed in ETV6-RUNX1 patients.37 From the examination of paired diagnostic and relapse samples, we demonstrated that P2RY8-CRLF2, IKZF1I, or PAX5, together with CDKN2A present at relapse in 3 patients, were not observed at diagnosis, although cells with a high incidence of iAMP21 were observed in both samples. Secondly, we used a FISH approach to study iAMP21 and associated abnormalities within the same cells. Although only observed in a small number of cases, the finding of mixed populations of iAMP21-positive cells both with and without IKZF1 or PAX5 deletions or P2RY8-CRLF2 indicated the presence of clonal heterogeneity. These observations provide conclusive evidence that, at least in some cases, deletions arise as secondary events to the formation of iAMP21. From the FISH analysis of 1 patient, 7219, we were able to demonstrate the clonal architecture of the diagnostic sample and to generate a model predicting the temporal order of genetic events: iAMP21 as the primary abnormality, followed by deletion of IKZF1, then the generation of P2RY8-CRLF2. Therefore, this limited sequential FISH analysis has contributed to our knowledge of the subclonal genetic architecture of leukemic cells.
In conclusion, this study has provided convincing evidence that chromosomal instability of chromosome 21 is the only recurrent abnormality among those so far investigated that is common to all patients with iAMP21. The lack of any other consistent abnormality outside of chromosome 21 that is detectable at the resolution of aCGH reinforces the assertion that iAMP21 is the likely primary genetic event. However, until whole-genome screens of such cases become available, it cannot be ruled out that some other, currently cryptic, abnormality precedes iAMP21. In addition, the observation that the frequency of secondary abnormalities (especially IKZF1 deletions) is broadly similar to other genetic subtypes, and that in this small study their presence was not related to time to relapse, strengthens our conclusion that the poor outcome of this subgroup is likely driven by iAMP21 itself. Because no individual gene has yet emerged as a candidate, the initiation of iAMP21 is likely hidden within the complex structural rearrangements of the abnormal chromosome 21. Whereas this conclusion may not be novel, the evidence is now more compelling. In the meantime, focus will be on improved diagnosis of iAMP21 to identify those patients in which modified therapy may improve outcome and to ensure that patients at reduced risk of relapse are not exposed to unnecessary intensive treatment.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Leukaemia and Lymphoma Research for funding this work and to the membership of the UK Cancer Cytogenetics Group for providing cytogenetic data and fixed cell suspensions. Primary childhood leukemia samples used in this study were provided by the Leukaemia and Lymphoma Research Childhood Leukaemia Cell Bank working with the laboratory teams in the Bristol Genetics Laboratory, Southmead Hospital (Bristol); Molecular Biology Laboratory, Royal Hospital for Sick Children (Glasgow); Molecular Hematology Laboratory, Royal London Hospital (London); and the Molecular Genetics Service and Sheffield Children's Hospital (Sheffield). This study could not have been performed without the dedication of the United Kingdom Medical Research Council Childhood and Adult Leukaemia Working Parties and their members, who designed and coordinated the clinical trials through which these patients were identified and treated. In particular, we acknowledge the contribution of the ALL97 and ALL2003 trial management groups: Dr Christopher Mitchell, Professor Ajay Vora, Professor Sally Kinsey, Professor Tim Eden, Dr Sue Richards, Dr Nick Goulden, Dr Clare Rowntree, and Dr Rachael Hough.
Authorship
Contribution: V.R. and C.J.H. designed the research and wrote the manuscript; H.P., L.J.R., C.S., H.E., J.I., L.J., D.M., L.M., H.M., S.R., H.R., P.S., A.V.M., and J.C.S. designed some aspects of the research, carried out the experiments, and/or analyzed the data; and all authors critically reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christine J. Harrison, PhD, FRCPath, Professor of Childhood Cancer Cytogenetics, Leukaemia Research Cytogenetics Group, Newcastle Cancer Centre at the Northern Institute for Cancer Research, Newcastle University, Level 5, Sir James Spence Institute, Royal Victoria Infirmary, Newcastle upon Tyne NE1 4LP, United Kingdom; e-mail: christine.harrison@newcastle.ac.uk.
References
Author notes
V.R. and H.P. contributed equally to this study.
J.C.S. and C.J.H. share senior authorship.