Abstract
To identify molecularly defined subgroups in multiple myeloma, gene expression profiling was performed on purified CD138+ plasma cells of 320 newly diagnosed myeloma patients included in the Dutch-Belgian/German HOVON-65/GMMG-HD4 trial. Hierarchical clustering identified 10 subgroups; 6 corresponded to clusters described in the University of Arkansas for Medical Science (UAMS) classification, CD-1 (n = 13, 4.1%), CD-2 (n = 34, 1.6%), MF (n = 32, 1.0%), MS (n = 33, 1.3%), proliferation-associated genes (n = 15, 4.7%), and hyperdiploid (n = 77, 24.1%). Moreover, the UAMS low percentage of bone disease cluster was identified as a subcluster of the MF cluster (n = 15, 4.7%). One subgroup (n = 39, 12.2%) showed a myeloid signature. Three novel subgroups were defined, including a subgroup of 37 patients (11.6%) characterized by high expression of genes involved in the nuclear factor kappa light-chain-enhancer of activated B cells pathway, which include TNFAIP3 and CD40. Another subgroup of 22 patients (6.9%) was characterized by distinct overexpression of cancer testis antigens without overexpression of proliferation genes. The third novel cluster of 9 patients (2.8%) showed up-regulation of protein tyrosine phosphatases PRL-3 and PTPRZ1 as well as SOCS3. To conclude, in addition to 7 clusters described in the UAMS classification, we identified 3 novel subsets of multiple myeloma that may represent unique diagnostic entities.
Introduction
Multiple myeloma (MM), a disease characterized by the accumulation of terminally differentiated antibody-secreting plasma cells (PCs), is an incurable malignancy with a median overall survival of 3 to 4 years. Disease sequelae include immunodeficiency, anemia, hypercalcemia, renal failure, and lytic bone lesions.1
On the basis of (cyto) genetics, myeloma can roughly be divided in nonhyperdiploid and hyperdiploid myeloma. Nonhyperdiploid myeloma is present in 40% of cases and is characterized by recurrent translocations involving the immunoglobulin heavy chain gene at 14q32, resulting in transcriptional activation of CCND1, CCND3, MAF, MAFB, or FGFR3/MMSET. Hyperdiploid myeloma is characterized by trisomies of multiple odd chromosomes (3, 5, 7, 9, 11, 15, 19, and 21).2-4 Together with t(11;14), hyperdiploidy confers a relatively favorable prognosis, whereas MAF, MAFB, or FGFR3/MMSET activation and deletion of chromosome 13 and/or 17 are associated with a poor prognosis.5-10
Several groups have reported gene expression profiles determined by RNA microarray technology in patients with newly diagnosed MM.11-16 Two major genetic classification systems have been developed, the translocation and cyclin D (TC) classification and the University of Arkansas for Medical Sciences (UAMS) molecular classification of myeloma. The TC classification distinguishes 8 subgroups on the basis of overexpression of genes deregulated by primary immunoglobulin H translocations and transcriptional activation of cyclin D genes.2 Use of the UAMS molecular classification of myeloma led to the identification of 7 tumor groups characterized by distinct gene expression profiles, including translocation clusters MS [t(4;14)], MF [t(14;16)/t(14;20), and CD-1/2 (t(11;14) and t(6;14)], as well as a hyperdiploid cluster (HY), a cluster with proliferation-associated genes (PR), and a cluster mainly characterized by a low percentage of bone disease (LB).15 Here, we report the hierarchical clustering determined by gene expression profiles in 320 primarily white, Northern European patients with newly diagnosed MM included in a multicenter phase 3 trial.
Methods
Patients
Bone marrow PC samples were obtained from newly diagnosed patients with MM who were included in a large multicenter, prospective, randomized phase 3 trial (Dutch-Belgian Cooperative Trial Group for Hemato-Oncology [HOVON]65/GMMG-HD4), trial EudraCT Nr 2004-000944-26. This trial included patients with Salmon & Durie stage II or III who were 18 to 65 years of age. Patients with amyloidosis or monoclonal gammopathy of undetermined significance were excluded. Informed consent to treatment protocols and sample procurement was obtained for all cases included in this study, in accordance with the Declaration of Helsinki. Use of diagnostic tumor material was approved by the institutional review board of Erasmus MC.
Myeloma cell purification and RNA isolation
PC purification of bone marrow samples from included patients was performed in 11 centers in The Netherlands, Germany, and Belgium that were equipped to perform PC purification. PCs were separated by the use of positive magnetic cell sorting selection with CD138 magnetic microbeads (Miltenyi Biotec B.V.). Next, purified samples were analyzed for purity and viability by flow cytometric analysis (FACSCalibur and CellQuest Software; BD Biosciences) with CD138-PE (Beckman Coulter), annexin-fluorescein isothiocyanate (NeXins Research), and 7-amino-actinomycin D (Beckman Coulter). Protocols for PC purification and fluorescence-activated cell sorting analysis were equal in all centers. Purified PCs were stored in RLT buffer at −80°C until collection. RNA isolation was performed at the Erasmus Medical Center and at the University of Heidelberg. Only samples with a monoclonal PC purity greater than 80% were used for analysis. RNA was isolated from purified PCs by the use of a DNA/RNA prep kit (QIAGEN). RNA concentration was measured by use of the NanoDrop spectrophotometer (Thermo Fisher Scientific). RNA quality and purity was assessed by use of the RNA 6000 pico or nano assay (Agilent 2100 Bioanalyzer; Agilent Technologies).
Gene expression profiling and array analysis
RNA processing, target labeling, and hybridization to gene expression arrays were performed exclusively in the Erasmus Medical Center. Biotin-labeled cRNA was obtained by the use of the 2-Cycle Eukaryotic Target Labeling Assay (Affymetrix). A total of 15 μg of fragmented, biotin-labeled cRNA was hybridized to Affymetrix GeneChip Human Genome U133 plus 2.0 arrays according to standard Affymetrix protocol (Affymetrix Inc)
Quality controls of arrays that used GeneChip Operating Software included scaling factor and percentage of genes present. Arrays with a scaling factor difference of less than 3 and more than 20% genes present were analyzed further. Raw data from selected gene expression arrays (CEL-files) were preprocessed by the use of GCRMA in Partek Genomics Suite, version 6.4 (Partek). Final quality control of arrays included relative log expression and normalized unscaled standard errors (NUSEs) from the AffyPLM package (http://www.bioconductor.org). Arrays showing a NUSE value greater than 1.05 and aberrant relative log expression plots were excluded from analysis. Microarray data presented in this work have been deposited in the Gene Expression Omnibus (National Center for Biotechnology Information) and are accessible through GEO Series accession number GSE19784 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19784).
Cluster analysis
GCRMA-normalized expression data were imported in Omniviz software version 6.1 (BioWisdom). In Omniviz, the exponential values were taken of the GCRMA-derived log2 intensity values, and because GeneChips do not reliably discriminate between values less than 30, all intensity values less than 30 were set to 30. The level of expression for every probe set was determined relative to the geometric mean and log transformed (base 2). The 5% (2730) most variable probe sets from the total were selected by the use of a cut-off of log2 geometric mean less than −5.12 or more than 5.12 (reflecting up- or down-regulation) in at least one patient (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Hierarchical clustering of average linkage with the centered correlation metric was performed by the use of BRB-array tools version 3.6.0 (http://linus.nci.nih.gov/BRB-ArrayTools.html). The dendrogram obtained was compared with translocation status, and robustness (R) indices (BRB-array tools) were calculated to give an indication concerning the reproducibility of the clusters. To determine expression signature of clusters, each cluster was compared with the remaining clusters by use of the Class Comparison option with the following settings: P less than .001 and false discovery rate less than 5% (BRB-array tools).
Prediction analysis of microarrays
To validate clusters, a method of nearest-shrunken centroid classification that uses prediction analysis of microarrays in R version 2.6.0 (PAMr package in R Version 2.6.0) was used.17 Validation of clusters was performed in an independent dataset, GSE2658, generated by the UAMS, which included 559 newly diagnosed MM patients. The dataset containing the 5% most variable genes, 2730 genes, was used (supplemental Table 1). Sensitivity (Sn), specificity (Sp), positive predictive values, and negative predictive values were calculated.
In addition, validation analysis to confirm all identified clusters was performed by use of the CEL files of 2 independent datasets, the APEX/SUMMIT/CREST dataset,18 and the UAMS dataset.15 CEL files were normalized by the use of our normalization methods, sample, and gene selection criteria as described.
An extensive description of the method of prediction analysis of translocations t(4;14), t(11;14), and t(14;16)/t(14;20) is outlined in the supplemental data (Document 1). In brief, samples with available fluorescence in situ hybridization (FISH) data were randomly divided in a training set (2/3) and a test set (1/3). Training set and test set were separately normalized. For the training set, the 5% most variable genes, 2730 genes, were generated by the use of the method described previously (supplemental Table 2). These 2730 probe sets were subsequently used in the test set. Percentage correctly classified samples, Sn, Sp, positive predictive value, and negative predictive value were calculated.
Cytogenetic analysis and FISH
FISH analysis was performed in 304 patients. In addition, karyotyping data were available for 119 patients. In nonpurified PC samples (n = 125) at least 200 interphase nuclei per sample were analyzed by the use of epi-fluorescence microscopy and image analysis software, with in several cases a preceding analysis of selected myeloma cells determined by light chain counterstaining or morphology. In CD138-purified PC samples (n = 179), 100 nuclei were evaluated by the use of an epifluorescence microscope (Leica Microsystems). Hybridization efficiency was validated on PCs obtained from bone marrow of a healthy donor; thresholds for gains, deletions, and translocations were set at 10%.
Interphase FISH analysis was performed as previously described.6,19 Detection of numerical changes was performed by the use of commercial 2-color probes chromosome loci 1q21/8p21, 11q23/13q14, 9q34/22q11, 6q21/15q22, and17p13/19q13 (Poseidon Probes; Kreatech) or by the use of alpha satellite probes for centromere regions of chromosome 9 and 11 (CEP 9 and CEP 11; Vysis; Abbott Molecular). The combination of trisomies 9, 11, and 15 was found to be predictive of hyperdiploidy.8 Hyperdiploid MM was defined by presence of trisomy of 2 of these chromosomes (trisomy 9 and 11, 11 and 15, or 9 and 15) or all of them (trisomy of chromosomes 9, 11, and 15), as determined by FISH and/or karyotyping data.
Translocations t(11;14)(q13;q32), t(4;14)(p16;q32), and (14;16)(q32;q23) were determined by the use of LSI IGH/CCND1, LSI IGH/FGFR3, and LSI IGH/MAF probes, respectively (Vysis; Abbott Molecular) or commercial 2-color probe sets for detection of translocations t(11;14)(q13;q32), t(4;14)(p16;q32) (both Poseidon Probes; Kreatech) and t(14;16) (q32;q23) (Vysis). A t(14;20)(q32;q12) with 14q32 IGH gene rearrangement was confirmed by FISH by the use of 14q32 immunoglobulin H rearrangement probe, LSI IGH DC, and whole chromosome paint 14 and 20 probes, wcp14 and wcp 20 (Vysis; Abbott Molecular). Conventional karyotyping was performed as described previously.20
Results
Identification of expression signatures
A total of 320 bone marrow aspirates from newly diagnosed patients were obtained upon inclusion in the HOVON65/GMMG-HD4 trial for gene expression profiling. Comparison of baseline clinical characteristics of this subset of patients showed no significant difference between characterized subset and the whole patient group in the trial (supplemental Table 3). The sample clustering presented by a dendrogram with 5 major branches and 11 clusters is shown in Figure 1. Translocation status and robustness (R) indices per cluster are shown in supplemental Table 4. The top 10 genes (P < .001, false discovery rate < 5%) showing the greatest fold change per cluster in comparison with remaining clusters are shown in Tables 1 and 2, and the top 50 genes are shown in supplemental Table 5. Of the 11 clusters found, 10 were characterized in detail. The remaining cluster, consisting of 9 samples with 41 differentially expressed genes, was excluded from analysis because no clear signature could be determined. Six of the identified clusters corresponded well to the published UAMS classification and were therefore named accordingly.15
Dendrogram and heatmap. Vertical dendrogram shows sample clustering with 5 major branches and 11 distinct clusters; the dendrogram is cut at 11 clusters. First column,11 clusters: CD2, CD-2 cluster; CD1, CD-1 cluster; CTA, CTA cluster; NFκB, NFκB cluster; NP, no clear profile; HY, HY cluster; PRL3, PRL3 cluster; PR, PR cluster; MF, MF cluster; MS, MS cluster; Myeloid, Myeloid cluster. Translocations are shown in the second column: t(11;14), yellow; t(4;14), blue; t(14;16) or t(14;20), red; no translocation, green; and not determined, white. The third column indicates hyperdiploidy: y, hyperdiploidy (yellow); n, no hyperdiploidy (green) and nd (not determined; white). Horizontal dendrogram shows clustering of genes. The heatmap shows the spectrum of expression values, the log2 expression value from the geometric mean for each gene is indicated by a color, with red representing positive expression (up-regulation) and blue representing negative expression (down-regulation) of a gene.
Dendrogram and heatmap. Vertical dendrogram shows sample clustering with 5 major branches and 11 distinct clusters; the dendrogram is cut at 11 clusters. First column,11 clusters: CD2, CD-2 cluster; CD1, CD-1 cluster; CTA, CTA cluster; NFκB, NFκB cluster; NP, no clear profile; HY, HY cluster; PRL3, PRL3 cluster; PR, PR cluster; MF, MF cluster; MS, MS cluster; Myeloid, Myeloid cluster. Translocations are shown in the second column: t(11;14), yellow; t(4;14), blue; t(14;16) or t(14;20), red; no translocation, green; and not determined, white. The third column indicates hyperdiploidy: y, hyperdiploidy (yellow); n, no hyperdiploidy (green) and nd (not determined; white). Horizontal dendrogram shows clustering of genes. The heatmap shows the spectrum of expression values, the log2 expression value from the geometric mean for each gene is indicated by a color, with red representing positive expression (up-regulation) and blue representing negative expression (down-regulation) of a gene.
Top 10 fold up-regulated genes
| Cluster/probe set . | Gene symbol . | Fold change . |
|---|---|---|
| CD-2 | ||
| 208711_s_at | CCND1 | 78.43 |
| 228592_at | MS4A1 | 52.20 |
| 208712_at | CCND1 | 34.84 |
| 223823_at | KCNMB2 | 21.04 |
| 235518_at | SLC8A1 | 17.57 |
| 220068_at | VPREB3 | 13.64 |
| 210356_x_at | MS4A1 | 13.49 |
| 228599_at | MS4A1 | 13.16 |
| 225842_at | PHLDA1 | 12.81 |
| 217418_x_at | MS4A1 | 11.31 |
| CD-1 | ||
| 230493_at | SHISA2 | 29.08 |
| 235278_at | MACROD2 | 17.77 |
| 210587_at | INHBE | 13.97 |
| 218589_at | P2RY5 | 12.52 |
| 206760_s_at | FCER2 | 12.17 |
| 207076_s_at | ASS1 | 11.14 |
| 223823_at | KCNMB2 | 11.07 |
| 206759_at | FCER2 | 9.78 |
| 221911_at | ETV1 | 9.33 |
| 225285_at | BCAT1 | 8.83 |
| CTA | ||
| 210387_at | HIST1H2BG | 7.59 |
| 206102_at | GINS1 | 5.01 |
| 208955_at | DUT | 4.94 |
| 238021_s_at | hCG_1815491 | 4.79 |
| 238762_at | MTHFD2L | 4.58 |
| 206834_at | HBB, HBD | 4.57 |
| 227212_s_at | PHF19 | 4.55 |
| 202016_at | MEST | 4.45 |
| 203213_at | CDC2 | 4.22 |
| 238022_at | hCG_1815491 | 4.16 |
| NFκB | ||
| 214230_at | CDC42 | 19.17 |
| 202643_s_at | TNFAIP3 | 11.73 |
| 211032_at | COBLL1 | 10.87 |
| 1557257_at | BCL10 | 10.20 |
| 1559249_at | ATXN1 | 9.52 |
| 208622_s_at | EZR | 8.62 |
| 230082_at | LOC100133660 | 8.57 |
| 1554229_at | C5orf41 | 8.05 |
| 238633_at | EPC1 | 7.96 |
| 1552542_s_at | TAGAP7 | 7.84 |
| HY | ||
| 222943_at | GBA3 | 8.28 |
| 219954_s_at | GBA3 | 5.78 |
| 219463_at | C20orf103 | 5.15 |
| 203153_at | IFIT1 | 5.08 |
| 214329_x_at | TNFSF10 | 5.01 |
| 202687_s_at | TNFSF10 | 4.80 |
| 212843_at | NCAM1 | 4.57 |
| 205051_s_at | KIT | 4.55 |
| 202688_at | TNFSF10 | 4.49 |
| 206609_at | MAGEC1 | 4.43 |
| PRL3 | ||
| 200953_s_at | CCND2 | 19.28 |
| 206574_s_at | PTP4A3 | 15.66 |
| 209695_at | PTP4A3 | 15.11 |
| 204469_at | PTPRZ1 | 9.73 |
| 227697_at | SOCS3 | 5.93 |
| 217865_at | RNF130 | 5.89 |
| 209183_s_at | C10orf10 | 4.40 |
| 218788_s_at | SMYD3 | 4.11 |
| 228051_at | KIAA1244 | 3.82 |
| 219195_at | PPARGC1A | 3.69 |
| PR | ||
| 232231_at | RUNX2 | 14.43 |
| 210432_s_at | SCN3A | 7.64 |
| 206640_x_at | [multiple gene symbols] | 7.21 |
| 203213_at | CDC2 | 7.04 |
| 206102_at | GINS1 | 6.80 |
| 208235_x_at | GAGE12F, GAGE12 | 6.76 |
| 201292_at | TOP2A | 6.74 |
| 207739_s_at | [multiple gene symbols] | 6.50 |
| 214070_s_at | ATP10B | 6.39 |
| 201291_s_at | TOP2A | 6.38 |
| MF | ||
| 200953_s_at | CCND2 | 17.72 |
| 212724_at | RND3 | 15.84 |
| 211986_at | AHNAK | 12.55 |
| 200951_s_at | CCND2 | 11.03 |
| 226875_at | DOCK11 | 6.95 |
| 202207_at | ARL4C | 6.77 |
| 205590_at | RASGRP1 | 6.70 |
| 204589_at | NUAK1 | 5.95 |
| 218935_at | EHD3 | 5.54 |
| 226961_at | PRR15 | 5.23 |
| MS | ||
| 204379_s_at | FGFR3 | 153.02 |
| 222777_s_at | WHSC1 | 45.72 |
| 200953_s_at | CCND2 | 35.57 |
| 217901_at | DSG2 | 23.53 |
| 201387_s_at | UCHL1 | 22.40 |
| 212190_at | SERPINE2 | 21.45 |
| 222778_s_at | WHSC1 | 20.47 |
| 217963_s_at | NGFRAP1 | 17.83 |
| 209053_s_at | WHSC1 | 16.52 |
| 212771_at | C10orf38 | 16.29 |
| Myeloid | ||
| 206111_at | [multiple gene symbols] | 12.54 |
| 205033_s_at | DEFA1, DEFA3, LOC7 | 10.01 |
| 215051_x_at | AIF1 | 9.24 |
| 201137_s_at | HLA-DPB1 | 8.33 |
| 207269_at | DEFA4 | 7.95 |
| 202917_s_at | S100A8 | 7.93 |
| 213975_s_at | LYZ | 7.84 |
| 205950_s_at | CA1 | 7.77 |
| 215193_x_at | [multiple gene symbols] | 7.66 |
| 203645_s_at | CD163 | 7.64 |
| Cluster/probe set . | Gene symbol . | Fold change . |
|---|---|---|
| CD-2 | ||
| 208711_s_at | CCND1 | 78.43 |
| 228592_at | MS4A1 | 52.20 |
| 208712_at | CCND1 | 34.84 |
| 223823_at | KCNMB2 | 21.04 |
| 235518_at | SLC8A1 | 17.57 |
| 220068_at | VPREB3 | 13.64 |
| 210356_x_at | MS4A1 | 13.49 |
| 228599_at | MS4A1 | 13.16 |
| 225842_at | PHLDA1 | 12.81 |
| 217418_x_at | MS4A1 | 11.31 |
| CD-1 | ||
| 230493_at | SHISA2 | 29.08 |
| 235278_at | MACROD2 | 17.77 |
| 210587_at | INHBE | 13.97 |
| 218589_at | P2RY5 | 12.52 |
| 206760_s_at | FCER2 | 12.17 |
| 207076_s_at | ASS1 | 11.14 |
| 223823_at | KCNMB2 | 11.07 |
| 206759_at | FCER2 | 9.78 |
| 221911_at | ETV1 | 9.33 |
| 225285_at | BCAT1 | 8.83 |
| CTA | ||
| 210387_at | HIST1H2BG | 7.59 |
| 206102_at | GINS1 | 5.01 |
| 208955_at | DUT | 4.94 |
| 238021_s_at | hCG_1815491 | 4.79 |
| 238762_at | MTHFD2L | 4.58 |
| 206834_at | HBB, HBD | 4.57 |
| 227212_s_at | PHF19 | 4.55 |
| 202016_at | MEST | 4.45 |
| 203213_at | CDC2 | 4.22 |
| 238022_at | hCG_1815491 | 4.16 |
| NFκB | ||
| 214230_at | CDC42 | 19.17 |
| 202643_s_at | TNFAIP3 | 11.73 |
| 211032_at | COBLL1 | 10.87 |
| 1557257_at | BCL10 | 10.20 |
| 1559249_at | ATXN1 | 9.52 |
| 208622_s_at | EZR | 8.62 |
| 230082_at | LOC100133660 | 8.57 |
| 1554229_at | C5orf41 | 8.05 |
| 238633_at | EPC1 | 7.96 |
| 1552542_s_at | TAGAP7 | 7.84 |
| HY | ||
| 222943_at | GBA3 | 8.28 |
| 219954_s_at | GBA3 | 5.78 |
| 219463_at | C20orf103 | 5.15 |
| 203153_at | IFIT1 | 5.08 |
| 214329_x_at | TNFSF10 | 5.01 |
| 202687_s_at | TNFSF10 | 4.80 |
| 212843_at | NCAM1 | 4.57 |
| 205051_s_at | KIT | 4.55 |
| 202688_at | TNFSF10 | 4.49 |
| 206609_at | MAGEC1 | 4.43 |
| PRL3 | ||
| 200953_s_at | CCND2 | 19.28 |
| 206574_s_at | PTP4A3 | 15.66 |
| 209695_at | PTP4A3 | 15.11 |
| 204469_at | PTPRZ1 | 9.73 |
| 227697_at | SOCS3 | 5.93 |
| 217865_at | RNF130 | 5.89 |
| 209183_s_at | C10orf10 | 4.40 |
| 218788_s_at | SMYD3 | 4.11 |
| 228051_at | KIAA1244 | 3.82 |
| 219195_at | PPARGC1A | 3.69 |
| PR | ||
| 232231_at | RUNX2 | 14.43 |
| 210432_s_at | SCN3A | 7.64 |
| 206640_x_at | [multiple gene symbols] | 7.21 |
| 203213_at | CDC2 | 7.04 |
| 206102_at | GINS1 | 6.80 |
| 208235_x_at | GAGE12F, GAGE12 | 6.76 |
| 201292_at | TOP2A | 6.74 |
| 207739_s_at | [multiple gene symbols] | 6.50 |
| 214070_s_at | ATP10B | 6.39 |
| 201291_s_at | TOP2A | 6.38 |
| MF | ||
| 200953_s_at | CCND2 | 17.72 |
| 212724_at | RND3 | 15.84 |
| 211986_at | AHNAK | 12.55 |
| 200951_s_at | CCND2 | 11.03 |
| 226875_at | DOCK11 | 6.95 |
| 202207_at | ARL4C | 6.77 |
| 205590_at | RASGRP1 | 6.70 |
| 204589_at | NUAK1 | 5.95 |
| 218935_at | EHD3 | 5.54 |
| 226961_at | PRR15 | 5.23 |
| MS | ||
| 204379_s_at | FGFR3 | 153.02 |
| 222777_s_at | WHSC1 | 45.72 |
| 200953_s_at | CCND2 | 35.57 |
| 217901_at | DSG2 | 23.53 |
| 201387_s_at | UCHL1 | 22.40 |
| 212190_at | SERPINE2 | 21.45 |
| 222778_s_at | WHSC1 | 20.47 |
| 217963_s_at | NGFRAP1 | 17.83 |
| 209053_s_at | WHSC1 | 16.52 |
| 212771_at | C10orf38 | 16.29 |
| Myeloid | ||
| 206111_at | [multiple gene symbols] | 12.54 |
| 205033_s_at | DEFA1, DEFA3, LOC7 | 10.01 |
| 215051_x_at | AIF1 | 9.24 |
| 201137_s_at | HLA-DPB1 | 8.33 |
| 207269_at | DEFA4 | 7.95 |
| 202917_s_at | S100A8 | 7.93 |
| 213975_s_at | LYZ | 7.84 |
| 205950_s_at | CA1 | 7.77 |
| 215193_x_at | [multiple gene symbols] | 7.66 |
| 203645_s_at | CD163 | 7.64 |
Per cluster, the top 10 up-regulated genes are shown. The first column shows the cluster and probe set IDs, the second column shows the gene symbols, and the third column indicates the fold change differences of per probe set in the specific cluster versus the remaining clusters.
CTA indicates cancer testis antigens; HY, hyperdiploid cluster; NFκB, nuclear factor kappa light-chain-enhancer of activated B cells; and PR, proliferation-associated genes.
Top 10 fold down-regulated genes
| Cluster/probe set . | Gene symbol . | Fold change . |
|---|---|---|
| CD-2 | ||
| 219463_at | C20orf103 | 0.10 |
| 200953_s_at | CCND2 | 0.13 |
| 205159_at | CSF2RB | 0.14 |
| 205968_at | KCNS3 | 0.15 |
| 226333_at | IL6R | 0.15 |
| 204489_s_at | CD44 | 0.16 |
| 201387_s_at | UCHL1 | 0.18 |
| 212063_at | CD44 | 0.18 |
| 238021_s_at | hCG_1815491 | 0.19 |
| 219954_s_at | GBA3 | 0.20 |
| CD-1 | ||
| 201005_at | CD9 | 0.08 |
| 205352_at | SERPINI1 | 0.17 |
| 205239_at | AREG, LOC727738 | 0.18 |
| 222392_x_at | PERP | 0.19 |
| 213737_x_at | GOLGA9P | 0.26 |
| 211071_s_at | MLLT11 | 0.26 |
| 227067_x_at | NOTCH2NL | 0.26 |
| 204647_at | HOMER3 | 0.27 |
| 208078_s_at | SNF1LK | 0.28 |
| CTA | ||
| 200643_at | HDLBP | 0.07 |
| 201024_x_at | EIF5B | 0.10 |
| 212586_at | CAST | 0.11 |
| 203675_at | NUCB2 | 0.11 |
| 200977_s_at | TAX1BP1 | 0.12 |
| 224567_x_at | MALAT1 | 0.12 |
| 211968_s_at | HSP90AA1 | 0.12 |
| 200595_s_at | EIF3A | 0.13 |
| 210645_s_at | TTC3 | 0.13 |
| 219221_at | ZBTB38 | 0.14 |
| NFκB | ||
| 206978_at | CCR2, FLJ78302 | 0.07 |
| 212731_at | ANKRD46 | 0.07 |
| 200768_s_at | MAT2A | 0.10 |
| 231576_at | 0.11 | |
| 202797_at | SACM1L | 0.12 |
| 209296_at | PPM1B | 0.12 |
| 213005_s_at | KANK1 | 0.13 |
| 201503_at | G3BP1 | 0.14 |
| 201664_at | SMC4 | 0.15 |
| 201098_at | COPB2 | 0.19 |
| HY | ||
| 200953_s_at | CCND2 | 0.12 |
| 219799_s_at | DHRS9 | 0.18 |
| 203186_s_at | S100A4 | 0.19 |
| 201029_s_at | CD99 | 0.19 |
| 225673_at | MYADM | 0.20 |
| 201666_at | TIMP1 | 0.22 |
| 224009_x_at | DHRS9 | 0.22 |
| 205229_s_at | COCH | 0.23 |
| 200951_s_at | CCND2 | 0.23 |
| 223952_x_at | DHRS9 | 0.24 |
| PRL3 | ||
| 201721_s_at | LAPTM5 | 0.04 |
| 212063_at | CD44 | 0.04 |
| 204489_s_at | CD44 | 0.08 |
| 216438_s_at | TMSB4X, TMSL3 | 0.16 |
| 208892_s_at | DUSP6 | 0.19 |
| 208690_s_at | PDLIM1 | 0.20 |
| 225282_at | SMAP2 | 0.28 |
| 201432_at | CAT | 0.34 |
| 201300_s_at | PRNP | 0.40 |
| PR | ||
| 215001_s_at | GLUL | 0.10 |
| 206150_at | CD27 | 0.11 |
| 209201_x_at | CXCR4 | 0.13 |
| 235400_at | FCRLA | 0.13 |
| 211919_s_at | CXCR4 | 0.14 |
| 205671_s_at | HLA-DOB | 0.15 |
| 209619_at | CD74 | 0.15 |
| 228284_at | TLE1 | 0.15 |
| 210889_s_at | FCGR2B | 0.16 |
| 235372_at | FCRLA | 0.16 |
| MF | ||
| 204602_at | DKK1 | 0.05 |
| 226702_at | CMPK2 | 0.06 |
| 202391_at | BASP1 | 0.10 |
| 203698_s_at | FRZB | 0.11 |
| 216576_x_at | IGKC, IGKV1-5, LOC647506 | 0.12 |
| 215176_x_at | LOC100130100 | 0.12 |
| 202688_at | TNFSF10 | 0.13 |
| 225681_at | CTHRC1 | 0.13 |
| 203697_at | FRZB | 0.13 |
| 242625_at | RSAD2 | 0.14 |
| MS | ||
| 208712_at | CCND1 | 0.12 |
| 203698_s_at | FRZB | 0.19 |
| 203697_at | FRZB | 0.19 |
| 228592_at | MS4A1 | 0.20 |
| 201721_s_at | LAPTM5 | 0.20 |
| 221969_at | 0.21 | |
| 206609_at | MAGEC1 | 0.22 |
| 204794_at | DUSP2 | 0.23 |
| 226068_at | SYK | 0.23 |
| 243780_at | 0.25 | |
| Myeloid | ||
| 200730_s_at | PTP4A1 | 0.42 |
| 218826_at | SLC35F2 | 0.43 |
| 210942_s_at | ST3GAL6 | 0.47 |
| 227189_at | CPNE5 | 0.48 |
| 206445_s_at | PRMT1 | 0.50 |
| 214359_s_at | HSP90AB1 | 0.51 |
| 209457_at | DUSP5 | 0.51 |
| 204790_at | SMAD7 | 0.51 |
| 211967_at | TMEM123 | 0.52 |
| 226612_at | FLJ25076 | 0.52 |
| Cluster/probe set . | Gene symbol . | Fold change . |
|---|---|---|
| CD-2 | ||
| 219463_at | C20orf103 | 0.10 |
| 200953_s_at | CCND2 | 0.13 |
| 205159_at | CSF2RB | 0.14 |
| 205968_at | KCNS3 | 0.15 |
| 226333_at | IL6R | 0.15 |
| 204489_s_at | CD44 | 0.16 |
| 201387_s_at | UCHL1 | 0.18 |
| 212063_at | CD44 | 0.18 |
| 238021_s_at | hCG_1815491 | 0.19 |
| 219954_s_at | GBA3 | 0.20 |
| CD-1 | ||
| 201005_at | CD9 | 0.08 |
| 205352_at | SERPINI1 | 0.17 |
| 205239_at | AREG, LOC727738 | 0.18 |
| 222392_x_at | PERP | 0.19 |
| 213737_x_at | GOLGA9P | 0.26 |
| 211071_s_at | MLLT11 | 0.26 |
| 227067_x_at | NOTCH2NL | 0.26 |
| 204647_at | HOMER3 | 0.27 |
| 208078_s_at | SNF1LK | 0.28 |
| CTA | ||
| 200643_at | HDLBP | 0.07 |
| 201024_x_at | EIF5B | 0.10 |
| 212586_at | CAST | 0.11 |
| 203675_at | NUCB2 | 0.11 |
| 200977_s_at | TAX1BP1 | 0.12 |
| 224567_x_at | MALAT1 | 0.12 |
| 211968_s_at | HSP90AA1 | 0.12 |
| 200595_s_at | EIF3A | 0.13 |
| 210645_s_at | TTC3 | 0.13 |
| 219221_at | ZBTB38 | 0.14 |
| NFκB | ||
| 206978_at | CCR2, FLJ78302 | 0.07 |
| 212731_at | ANKRD46 | 0.07 |
| 200768_s_at | MAT2A | 0.10 |
| 231576_at | 0.11 | |
| 202797_at | SACM1L | 0.12 |
| 209296_at | PPM1B | 0.12 |
| 213005_s_at | KANK1 | 0.13 |
| 201503_at | G3BP1 | 0.14 |
| 201664_at | SMC4 | 0.15 |
| 201098_at | COPB2 | 0.19 |
| HY | ||
| 200953_s_at | CCND2 | 0.12 |
| 219799_s_at | DHRS9 | 0.18 |
| 203186_s_at | S100A4 | 0.19 |
| 201029_s_at | CD99 | 0.19 |
| 225673_at | MYADM | 0.20 |
| 201666_at | TIMP1 | 0.22 |
| 224009_x_at | DHRS9 | 0.22 |
| 205229_s_at | COCH | 0.23 |
| 200951_s_at | CCND2 | 0.23 |
| 223952_x_at | DHRS9 | 0.24 |
| PRL3 | ||
| 201721_s_at | LAPTM5 | 0.04 |
| 212063_at | CD44 | 0.04 |
| 204489_s_at | CD44 | 0.08 |
| 216438_s_at | TMSB4X, TMSL3 | 0.16 |
| 208892_s_at | DUSP6 | 0.19 |
| 208690_s_at | PDLIM1 | 0.20 |
| 225282_at | SMAP2 | 0.28 |
| 201432_at | CAT | 0.34 |
| 201300_s_at | PRNP | 0.40 |
| PR | ||
| 215001_s_at | GLUL | 0.10 |
| 206150_at | CD27 | 0.11 |
| 209201_x_at | CXCR4 | 0.13 |
| 235400_at | FCRLA | 0.13 |
| 211919_s_at | CXCR4 | 0.14 |
| 205671_s_at | HLA-DOB | 0.15 |
| 209619_at | CD74 | 0.15 |
| 228284_at | TLE1 | 0.15 |
| 210889_s_at | FCGR2B | 0.16 |
| 235372_at | FCRLA | 0.16 |
| MF | ||
| 204602_at | DKK1 | 0.05 |
| 226702_at | CMPK2 | 0.06 |
| 202391_at | BASP1 | 0.10 |
| 203698_s_at | FRZB | 0.11 |
| 216576_x_at | IGKC, IGKV1-5, LOC647506 | 0.12 |
| 215176_x_at | LOC100130100 | 0.12 |
| 202688_at | TNFSF10 | 0.13 |
| 225681_at | CTHRC1 | 0.13 |
| 203697_at | FRZB | 0.13 |
| 242625_at | RSAD2 | 0.14 |
| MS | ||
| 208712_at | CCND1 | 0.12 |
| 203698_s_at | FRZB | 0.19 |
| 203697_at | FRZB | 0.19 |
| 228592_at | MS4A1 | 0.20 |
| 201721_s_at | LAPTM5 | 0.20 |
| 221969_at | 0.21 | |
| 206609_at | MAGEC1 | 0.22 |
| 204794_at | DUSP2 | 0.23 |
| 226068_at | SYK | 0.23 |
| 243780_at | 0.25 | |
| Myeloid | ||
| 200730_s_at | PTP4A1 | 0.42 |
| 218826_at | SLC35F2 | 0.43 |
| 210942_s_at | ST3GAL6 | 0.47 |
| 227189_at | CPNE5 | 0.48 |
| 206445_s_at | PRMT1 | 0.50 |
| 214359_s_at | HSP90AB1 | 0.51 |
| 209457_at | DUSP5 | 0.51 |
| 204790_at | SMAD7 | 0.51 |
| 211967_at | TMEM123 | 0.52 |
| 226612_at | FLJ25076 | 0.52 |
Per cluster, the top 10 down-regulated genes are shown. The first column shows the cluster and probe set IDs, the second column shows the gene symbols, and the third column indicates the fold change differences of per probe set in the specific cluster versus the remaining clusters.
CTA indicates cancer testis antigens; HY, hyperdiploid cluster; NFκB, nuclear factor kappa light-chain-enhancer of activated B cells; and PR, proliferation-associated genes.
Samples harboring t(11;14) and/or overexpression of CCND1 were divided into 2 clusters, CD-1 and CD-2. A relatively low frequency of t(11;14) (33%) was found in the CD-1 cluster in our study, which is low compared with previous reports.15 Still, this cluster was characterized by high CCND1 expression and by overexpression of argininosuccinate synthetase 1 ASS1, inhibin beta E INHBE, and nidogen 2 NID2 as has been described previously. B-cell markers MS4A1 (CD20), VPREB3, CD79A, and BANK1 defined cluster CD-2 (Table 1; supplemental Table 5). CD20 expression has been associated with presence of t(11;14),21 which is consistent with the high percentage of t(11;14) observed in this cluster in comparison with cluster CD-1.
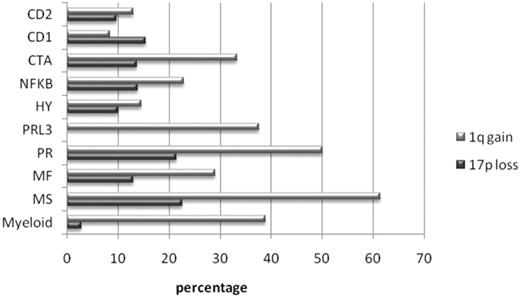
The MS cluster was characterized by translocation t(4;14), deregulating FGFR3 and MMSET, present in 96% of patients in this cluster. Other notable overexpressed genes include desmoglein DSG2, CCND2, selectin L (lymphocyte adhesion molecule 1) SELL, and serpin peptidase inhibitors SERPINE2 and SERPINI1 (Table 1; supplemental Table 5). This cluster showed a significantly greater percentage of patients with a 1q21 amplification (61%, compared with 8% to 50% in the remaining clusters; P < .001; Figure 2).
1q gain and 17p loss. Percentage of patients per cluster showing 1q gain (dark gray bar) and 17p loss (light gray bar).
1q gain and 17p loss. Percentage of patients per cluster showing 1q gain (dark gray bar) and 17p loss (light gray bar).
The MF cluster contained 32 samples, of which 7 harbored a confirmed t(14;16) or t(14;20). c-MAF, which is deregulated by t(14;16), and MAFB, deregulated by t(14;20), were observed only in a subset of patients, which clustered separately within this MF cluster. The remaining samples in the MF cluster clustered with these samples on the basis of overexpression of downstream targets of MAFB and/or c-MAF: RND3, CCND2, and ITGB7 (supplemental Figure 1).22 FRZB and DKK1, both WNT inhibitors of which the presence is associated with osteolytic lesions in myeloma patients, were among the top down-regulated genes (Table 1; supplemental Table 5).23,24 Our analysis of both subsets separately revealed an even stronger signature for the MF subcluster (supplemental Table 6). Clinical features such as elevated lactate dehydrogenase and thrombocytopenia were predominantly present in the MF subcluster and significantly greater in comparison with the remaining clusters, 47% versus 0% to 46% (P = .01) and 35% versus 0% to 21% (P < .001; supplemental Table 7). The remaining subset of 15 samples lacking translocations and clustering together only on the basis of downstream targets showed a gene signature with the top overexpressing genes corresponding to those overexpressed in the UAMS LB cluster, CST6, specific for the UAMS LB cluster, as well as RASGRP1 and PHACTR3 (supplemental Table 6). The MF cluster showed the lowest percentage of patients with bone lesions, 52% versus 62% to 100% in the remaining clusters (P = .004). This percentage was even lower in the LB subcluster, 50% versus 53% to 100% (P = .04; supplemental Table 7).
Six clusters were characterized by high frequencies of hyperdiploidy, ranging from 57% to 94% (supplemental Table 8). One of these clusters showed up-regulation of erythroid and myeloid markers as well as genes involved in cell-mediated immune response, humoral immune response, and antigen presentation. This cluster was indicated as the myeloid cluster. No distinct clinical features characterized this cluster, as was observed in the UAMS classification regarding the low percentage of patients having an immunoglobulin A subtype, β2M, and renal injury. However, bone marrow PC percentage before and after PC purification was significantly lower in this cluster in comparison with the remaining clusters, 30% versus 50% (P = .008) and 87% versus 91% (P < .001), respectively. The lower level of bone marrow plasmacytosis at diagnosis also was observed in the UAMS myeloid cluster.
The HY cluster showed hyperdiploidy in 94% of cases. This group was characterized by up-regulation of death receptor TNFSF10 (TRAIL); interferon-induced genes such as IFIT1, IFIT3, and IFI27; WNT antagonists FRZB and DKK1; glucosidase; beta; acid 3 (cytosolic) GBA3; and MYC proto-oncogene.
Two predominantly hyperdiploid clusters showed up-regulation of cancer testis antigens (CTA; supplemental Table 8). These include MAGEA3, MAGEA6F, MAGEA12, PAGE1, and GAGE12F. The presence calls of some CTA genes have been reported to correlate with significantly shorter event-free survival, such as CTAG1B, CTAG2, MAGEA1, MAGEA2, MAGEA3, and MAGEA6.25 The latter 2 were among the top 50 up-regulated genes in both clusters. In addition, cases with the 15% highest values of the high-risk index were predominantly observed in these clusters (P < .001). The high-risk index is determined on the basis of the published 17 gene model, which has been linked to early disease-related death (supplemental Figure 2).26 The difference between these 2 clusters was determined on the basis of overexpression of genes involved in cell cycle and proliferation in one of the clusters (Table 1; supplemental Table 5), with a significantly greater proliferation index (PI), on the basis of the calculated median expression of 11 genes associated with proliferation: TOP2A, BIRC5, CCNB2, NEK2, ANAPC7, STK6, BUB1, CDC2, C10orf3, ASPM, and CDCA1 (P < .001; supplemental Figure 3).27 This cluster was named PR cluster, described before by Zhan et al.15 The other CTA overexpressing cluster was mainly characterized by CTA genes and therefore named CTA cluster. Overlapping characteristics between the CTA and PR cluster were the overexpression of Aurora kinase A (AURKA), recently reported to be associated with a greater proliferation rate and poor outcome, which was significantly greater in both clusters in comparison with the remaining clusters (P < .001) and even greater in the PR compared with the CTA cluster (P = .2; supplemental Figure 4).28,29 Also BIRC5, another recently described gene of which the presence call has been associated with lower event-free survival and overall survival (OS) in newly diagnosed MM patients, was observed among the top 50 up-regulated genes in PR and CTA cluster.30 The CTA cluster has not been described as a distinct entity before and is therefore proposed as a new cluster.
The second new cluster was characterized by clear differential expression of genes involved in the nuclear factor κB (NFκB) pathway. Highly expressed NFκB genes include BCL10, TNFAIP3, IL8, GADD45B, NFKNIE, TNFIP1, NFKBIZ, IL2RG, CD40, and CD74 (Table 1; supplemental Table 5). In addition, the NFκB index as reported by Keats et al on the basis of the mean expression level of 4 probe sets corresponding to CD74, IL2RG, and TNFAIP3 (2×), as well as the NFκB index published by Annunziata et al, based on the mean expression of 11 probe sets (BIRC3, TNFAIP3, NFKB2, IL2RG, NFKBIE, RELB, NFKBIA, CD74, PLEK, MALT1, and WNT10A) were significantly greater in this cluster compared with the other clusters (P < .001; Figure 3A-B).31,32
NFκB index and regulators of NFκB activity. (A) NFκB index determined by Annunziata et al31 on the basis of the mean expression of 11 genes (BIRC3, TNFAIP3, NFKB2, IL2RG, NFKBIE, RELB, NFKBIA, CD74, PLEK, MALT1, WNT10A). (B) NFκB index determined by Keats et al32 on the basis of the mean expression of 4 genes (CD74, IL2RG, and TNFAIP3, 2×). Expression (log2) per cluster of negative regulators of the NFκB pathway: (C) TRAF3 and (D) CYLD. Expression (log2) of positive regulators of NFκB pathway: (E) CD40 and (F) NIK.
NFκB index and regulators of NFκB activity. (A) NFκB index determined by Annunziata et al31 on the basis of the mean expression of 11 genes (BIRC3, TNFAIP3, NFKB2, IL2RG, NFKBIE, RELB, NFKBIA, CD74, PLEK, MALT1, WNT10A). (B) NFκB index determined by Keats et al32 on the basis of the mean expression of 4 genes (CD74, IL2RG, and TNFAIP3, 2×). Expression (log2) per cluster of negative regulators of the NFκB pathway: (C) TRAF3 and (D) CYLD. Expression (log2) of positive regulators of NFκB pathway: (E) CD40 and (F) NIK.
On the basis of these characteristics, this cluster was termed NFκB cluster. Regulators of the NFκB pathway were further analyzed. CD40 and NIK (NFκB-inducing kinase) expression are both involved in activation of NFκB signaling. Only CD40 expression was significantly greater (P < .001), whereas the tumor necrosis factor receptor-associated factor 3 TRAF3, a negative NFκB regulator, showed significantly lower expression in the NFκB cluster (P = .004; Figure 3C,E).
The third new cluster consisted of 9 cases, and only 27 genes were differentially expressed, including overexpression of protein tyrosine phosphatase PTP4A3 (PRL3), protein tyrosine phosphatase, receptor-type, Z polypeptide 1 PTPRZ1, and suppressor of cytokine signaling 3 SOCS3. In lieu of any other characteristic, this cluster was termed PRL3 cluster. Chromosomal characteristics include hyperdiploidy in 75% of patients in this cluster; 1q gain was observed in 38% of patients. However, no 17p loss was observed. Strikingly, all patients in this cluster exhibited bone lesions. Furthermore, this cluster had the greatest percentage of patients in International Staging System stage I, 67% versus 19% to 57% in remaining clusters (P = .062). Expression levels of certain important genes in different clusters, such as MMSET, FGFR3, CCND1, INHBE, ASS1, VPREB3, MS4A1, NUAK1, and RND3 were successfully verified by quantitative reverse transcription polymerase chain reaction (supplemental Figure 5).
Validation in independent datasets and comparison with TC and UAMS classification
To validate clusters described here, we used the dataset upon which the UAMS classification is based (GSE2658). We performed prediction analysis of microarrays analysis of corresponding clusters using the UAMS cluster definitions (Table 3; supplemental Table 9).15 High Sn and Sp values were found for the classifiers of clusters CD-2, MS, MF, and HY, with Sn varying from 84% to 97% and Sp from 91% to 100%. Lower Sn was observed with classifiers for clusters CD-1 and PR. The classifier for the myeloid cluster consisting of 87 probe sets yielded the lowest Sn and Sp. The CTA, NFκB, and PRL3 cluster were novel clusters and could therefore not be validated by use of the UAMS cluster definitions.
Validation of clusters: PAM analysis generating classifiers for clusters validated in independent dataset GSE2658
| . | CD1 cluster . | CD2 cluster . | MF cluster . | MS cluster . | PR cluster . | HY cluster . | Myeloid cluster . |
|---|---|---|---|---|---|---|---|
| Samples in training set* | 13 | 34 | 32 | 33 | 15 | 77 | 39 |
| Samples in validation set (GSE2568)† | 28 | 60 | 37 | 68 | 47 | 116 | 145 |
| No. of probes in classifier | 106 | 45 | 35 | 8 | 86 | 56 | 87 |
| PPV, % | 57.60 | 80.60 | 97.10 | 94.30 | 61.70 | 76.70 | 58.50 |
| NPV, % | 97.60 | 98.30 | 98.90 | 99.40 | 95.10 | 94.60 | 81.70 |
| Sn, % | 67.90 | 90.00 | 89.20 | 97.10 | 61.70 | 84.80 | 42.80 |
| Sp, % | 96.40 | 96.30 | 99.70 | 98.80 | 95.10 | 91.30 | 89.40 |
| . | CD1 cluster . | CD2 cluster . | MF cluster . | MS cluster . | PR cluster . | HY cluster . | Myeloid cluster . |
|---|---|---|---|---|---|---|---|
| Samples in training set* | 13 | 34 | 32 | 33 | 15 | 77 | 39 |
| Samples in validation set (GSE2568)† | 28 | 60 | 37 | 68 | 47 | 116 | 145 |
| No. of probes in classifier | 106 | 45 | 35 | 8 | 86 | 56 | 87 |
| PPV, % | 57.60 | 80.60 | 97.10 | 94.30 | 61.70 | 76.70 | 58.50 |
| NPV, % | 97.60 | 98.30 | 98.90 | 99.40 | 95.10 | 94.60 | 81.70 |
| Sn, % | 67.90 | 90.00 | 89.20 | 97.10 | 61.70 | 84.80 | 42.80 |
| Sp, % | 96.40 | 96.30 | 99.70 | 98.80 | 95.10 | 91.30 | 89.40 |
Per cluster, the number of samples and total number of samples in the training set and test set are shown, the number of probe sets in the classifier, and the PPV, NPV, Sn, and Sp.
NPV indicates negative predictive value; PAM, prediction analysis of microarrays; PPV, positive predictive value; PR, proliferation-associated genes; Sn, sensitivity; and Sp, specificity.
The total of all samples in the training set was 320.
The total of all samples in the validation set was 414 except in the Myeloid cluster, where the total was 559.
In addition, our clustering was compared with the TC classification,12 and UAMS classification.15 To this end, TC criteria were used to assign the samples to TC classes and the published top 50 up-regulated and top 50 down-regulated probe sets that defined the 7 UAMS clusters to cluster our dataset (Tables 4 and 5). The MF subcluster, as defined previously, corresponded well to the Maf TC class; the MS cluster corresponded well to the 4p16 TC class. Samples from our CD-1/2 clusters corresponded to 11q13 and D1 classes.
Confusion matrices comparing with TC classification: H65 samples assigned to TC classes on the basis of TC criteria
| . | 11q13 . | D1 . | D2 . | D1 + D2 . | 6p21 . | maf . | 4p16 . | None . | Class error rate . |
|---|---|---|---|---|---|---|---|---|---|
| CD-1 | 4 | 6 | 1 | 2 | 0 | 0 | 0 | 0 | 0.69 |
| CD-2 | 25 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0.26 |
| MS | 0 | 0 | 0 | 0 | 0 | 0 | 33 | 0 | 0.00 |
| MF | 0 | 0 | 5 | 0 | 0 | 12 | 0 | 0 | 0.29 |
| HY | 1 | 66 | 0 | 6 | 0 | 2 | 0 | 2 | NA |
| PR | 0 | 8 | 5 | 0 | 1 | 0 | 0 | 1 | NA |
| LB | 0 | 3 | 9 | 1 | 0 | 1 | 0 | 1 | NA |
| NFκB | 1 | 23 | 6 | 1 | 0 | 1 | 1 | 4 | NA |
| CTA | 1 | 8 | 8 | 2 | 1 | 0 | 0 | 2 | NA |
| PRL3 | 0 | 4 | 5 | 0 | 0 | 0 | 0 | 0 | NA |
| Myeloid | 1 | 13 | 13 | 5 | 0 | 0 | 6 | 1 | NA |
| NA | 0 | 5 | 3 | 0 | 1 | 0 | 0 | 0 | NA |
| . | 11q13 . | D1 . | D2 . | D1 + D2 . | 6p21 . | maf . | 4p16 . | None . | Class error rate . |
|---|---|---|---|---|---|---|---|---|---|
| CD-1 | 4 | 6 | 1 | 2 | 0 | 0 | 0 | 0 | 0.69 |
| CD-2 | 25 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0.26 |
| MS | 0 | 0 | 0 | 0 | 0 | 0 | 33 | 0 | 0.00 |
| MF | 0 | 0 | 5 | 0 | 0 | 12 | 0 | 0 | 0.29 |
| HY | 1 | 66 | 0 | 6 | 0 | 2 | 0 | 2 | NA |
| PR | 0 | 8 | 5 | 0 | 1 | 0 | 0 | 1 | NA |
| LB | 0 | 3 | 9 | 1 | 0 | 1 | 0 | 1 | NA |
| NFκB | 1 | 23 | 6 | 1 | 0 | 1 | 1 | 4 | NA |
| CTA | 1 | 8 | 8 | 2 | 1 | 0 | 0 | 2 | NA |
| PRL3 | 0 | 4 | 5 | 0 | 0 | 0 | 0 | 0 | NA |
| Myeloid | 1 | 13 | 13 | 5 | 0 | 0 | 6 | 1 | NA |
| NA | 0 | 5 | 3 | 0 | 1 | 0 | 0 | 0 | NA |
Comparing HOVON-65/GMMG-HD4 and TC classification: HOVON-65/GMMG-HD4 samples are assigned to TC classes on the basis of expression of CCND1, MMSET, FGFR3, ITGB7, c-MAF, and CCND3 expression according to TC criteria. In lieu of normal bone marrow samples, for assignment of samples to D1, D1 + D2, and D2 classes, the threshold expression value of 30, as explained in “Methods,” was used as a reference value for normal bone marrow. Class error rate is depicted in the last column.
CTA indicates cancer testis antigens; HOVON, Dutch-Belgian Cooperative Trial Group for Hemato-Oncology; HY, hyperdiploid cluster; LB, low percentage of bone disease; NA, class error rate not determined because there were no corresponding subgroups; NFκB, nuclear factor kappa light-chain-enhancer of activated B cells; PR, proliferation-associated genes; and TC, translocation and cyclin D.
Confusion matrices comparing with UAMS classification: HOVON 65 samples clustered with 50 up- and 50 down-regulated UAMS probe sets
| . | CD-1 . | CD-2 . | MS . | MF . | HY . | PR . | LB . | NFκB . | Class error rate . |
|---|---|---|---|---|---|---|---|---|---|
| CD-1 | 8 | 0 | 0 | 0 | 1 | 0 | 4 | 0 | 0.38 |
| CD-2 | 1 | 28 | 0 | 0 | 1 | 0 | 4 | 0 | 0.18 |
| MS | 0 | 0 | 31 | 0 | 0 | 1 | 0 | 1 | 0.06 |
| MF | 0 | 0 | 0 | 17 | 0 | 0 | 0 | 0 | 0.00 |
| HY | 1 | 0 | 0 | 2 | 63 | 4 | 6 | 1 | 0.18 |
| PR | 2 | 0 | 0 | 2 | 2 | 9 | 0 | 0 | 0.40 |
| LB | 0 | 0 | 0 | 2 | 0 | 0 | 13 | 0 | 0.13 |
| NFκB | 0 | 8 | 0 | 1 | 1 | 1 | 0 | 26 | 0.30 |
| CTA | 0 | 5 | 0 | 0 | 5 | 8 | 0 | 4 | NA |
| PRL3 | 0 | 0 | 0 | 0 | 0 | 4 | 3 | 2 | NA |
| Myeloid | 4 | 0 | 7 | 12 | 4 | 5 | 1 | 6 | NA |
| NA | 3 | 0 | 0 | 0 | 1 | 0 | 5 | 0 | NA |
| . | CD-1 . | CD-2 . | MS . | MF . | HY . | PR . | LB . | NFκB . | Class error rate . |
|---|---|---|---|---|---|---|---|---|---|
| CD-1 | 8 | 0 | 0 | 0 | 1 | 0 | 4 | 0 | 0.38 |
| CD-2 | 1 | 28 | 0 | 0 | 1 | 0 | 4 | 0 | 0.18 |
| MS | 0 | 0 | 31 | 0 | 0 | 1 | 0 | 1 | 0.06 |
| MF | 0 | 0 | 0 | 17 | 0 | 0 | 0 | 0 | 0.00 |
| HY | 1 | 0 | 0 | 2 | 63 | 4 | 6 | 1 | 0.18 |
| PR | 2 | 0 | 0 | 2 | 2 | 9 | 0 | 0 | 0.40 |
| LB | 0 | 0 | 0 | 2 | 0 | 0 | 13 | 0 | 0.13 |
| NFκB | 0 | 8 | 0 | 1 | 1 | 1 | 0 | 26 | 0.30 |
| CTA | 0 | 5 | 0 | 0 | 5 | 8 | 0 | 4 | NA |
| PRL3 | 0 | 0 | 0 | 0 | 0 | 4 | 3 | 2 | NA |
| Myeloid | 4 | 0 | 7 | 12 | 4 | 5 | 1 | 6 | NA |
| NA | 3 | 0 | 0 | 0 | 1 | 0 | 5 | 0 | NA |
Comparing HOVON-65/GMMG-HD4 and UAMS classification: The top 50 up- and top 50 down-regulated genes of all 7 UAMS clusters were used to cluster the HOVON-65/GMMG-HD4 samples. NFκB is marked because the gene signature was very weak, on the basis of the expression of BCL10 and high NFκB index.
CTA indicates cancer testis antigens; HOVON, Dutch-Belgian Cooperative Trial Group for Hemato-Oncology; HY, hyperdiploid cluster; LB, low percentage of bone disease; NA, class error rate not determined because there were no corresponding subgroups; NFκB, nuclear factor kappa light-chain-enhancer of activated B cells; PR, proliferation-associated genes; and UAMS, University of Arkansas for Medical Sciences.
Because of the limited nature of the TC classification, the classes did not compare with any of our other clusters. Regarding the UAMS classification, we confirmed the 7 described clusters. In addition, we identified a cluster showing a high NFκB index and overexpression of BCL10, which was observed among the top up-regulated genes in our NFκB cluster. Furthermore, we observed that HOVON65/GMMG-HD4 samples originally present in the NFκB cluster were now shifted to this extra cluster, which therefore probably represents the NFκB cluster.
For additional validation of our classification, including the novel described clusters, we used 2 independent datasets, ie, the UAMS data and a separate set of data from relapsed MM cases included in the APEX/SUMMIT/CREST trials to which we applied our normalization methods and gene selection criteria.18
From the UAMS dataset, 548 CEL files were made available. After performing quality control with NUSE, 10 arrays were excluded. The 2730 most variable genes of the remaining 538 samples were selected as described. A total of 1255 genes overlapped with the HOVON65/GMMG-HD4 gene set. Clustering resulted in the identification of the translocation clusters, HY, PR, and myeloid cluster (supplemental Figure 6). We identified an NFκB cluster with up-regulation of genes involved in the NFκB pathway such as TNFAIP3, CFLAR, NFKB2, PLEK, IL2RG, and CD74 and a high NFκB index, and additionally genes up-regulated in the UAMS LB cluster such as CST6, PHACTR3, RASGRP1, IL6R, BIK, and EDN1. This cluster clustered next to the MF cluster with subsequent up-regulation of RND3, AHNAK, CCND2, and ARL4C. Down-regulated genes included CCR2, TNFSF10, DKK1, FRZB, and interferon-induced genes. This cluster consists of UAMS LB and contaminated samples. Furthermore we identified a PRL3 cluster on the basis of overexpression of PRL3 and SOCS3. No separate CTA cluster was identified. On the basis of the 100 up/down-regulated genes characterizing the CTA cluster, we observed that 7% samples (n = 37) with highest/lowest expression of these genes were found mainly within the UAMS PR cluster (n = 15), MS (n = 5), HY (n = 5) and contaminated cluster (n = 5; data not shown).
The APEX/SUMMIT/CREST dataset consisted of 264 gene expression profiles of relapsed MM patients; all of the U133A and B arrays used showed good NUSE values. Gene selection by the criteria used in the present study yielded 2248 probe sets. The overlap with HOVON65/GMMG-HD4 gene set was 1002 genes. Again, the translocation clusters, HY, PR, and myeloid cluster were identified (supplemental Figure 7). In addition we detected an NFκB cluster with up-regulation of NFκB related genes such as TNFAIP3, IL2RG, CFLAR, NFKBIA, LMNA, and KLF6 but also genes up-regulated in the UAMS LB cluster, such as PHACTR3, RASGRP1, IL6R, and CST6 and genes frequently up-regulated in the MF cluster, such as AHNAK, CCND2, and ARL4C. Furthermore, we identified a PRL3 cluster on the basis of overexpression of CCND2, PRL3, and PTPRZ1 and a CTA-like cluster. The CTA like cluster was defined by a different CTA profile than observed in the CTA cluster in our dataset, with up-regulation of SSX3, SSX4B, and MAGE2B.
A classifier for translocations
Samples with available FISH data were used to develop class predictors for translocations. Results are shown in Table 6 and more in detail in supplemental Table 10. For translocation t(11;14) the lowest classification error generated a classifier of only 5 probe sets among which multiple probe sets of CCND1 and KCNMB2, yielding a Sn of 83% and Sp of 97%. For translocation t(4;14) a 25-probe set classifier generated a Sn of 100% and Sp of 97%. Because samples with t(14;16) and t(14;20) clustered together, a combined t(14;16)/t(14;20) classifier of 18 probe sets was generated, which yielded a Sn of 100% and Sp of 99%.
Validation of translocations: PAM analysis generating classifiers to predict translocations t(11;14), t(4;14), and t(14;16)/t(14;20)
| . | t(4;14) . | t(11;14) . | t(14;16)/t(14;20) . |
|---|---|---|---|
| Samples in training set | |||
| Translocations | 26 | 24 | 5 |
| Total | 153 | 143 | 143 |
| Samples in test set | |||
| Translocations | 11 | 13 | 4 |
| Total | 80 | 75 | 73 |
| No. of probes in classifier | 25 | 5 | 18 |
| PPV, % | 84.6 | 83.3 | 80.0 |
| NPV, % | 100.0 | 96.8 | 100.0 |
| Sn, % | 100.0 | 83.3 | 100.0 |
| Sp, % | 97.1 | 96.8 | 98.5 |
| Correctly classified, % | 97.5 | 94.7 | 98.6 |
| . | t(4;14) . | t(11;14) . | t(14;16)/t(14;20) . |
|---|---|---|---|
| Samples in training set | |||
| Translocations | 26 | 24 | 5 |
| Total | 153 | 143 | 143 |
| Samples in test set | |||
| Translocations | 11 | 13 | 4 |
| Total | 80 | 75 | 73 |
| No. of probes in classifier | 25 | 5 | 18 |
| PPV, % | 84.6 | 83.3 | 80.0 |
| NPV, % | 100.0 | 96.8 | 100.0 |
| Sn, % | 100.0 | 83.3 | 100.0 |
| Sp, % | 97.1 | 96.8 | 98.5 |
| Correctly classified, % | 97.5 | 94.7 | 98.6 |
Per translocation, the number of samples harboring the specific translocation and total number of samples in the training set and test set, the number of probe sets in the classifier, and the PPV, NPV, Sn, and Sp are shown.
NPV indicates negative predictive value; PAM, prediction analysis of microarrays; PPV, positive predictive value; Sn, sensitivity; and Sp, specificity.
Discussion
Gene expression profiling was performed on 320 bone marrow PCs obtained at diagnosis from primarily white North European patients included in the HOVON-65/GMMG-HD4 trial. The objective of this randomized, open-label, phase 3 trial was to evaluate the efficacy of bortezomib in newly diagnosed MM cases.33
Unsupervised hierarchical clustering resulted in a subdivision in 10 clusters, of which 3 novel clusters have not been described previously. These are the NFκB, CTA, and PRL3 clusters. The NFκB cluster was characterized by hyperdiploidy in 66% of cases, demonstrated clear differential expression of genes involved in the NFκB pathway. A subgroup characterized by genes involved in NFκB signaling and antiapoptosis was previously reported in an analysis restricted to hyperdiploid myeloma samples.16 NFκB signaling is crucial in the pathogenesis of myeloma,31,32 involving both inactivating and activating mutations that primarily result in constitutive activation of the noncanonical NFκB pathway.31,32 Cases with high expression values of probe sets corresponding to NFκB genes CD74, IL2RG, and TNFAIP3 show a better response to bortezomib but no change in progression-free survival (PFS), whereas patients with low TRAF3 expression show a better response to bortezomib and a prolonged PFS.32 The NFκB index determined by CD74, IL2RG, and TNFAIP3 was significantly greater in our NFκB cluster. In keeping with this finding, negative regulators of NFκB signaling showed reduced expression, for instance, TRAF3, whereas genes involved in stimulating NFκB activity, for instance, CD40, were found to be increased. The unexpectedly high expression of CYLD in the NFκB cluster, a negative regulator of the NFκB pathway, may be explained by the effect of inflammation stimuli, such as tumor necrosis factor-α, that activate the NFκB pathway, which in turn could induce CYLD.34 High NFκB activity also characterized the MF cluster, in which predominantly overexpression of NIK was observed. In conclusion, various mechanisms appear to be responsible for the high NFκB activity observed in the NFκB and MF cluster.
The second distinct novel subgroup observed here is the CTA cluster. This resembles the PR cluster concerning the presence of poor prognostic markers such as CTA genes, the highest percentage of patients with an extreme high-risk index, and overexpression of AURKA and BIRC5. Although proliferation associated genes such as AURKA and BIRC5 as well as cell cycle genes such as CDC2 and CDC42 were among the top up-regulated genes in the CTA cluster, the CTA cluster showed a significantly lower PI in comparison with the PR cluster. An explanation could be the absence of several genes representing the PI. Alternatively, the fold change difference of present genes between the CTA cluster and the remaining clusters was lower than the fold change difference in PR cluster versus remaining clusters. Besides features such as a greater percentage of 1q gain and a significantly greater PI, no clinical features distinguished the CTA from the PR cluster. This CTA cluster might represent a group of samples going through a transition phase from hyperdiploid myeloma to a PR signature. Evidence for this comes from the comparison with the UAMS classification, in which CTA characterizing genes did not cluster samples to one cluster but were found among samples in the UAMS PR, MS, HY, and contaminated cluster.
Overexpression of protein tyrosine phosphatases PRL3, PTPRZ1, and SOCS3 characterized the third novel cluster, the PRL3 cluster. Greater PRL3 expression was found in bone marrow PCs from patients with newly diagnosed monoclonal gammopathies than in PCs from healthy donors and significantly greater in the UAMS PR, LB and MS groups. Silencing of PRL3 by siRNA impaired SDF-1–induced migration of MM cells, but no influence on cell-cycle distribution or cell proliferation was observed.35 PTPRZ1 is involved in the regulation of protein phosphorylation and plays a critical function in signal transduction, cell growth, differentiation, and oncogenesis.36,37 SOCS3 is a cytokine-inducible negative regulator of cytokine signaling. The expression of this gene is induced by various cytokines, including interleukin-6 (IL-6), IL-10, and interferon gamma (IFN-γ). Transfection of myeloma cell lines with SOCS3 showed protection from growth suppression by IFN-α. IL-6 induced by IFN-α may play an important role in the growth and survival of myeloma cells, and up-regulated SOCS3 by IL-6 may be at least partially responsible for the IL-6–mediated inhibition of IFN-α signaling in myeloma cells.38-40 PRL-3 overexpression in mammalian cells was reported to inhibit angiotensin-II–induced cell calcium mobilization and promote cell growth. Absence of poor prognostic factors such as 17p loss, combined with low values for high-risk index, proliferation index, and AURKA expression, suggests patients within this cluster may have less severe disease (P ≤ .001). Indeed, the frequency of International Staging System I was markedly greater in this cluster than in the other clusters.
Comparison with existing classifications confirmed the 7 clusters described in the UAMS classification, CD-1, CD-2, MS, MF, HY, PR, and LB.15 Furthermore, Zhan et al15 reported a group of cases with a myeloid signature that was excluded from further analyses. The patients in this so-called contaminated cluster showed less disease activity and performed better on treatment, with significantly prolonged event-free survival and OS. We retained the group of patients with a myeloid signature in the gene expression analysis. These samples clustered together, clinically characterized by a significantly lower level of bone marrow plasmacytosis.
We validated our classification by applying our sample and gene selection criteria to 538 UAMS raw data files representing newly diagnosed MM cases and 264 APEX/SUMMIT/CREST raw data files representing relapsed MM cases. Sample clustering resulted in confirmation of clusters CD1, CD2, MS, MF, HY, PR, and in addition a myeloid cluster in both datasets. In both sets we observed a combined NFκB/LB cluster, showing overexpression of genes involved in the NFκB signaling pathway, but also of PHACTR3, RASGRP1, IL6R, and downstream targets of MAF/MAFB. In our clustering, LB samples were found as a subcluster of the MF cluster, on the basis of expression of MAFB and c-MAF downstream targets. This LB subcluster might represent a subgroup corresponding to an earlier stage of disease, as suggested by the lack of poor prognostic markers, such as thrombocytopenia and elevated LDH. The HOVON65/GMMG-HD4 NFκB cluster and LB subcluster were observed as 2 distinct clusters; except for a high NFκB index, no overlap in differentially expressed genes or percentage of bone lesions was observed. Merging of these 2 clusters in independent datasets might be possible on the basis of a weaker expression of NFκB-related genes showing lower fold changes relative to LB cluster genes and MAF/MAFB downstream targets. However, the presence of a cluster mainly characterized by an NFκB index cannot be disputed.
We also confirmed the PRL3 cluster on the basis of the overexpression of at least PRL3 among the top 10 genes showing the highest fold change difference, and SOCS3, CCND2, and/ or PTPRZ1. A CTA-like cluster was found in the APEX dataset characterized by a different CTA expression profile compared with our CTA cluster. No CTA cluster was detected in the UAMS dataset; samples with a CTA signature clustered within the PR, HY, MS, and myeloid cluster. Although the CTA cluster showed a high robustness index of 0.77 (range, 0-1) in our dataset, we were not able to consistently confirm this cluster. Population as well as technical differences might play a role in this. Despite these differences as well as differences in myeloma status, PC selection procedures, and technical procedures, we were able to divide myeloma patients into 9 robust and consistent clusters. Naturally, changing the gene list does influence the size and composition of clusters. The 5% most variable gene list was selected on the basis of the division of translocations over the clusters
In the UAMS classification, the PR, MS, and MF clusters were defined as high-risk groups with a significantly lower PFS and OS.15 In agreement to this report, we demonstrated associations between clusters PR, MS, MF, the novel cluster CTA, and poor prognostic factors, such as increased high-risk index and elevated LDH. The authors of future analyses will evaluate the prognostic impact of the current defined clusters in the HOVON65/GMMG-HD4 trial.
Finally, the ability to predict the primary translocations is important for diagnostic purposes. Because these classifiers were found to be robust, the development of methods that complement or even replace FISH techniques will be relevant and subject to future studies. In conclusion, the classification described here showed good correlation to the previously described classifications in MM. Yet, 3 new clusters were identified, one of which signifies the involvement of NFκB signaling in MM.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank J. Shaughnessy from the University of Arkansas for Medical Sciences (UAMS), Little Rock, for providing the CEL files from the UAMS dataset.
This work was supported by a clinical research grant from the European Hematology Association, EMCR Translational Research Grant, Janssen Orthobiotech, and MSCNET.
Authorship
Contribution: A. Broyl collected data, performed research, analyzed and interpreted the data, and wrote the manuscript; D.H. designed research and critically reviewed the manuscript; H.L. is an investigator for H65/GMMG-HD4 and reviewed the manuscript; Y.d.K. collected data and performed research; J.P. participated in data analysis and interpretation; A.J. performed cytogenetics and FISH; U.B. is the coordinating data manager for the H65/GMMG-HD4 trial; A. Buijs performed cytogenetics and FISH and is responsible for central review of cytogenetics; M.S.-K. performed cytogenetics and FISH and is responsible for central review of cytogenetics; H.B.B. performed cytogenetics and FISH; E.V. is an investigator for H65/GMMG-HD4 and reviewed the manuscript; S.Z. is an investigator for H65/GMMG-HD4 and reviewed the manuscript; M.-J.K. is an investigator for H65/GMMG-HD4 and reviewed the manuscript; B.v.d.H. performed statistical analysis and participated in trial data preparation; L.e.J. is a coordinating data manager for the H65/GMMG-HD4 trial; G.M. provided the CEL files from the APEX/SUMMIT/CREST trials for analysis and critically reviewed the manuscript; H.G. designed research and is a trial coordinator for GMMG group for H65/GMMG-HD4; M.v.D. participated in the data analysis and interpretation and critically reviewed the manuscript; and P.S. designed research and is a principal investigator of the H65/GMMG-HD4 trial and the performed research.
Conflicts-of-interest disclosure: G.M. has declared a financial interest in Millennium Pharmaceuticals Inc, whose product was object of study in the HOVON 65/ GMMG-HD4. G.M. is currently employed by Millennium Pharmaceuticals Inc. P.S. has served on advisory boards of Johnson & Johnson and Millennium Pharmaceuticals. H.G. has served on advisory boards of Johnson & Johnson. The remaining authors declare no competing financial interests.
Correspondence: A. Broyl, Department of Hematology, Rm No. Ee1300, Erasmus University Medical Centre, Dr. Molewaterplein 50, 3015 GE Rotterdam, The Netherlands; e-mail: a.broyl@erasmusmc.nl.