Ikaros plays an important role in the control of differentiation and proliferation of all lymphoid lineages. The expression of short isoforms lacking DNA-binding motifs alters the differentiation capacities of hematopoietic progenitors, arresting lineage commitment. We sought to determine whether molecular abnormalities involving the IKZF1 gene were associated with resistance to tyrosine kinase inhibitors (TKIs) in Ph+ acute lymphoblastic leukemia (ALL) patients. Using reverse-transcribed polymerase chain reaction, cloning, and nucleotide sequencing, only the non–DNA-binding Ik6 isoform was detected in 49% of Ph+ ALL patients. Ik6 was predominantly localized to the cytoplasm versus DNA-binding Ik1 or Ik2 isoforms, which showed nuclear localization. There was a strong correlation between nonfunctional Ikaros isoforms and BCR-ABL transcript level. Furthermore, patient-derived leukemia cells expressed oncogenic Ikaros isoforms before TKI treatment, but not during response to TKIs, and predominantly at the time of relapse. In vitro overexpression of Ik6 strongly increased DNA synthesis and inhibited apoptosis in TKI-sensitive cells. Genomic sequence and computational analyses of exon splice junction regions of IKZF1 in Ph+ ALL patients predicted several mutations that may alter alternative splicing. These results establish a previously unknown link between specific molecular defects that involve alternative splicing of the IKZF1 gene and the resistance to TKIs in Ph+ ALL patients.
Introduction
The Philadelphia (Ph) chromosome, the result of a reciprocal translocation fusing the abelson (ABL) proto-oncogene from chromosome 9 with the breakpoint cluster region (BCR) sequences on chromosome 22, represents the most common cytogenetic abnormality detected in human leukemias. It is considered the hallmark of chronic myeloid leukemia (CML) but is detectable in 20% to 30% of adult acute lymphoblastic leukemia (ALL), with the incidence rising to more than 50% of patients 50 years of age or older.1 The BCR-ABL–encoded isoforms p210 and p190 recruit and activate multiple pathways that transduce oncogenic signals, leading to increased survival, enhanced proliferation, impaired migration and adhesion, and arrested differentiation of hematopoietic progenitors.2,3 The development of imatinib mesylate,4 a specific BCR-ABL tyrosine kinase inhibitor (TKI), has enhanced the management of Ph+ ALL patients, especially in elderly adults.5,,–8 Despite initial hematologic responses, Ph+ ALL patients treated with imatinib eventually relapse and die of their diseases.9,–11 The main mechanism of acquired resistance is the acquisition of point mutations in the ABL kinase domain that impair imatinib binding to various degrees. Other causes have pharmacokinetic or pharmacodynamic12 bases or are the result of the development of other secondary genetic abnormalities.13 Because these mechanisms are common with CML, in Ph+ ALL patients BCR-ABL–independent pathways may drive both the disease and TKI resistance. Pre-mRNA splicing, an important determinant of the protein repertoire in human cells but also a natural source of cancer-causing errors in gene expression, may have a role in Ph+ ALL. Human leukemia shows a heterogeneous pattern of spliced isoforms of Ikaros (IKZF1, Lyf-1) proteins, which are critical for the development of lymphocytes and other hematopoietic lineages.14,15 The IKZF1 gene is composed of 8 exons, of which 7 exons (2-8) are coding16 and allow different isoforms to be generated by alternative splicing. All isoforms contain 2 C-terminal zinc fingers, which mediate functionally indispensable protein-protein interactions between Ikaros family members. The isoforms differ in the number of N-terminal zinc finger motifs, which dictate sequence specificity and DNA affinity. Only the isoforms with at least 3 of 4 of the N-terminal zinc fingers are capable of binding to DNA sites that contain the GGGGA or AGGAA motif. Overexpression of dominant-negative Ikaros isoforms is associated with a decrease in expression of the normally predominant long isoforms, as previously reported in cases of T- and B-cell childhood ALL,17,–19 as well as during blast crisis of CML,20 suggesting a role for Ikaros as a tumor suppressor gene in human B-cell malignancies. In this study, we analyzed the differential expression patterns of Ikaros isoforms in Ph+ ALL patients treated with imatinib and dasatinib. We investigated a potential correlation between the different Ikaros isoforms expressed by alternative splicing and the BCR-ABL transcript levels. In addition, we determined whether the molecular abnormalities involved in the IKZF1 gene splicing could be associated with resistance to TKIs. We found a strong correlation (P < .001) between the non–DNA-binding Ik6 isoform and the BCR-ABL transcript levels, and at the time of resistance Ik6 was the major isoform expressed, suggesting its possible role in the loss of response to TKIs. Because alternative splicing of exons requires the presence of splicing cis-elements present in the exon and within the flanking introns, a genomic sequence analysis was performed in search of mutations in the regions surrounding the splice sites. Some mutations were identified and predicted by computational analyses to alter cis-splicing elements
Methods
Patients and cell lines
The study was approved by the ethical committee of the Institute of Hematology and Medical Oncology L. and A. Seràgnoli. Informed consent was obtained in accordance with the Declaration of Helsinki. Expression patterns of Ikaros isoforms were retrospectively studied in bone marrow and peripheral blood samples collected after informed consent from 47 adult patients with Ph+ ALL: 16 treated with imatinib, 31 treated with dasatinib; all of these were after imatinib failure (n = 8) and as front-line treatment (n = 23). The median age of the patients was 55 years (range, 18-76 years). Diagnosis of all ALL cases was made on the basis of morphologic, biochemical, and immunologic features of the leukemic cells. In addition, the human lymphoblastoid SD-1, the human B-cell precursor leukemia SUP-B15, and the human B-cell precursor leukemia BV-173 cell lines were included in the analysis. Human cell lines were obtained from DMSZ (DeutsheSammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany) and maintained in culture following the DMSZ recommendations. The cells were incubated at 37°C in a humidified atmosphere flushed with 5% CO2. Ph + leukemic cells were treated with 10 μM imatinib (Novartis, Basel, Switzerland), 1 μM nilotinib (Novartis), and 1 μM dasatinib (Bristol-Myers Squibb, Princeton, NJ) for 18 hours.
RNA isolation and reverse-transcribed polymerase chain reaction analysis
Mononuclear cells were separated by Ficoll-Hypaque density gradient centrifugation, and samples were stored at −190°C in RPMI 1640 with 20% fetal bovine serum (FBS) and 10% dimethylsulfoxide or in guanidine thiocyanate at −80°C, as needed. Total cellular RNA was extracted from cells using the RNeasy total RNA isolation kit (QIAGEN, Valencia, CA) according to the instructions of the manufacturer. Of the total RNA sample, 1 μg was used for cDNA synthesis in the reverse transcriptase reaction (RT) as previously described.21 Polymerase chain reaction (PCR) using primers specific for exon 2 (Ik1: 5′-CACATAACCTGAGGACCATG-3′) and exon 8 (Ik2: 5′-AGGGCTTTAGCTCATGTGGA-3′) of IKZF1 was performed using 5 μL cDNA, 5 μL Amplitaq Buffer II (10×, Applied Biosystems, Foster City, CA), 5 μL Amplitaq MgCl2 (25 mM, Applied Biosystems), 1 μL dNTP Mix (2.5 mM, GE Healthcare, Little Chalfont, United Kingdom), 25 pmol of each primer, 0.5 μL of the AmpliTaq Gold (5 U/μL, Applied Biosystems), and distilled water in a final volume of 50 μL. PCR conditions were: 95°C for 5 minutes for denaturation, then 95°C for 30 seconds, 57°C for 30 seconds, 72°C for 90 seconds, repeated for 35 cycles, followed by 72°C for 7 minutes. To increase the sensitivity and to better characterize the specific Ikaros isoforms, Ik3 (5′-ATGGATGCTGATGAGGGTCAAGAC-3′) and Ik4 (5′-GATGGCTTGGTCCATCACGTGG-3′) primers were also used with the following conditions: 95°C for 5 minutes, then 95°C for 30 seconds, 62°C for 30 seconds, 72°C for 30 seconds, repeated for 35 cycles, followed by 72°C for 7 minutes. The RNA integrity was confirmed by PCR amplification of the GAPDH mRNA, which is expressed ubiquitously in human hematopoietic cells.
Genomic analysis
Genomic DNA was isolated using QIAamp DNA Blood Mini Kit (QIAGEN). The genomic sequence surrounding the predominant splice donor and acceptor sites at the exon 2/3, exon 3/4, exon 7/8 splice junctions from leukemic patient samples, and cell lines were performed in search of mutations.
To amplify the region surrounding the splice junction of IKZF1 exons 2 and 3 for the 5′ splice site, we used a primer sense from exon 2 (F1: 5′-ATGGATGCTGATGAGGGTCAAGAC-3′) and an antisense primer (R1: 5′-gccagtccaacaaaaccgcac-3′) within the intronic region. For the 3′ splice site, we used F2: 5′-ctctaagcagaatacctggtg-3′and R2: 5′-CTCATCTGGAGTATCGCTTAC-3′ on exon 3. For the 3′ splice site of the exon 3/4 junction, we used F3: 5′-cggaaatctgaaagggaactg-3′ and R3: 5′-GATGAAGAGAATGGGCGTGC-3′ on exon 4. To amplify the region surrounding the splice junction of IKZF1 exons 7 and 8, F4: 5′-CGTGCTGGACAGACTAGCAAG-3′ on exon 7 and R4: 5′-ctaggagcattgcccagagtag-3′ were used for the 5′ splice site. The 3′ splice site was tested using the intronic primer F5: 5′-cgggcctgccaactacagag-3′ and R5: 5′-TCCCACGTGATGGACCAAGCCATC-3′ on exon 8. PCR was performed using 100 ng genomic DNA in a 50-μL reaction as described in “RNA isolation and reverse-transcribed polymerase chain reaction analysis.” The long-range PCR cycling parameters were as follows: 95°C for 5 minutes (complete denaturation), followed by 30 cycles at 95°C for 30 seconds, 61°C for 30 seconds, extension at 72°C for 70 seconds, with an additional extension at the end for 7 minutes (for F1-R1 and F4-R4); 95°C for 5 minutes (complete denaturation), followed by 35 cycles at 95°C for 30 seconds, 57°C for 30 seconds, extension at 72°C for 80 seconds, with an additional extension at the end for 7 minutes (for F2-R2, F3-R3, F5-R5). Generation of any cryptic splice site for any nucleotide variation was assessed using the RESCUE22 (relative enhancer and silencer classification by unanimous enrichment) approach. The RESCUE-ESE23,24 (exonic splicing enhancer) approach was used to predict whether exonic mutations disrupt putative exon splicing enhancers (http://genes.mit.edu/burgelab/rescue-ese/), whereas the RESCUE-ISE22 (intronic splicing enhancer) approach was applied to predict which intronic motifs may enhance or repress exon splicing (http://genes.mit.edu/acescan2/index.html).
Sequencing analysis
PCR products were purified with QIAquick PCR purification kit (QIAGEN) or QIAquick gel extraction kit according to the manufacturer's protocol and were directly sequenced using an ABI PRISM 3730 automated DNA sequencer (Applied Biosystems) and a Big Dye Terminator DNA sequencing kit (Applied Biosystems). In some cases, the PCR products were subcloned into the PCR2.1-TOPO vector using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA). The cloned PCR products were purified and sequenced as described in this section.
Western blot analysis
Cells were lysed with sample buffer (2% sodium dodecyl sulfate in 125 mM of Tris HCl, pH 6.8). Cell lysates were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis on 12% gels and then transferred to nitrocellulose membranes (GE Healthcare). The blots were incubated for 60 minutes in Odyssey blocking buffer before incubation overnight (4°C) with polyclonal anti-Ikaros antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Blotted proteins were detected and quantified using the Odyssey infrared imaging system LI-COR.
Subcellular localization studies using confocal laser scanning microscopy
The subcellular localization of Ikaros protein(s) was examined through immunofluorescence and confocal laser scanning microscopy. Cytospins were prepared using cell lines and leukemic cells from patients with Ph+ ALL as previously described.25 Representative digital images were processed using Photoshop software (Adobe Systems, Mountain View, CA).
Monitoring of BCR-ABL transcript levels
BCR-ABL transcript levels were detected at diagnosis and during follow-up using a standardized real-time quantitative PCR (RQ-PCR) method that was established within the framework of the EU Concerted Action.26 RQ-PCR was performed on an ABI PRISM 7900 Sequence Detector (PerkinElmer Life and Analytical Sciences, Waltham, MA). The quantification principles and procedure using the TaqMan probe have been previously described.21 All real time reverse-transcribed polymerase chain reaction (RT-PCR) experiments were performed in duplicate.
Construction of pcDNA-Ik6 expression vector
The complete Ik6 coding sequence was amplified by PCR; PCR products were isolated from agarose gel, purified using a Spin Column Kit (QIAGEN) and restriction digested with EcoRI and XbaI. The digested fragment was cloned into the pcDNA3.1 expression vector (Invitrogen, Carlsbad, CA) treated with EcoRI and XbaI. The recombinant plasmid carrying the Ik6 gene was transfected into Escherichia coli DH5-α strain and grown in LB (Luria Bertani) medium supplemented with ampicillin (100 μg/mL) overnight at 37°C. Preparation of plasmid DNA was performed using the QiafilterTM Plasmid Maxi Kit (QIAGEN) according to the manufacturer's instructions.
Transfection with pcDNA-Ik6
For transfection, 1 × 106 SUP-B15 cells were collected on day 2, phosphate-buffered saline (PBS)–washed, and resuspended in 1 mL serum-free medium. Cells were transiently transfected with 16 μg pcDNA-IK6 using the geneJammer (Invitrogen)-mediated DNA transfection technique following the manufacturer's instructions. In addition, a green fluorescent protein (GFP) empty vector (16 μg pEGFP) was used as the internal control. GFP expression was detected within 48 hours after transfection using a fluorescent microscope (Qfluoro software; Leica Microsystem, Deerfield, IL). GFP-positive cells were sorted.
Apoptosis assay
Apoptosis was evaluated by flow cytometry for the detection of annexin V–positive cells. Transfected and treated cells were labeled with annexin V conjugated with fluorescein isothiocyanate and propidium iodide. Briefly, cells were washed once in PBS and once in 1× binding buffer, then 5 μL each of annexin V–fluorescein isothiocyanate and propidium iodide was added to the cells. Cells were incubated at room temperature for 15 minutes, after which 300 μL 1× binding buffer was added and cells were analyzed by flow cytometry. Early apoptotic cells were defined as weakly annexin V–positive and propidium iodide–negative, whereas late apoptotic cells were those strongly annexin V–positive, including those cells that were both annexin V– and propidium iodide–positive. Experiments were performed in triplicate.
Proliferation assay by incorporation of 3H thymidine
Cells were seeded at 10 × 104 concentration in RPMI with 10% FBS for 18 hours and then starved. After 12 hours, 10% FBS is added. After 6 hours, 1 μCi/mL 3H thymidine was added (GE Healthcare). After 24 hours of incubation, the 3H thymidine was removed. Cells were then washed with PBS and 5% trichloric acetic acid and resuspended with NaOH. The amount of incorporated 3H thymidine was detected using a β counter. Experiments were performed in triplicate.
Clonogenic assay or colony formation assay
Cells (5 × 105) were plated in methylcellulose (Methocult 4230, StemCell Technologies, Vancouver, BC) supplemented with 10 ng/mL granulocyte colony-stimulating factor, 10 ng/mL granulocyte-macrophage colony-stimulating factor, 10 ng/mL interleukin-3, and 300 U/mL erythropoietin. Cells were plated in 35-mm Petri dishes and, for each sample, 4 dishes were prepared. Cells were maintained at 37°C in a fully humidified incubator with 5% CO2 for 14 days. The growth of hematopoietic colonies was assessed by counting with optic microscopy after 14 days of culture. The experiment was repeated 3 times.
Definitions
Hematologic complete remission was defined as: bone marrow cellularity of at least 20% and containing less than 5% blast cells, peripheral blood smears without blasts, no evidence of extramedullary involvement from leukemia, and polymorphonuclear cells more than 1.5 × 109/L and platelets more than 100 × 109/L. Cytogenetic response (CgR) was classified in complete (CCgR, 0% Ph+ metaphases), partial (PCgR, 1%-35% Ph+ metaphases), minor (MCgR, 36%-94% Ph+ metaphases), and none or minimal response (NCgR, > 95% abnormal metaphases). Hematologic relapse was defined as the reappearance of more than 5% bone marrow blasts or in peripheral blood in hematologic complete remission patients confirmed with a second bone marrow aspirate one week after the first test. Cytogenetic relapse was defined whenever a complete or a partial CgR was lost to minor or minimal/none.
Statistical analysis
To estimate whether the difference in the levels of BCR-ABL transcripts were statistically significant between different patient groups, we performed a nonparametric Mann-Whitney test, and a P value less than .05 was considered statistically significant. Comparisons of frequencies were made with the χ2 test or the Fisher exact test, as appropriate. All statistical calculations were performed and graphs constructed using GraphPad Prism 4 (GraphPad, San Diego, CA).
Results
The non–DNA-binding isoform Ik6 is highly expressed in ALL Ph+ patients
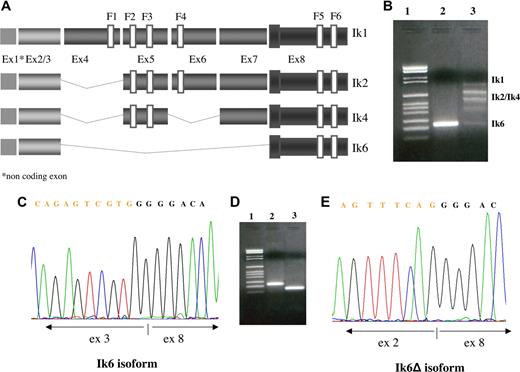
RT-PCR and nucleotide sequencing were used to examine leukemia cells from patients with Ph+ ALL and human cell lines for the expression of different IKZF1 transcripts. Primers derived from exons 2 and 8 generated multiplied bands corresponding to the alternatively spliced products of the Ikaros pre-mRNA transcript. The largest band we found corresponded to the Ik1 isoform (945 bp with our insider primers) containing exons 2 to 8 and encoding 4 Cys-2 to His-2 zinc fingers at its N-terminus (F1, F2, F3, and F4) and 2 at its C-terminus (F5 and F6). The band of 684 bp was identified as the Ik2 isoform, sharing 3 of the 4 N-terminal zinc fingers (F2, F3 and F4), whereas the band of 558 bp corresponded to the Ik4 isoform, in which exon 4 and exon 6 were lost. The smallest isoform was Ik6 (255 bp), in which all N-terminal zinc fingers involved in DNA-binding were lost and exon 3 was juxtaposed with exon 8 (Figure 1A). The majority of the analyzed samples expressed several isoforms of Ikaros at the same time (Figure 1B lane 3). In 43 of 47 (91%) patients, the Ik6 isoform was detected at least once; and in 23 patients (49% of the entire study population), it was the predominant isoform (Figure 1B lane 2). In the remaining patients, Ik6 was associated with Ik1, Ik2, and Ik4 isoforms in 20 patients (43%). The expression of only the functional DNA-binding isoforms was detected in 4 (9%) Ph+ ALL patients. In one patient (male, 43 years, resistant to imatinib), we found only a novel Ik6 isoform (Ik6Δ), previously unreported, in which exon 2 was juxtaposed with exon 8 maintaining the frame (Figure 1D lane 3 and Figure 1E). The BV-173 cell line was positive for the Ik6 isoform, whereas the SD-1 cell line showed expression of only the DNA-binding isoforms. Our findings from RT-PCR and sequencing analysis clearly demonstrated elevated expression of the Ik6 isoform in patients with Ph+ ALL, associated with a decrease in expression of the functional predominant long isoforms (Ik1 and Ik2).
Expression of different Ikaros isoforms. (A) Schematic diagram of the full-length Ikaros cDNA and the different isoforms produced in our samples by alternative splicing. (F) N-terminal zinc-fingers show DNA-binding activity, and C-terminal zinc fingers mediate dimerization of the protein. Ex indicates exon. (B) Bands generated by RT-PCR using primers derived from exons 2 and 8 and corresponding to the alternatively spliced products of the Ikaros pre-mRNA transcript. The left lane is the molecular size marker, marker VI Roche; lane 2, Ik6 expression; lane 3, coexpression of Ik1, Ik2, Ik4, and Ik6. (C) Sequence analysis of the Ik6 isoform in which exon 3 is juxtaposed with exon 8 and the novel Ik isoform (Ik6Δ) previously unreported, in which exon 3 is directly juxtaposed with exon 8 maintaining the frame (E). (D) Bands generated by RT-PCR; in lane 1 is the molecular size marker, marker VI Roche; in lane 2 is Ik6 (255 bp); in lane 3 is Ik6Δ (135 bp).
Expression of different Ikaros isoforms. (A) Schematic diagram of the full-length Ikaros cDNA and the different isoforms produced in our samples by alternative splicing. (F) N-terminal zinc-fingers show DNA-binding activity, and C-terminal zinc fingers mediate dimerization of the protein. Ex indicates exon. (B) Bands generated by RT-PCR using primers derived from exons 2 and 8 and corresponding to the alternatively spliced products of the Ikaros pre-mRNA transcript. The left lane is the molecular size marker, marker VI Roche; lane 2, Ik6 expression; lane 3, coexpression of Ik1, Ik2, Ik4, and Ik6. (C) Sequence analysis of the Ik6 isoform in which exon 3 is juxtaposed with exon 8 and the novel Ik isoform (Ik6Δ) previously unreported, in which exon 3 is directly juxtaposed with exon 8 maintaining the frame (E). (D) Bands generated by RT-PCR; in lane 1 is the molecular size marker, marker VI Roche; in lane 2 is Ik6 (255 bp); in lane 3 is Ik6Δ (135 bp).
Ik6 protein is expressed in an abnormal subcellular compartmentalization
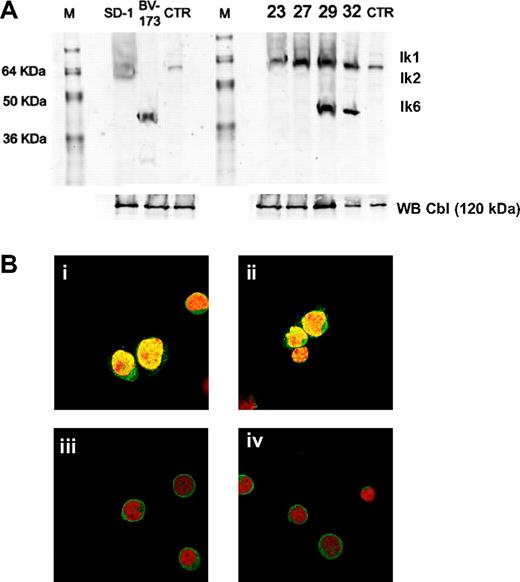
Ikaros protein expression was studied in cell lines and primary leukemic cells from patients with Ph+ ALL by Western blot analysis. Patients who expressed full-length Ikaros, as determined by RT-PCR analysis, showed a 57-kDa immunoreactive protein that corresponded in size to Ik1 and a 47-kDa immunoreactive protein that corresponded to Ik2. In contrast, we confirmed the presence of a smaller immunoreactive protein band of approximately 37 to 40 kDa, which corresponded in size to Ik6, in patients who were positive for this non–DNA-binding isoform from RT-PCR (Figure 2A). The absence of an abundant Ik1 or Ik2 was not caused by a generalized proteolytic degradation because a 120-kDa Cbl protein was detected by Western blot analysis in the same whole cell lysates. These results confirmed the aberrant expression of Ikaros proteins as previously determined by RT-PCR. In addition, we compared the subcellular compartmentalization of Ikaros proteins in leukemic cells from patients expressing the DNA-binding isoforms Ik1 and Ik2 to those of patients expressing the non–DNA-binding isoform Ik6. The nuclei, but not the cytoplasm, of patients with Ik1 and Ik2 isoforms were stained brightly by the anti-Ikaros antibody, as evidenced by a distinct punctuate green fluorescent staining pattern (Figure 2Bi,ii). In contrast to this nuclear localization, Ikaros proteins were expressed predominantly in the cytoplasm of leukemic cells from patients expressing the Ik6 isoform (Figure 2Biii,iv).
Ikaros protein expression and compartmentalization. (A) Anti-Ikaros Western blots of whole cell lysates from leukemic cell lines (SD-1 and BV-173), leukemic bone marrow mononuclear cells from Ph+ ALL at diagnosis (ID 23, ID 27, ID 29, and ID 32). As a control, we used a Jurkat cell nuclear extract. The positions corresponding to the migration patterns of Ik-1 (∼ 57 kDa), Ik-2 (∼ 47 kDa), and Ik-6 (∼ 37 kDa) proteins are indicated. Anti-Cbl Western blots of the whole cell lysates were performed as controls. (B) Expression and subcellular localization of Ikaros proteins in leukemic cells from Ph+ ALL patients. In all images, cells were stained with an Ikaros antibody (green) and with propidium iodide (red) to visualize the DNA. (Bi,ii) Confocal images of leukemic cells from patients expressing full-length Ikaros isoforms showed the characteristic multifocal nuclear localization pattern of Ikaros. (Biii,iv) Confocal images of leukemic cells express the Ik6 isoform and show cytoplasmic expression of Ikaros (eg, bright green fluorescent rim surrounding the toto-labeled red nuclei).
Ikaros protein expression and compartmentalization. (A) Anti-Ikaros Western blots of whole cell lysates from leukemic cell lines (SD-1 and BV-173), leukemic bone marrow mononuclear cells from Ph+ ALL at diagnosis (ID 23, ID 27, ID 29, and ID 32). As a control, we used a Jurkat cell nuclear extract. The positions corresponding to the migration patterns of Ik-1 (∼ 57 kDa), Ik-2 (∼ 47 kDa), and Ik-6 (∼ 37 kDa) proteins are indicated. Anti-Cbl Western blots of the whole cell lysates were performed as controls. (B) Expression and subcellular localization of Ikaros proteins in leukemic cells from Ph+ ALL patients. In all images, cells were stained with an Ikaros antibody (green) and with propidium iodide (red) to visualize the DNA. (Bi,ii) Confocal images of leukemic cells from patients expressing full-length Ikaros isoforms showed the characteristic multifocal nuclear localization pattern of Ikaros. (Biii,iv) Confocal images of leukemic cells express the Ik6 isoform and show cytoplasmic expression of Ikaros (eg, bright green fluorescent rim surrounding the toto-labeled red nuclei).
Ik6 expression is correlated with the percentage of blast cells
In some cases, we detected in our samples by RT-PCR and Western blot analysis the expression of the non–DNA-binding isoform Ik6 both alone and in association with functional Ik1 and Ik2 isoforms. Because these samples contained a mixed population of cells with a variable percentage of blast cells, it is not clear whether the larger isoforms result from normal cells present in the samples or from blast cells that have acquired a mutation in one IKZF1 allele that affects isoform expression. To address this issue, we correlated the different expression patterns of Ikaros isoforms to the percentage of blast cells in each sample (Table 1). We observed that the median percentage of blast cells in patients who expressed the Ik6 isoform alone was 90% (range, 54%-100%) versus 57% (range, 5%-94%) of patients coexpressing both the short Ik6 isoform and functional Ik1 and Ik2 isoforms (P = .001). Our hypothesis was that the coexpression of functional Ik1, Ik2, and Ik6 is the result of normal cells present in the samples that express larger isoforms, whereas blast cells only express the short Ik6 isoform. Furthermore, to better understand whether the non–DNA-binding Ik6 could be specific for blast cells, we also analyzed the expression of Ikaros isoforms in samples from normal volunteers (n = 10) and from patients with CML in remission (n = 10). As expected, in these patients we never detected the expression of Ik6 alone, using RT-PCR analysis.
Correlation between the different expression patterns of Ikaros isoforms and the percentage of blast cells
| Sample . | No. (%) . | Blast cell percentage, median (range) . |
|---|---|---|
| Patients expressing Ik6/Ik6Δ alone | 23 (49) | 94 (54-100) |
| Patients expressing Ik1, Ik2/Ik4 alone | 4 (8) | 95 (85-100) |
| Patients coexpressing Ik6 and Ik1, Ik2/Ik4 | 20 (43) | 57 (5-94) |
| Sample . | No. (%) . | Blast cell percentage, median (range) . |
|---|---|---|
| Patients expressing Ik6/Ik6Δ alone | 23 (49) | 94 (54-100) |
| Patients expressing Ik1, Ik2/Ik4 alone | 4 (8) | 95 (85-100) |
| Patients coexpressing Ik6 and Ik1, Ik2/Ik4 | 20 (43) | 57 (5-94) |
Ik6 expression strongly correlated with BCR-ABL transcript level in Ph+ ALL patients
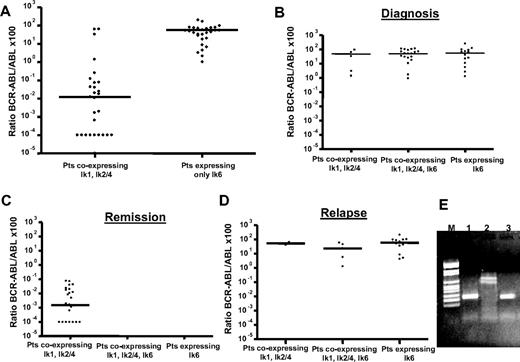
After we showed a correlation between the percentage of blast cells and expression of the Ik6 isoform in the sample, the next step was to investigate whether this alteration depended on Bcr-Abl activity. For this purpose, BCR-ABL transcript levels were monitored in all patients at different times during therapy with imatinib or dasatinib. First, we demonstrated that dominant Ik6 does not depend on the type of BCR-ABL transcript rearrangement because it can be detected both in patients carrying a p210 (9 patients) and in patients with a p190 (14 patients) oncoprotein. Second, we found a strongly statistically significant difference between the BCR-ABL/ABL ratios for patients who expressed the DNA-binding isoforms and those patients who expressed only the Ik6 dominant-negative isoform (Figure 3A). The median ratio of BCR-ABL/ABL for the first group was 0.01 (range, 64.49-0.0001) versus 61.26 (range, 251.62-1.21) for the second group (P < .001). These results showed that the expression of Ik6 strongly correlated with the BCR-ABL transcript levels.
Comparison between the BCR-ABL/ABL ×100 ratios of the patients who expressed the Ik1, Ik2 DNA-binding isoforms alone and patients expressing the Ik6 dominant-negative isoform alone, independent of the clinical disease status. (A)There was a strong correlation between Ik6 expression and BCR-ABL transcript levels (P < .001). Expression pattern of Ikaros isoforms at different checkpoints during treatment with tyrosine kinase inhibitor: at diagnosis (B), during remission (C), and at the time of relapse (D) (both hematologic/cytogenetic and molecular). In patients with relapse and high levels of BCR-ABL transcript, Ik6 was the major isoform expressed. (E) RT-PCR analysis at baseline (lane 1), during remission (lane 2), and at the time of relapse (lane 3).
Comparison between the BCR-ABL/ABL ×100 ratios of the patients who expressed the Ik1, Ik2 DNA-binding isoforms alone and patients expressing the Ik6 dominant-negative isoform alone, independent of the clinical disease status. (A)There was a strong correlation between Ik6 expression and BCR-ABL transcript levels (P < .001). Expression pattern of Ikaros isoforms at different checkpoints during treatment with tyrosine kinase inhibitor: at diagnosis (B), during remission (C), and at the time of relapse (D) (both hematologic/cytogenetic and molecular). In patients with relapse and high levels of BCR-ABL transcript, Ik6 was the major isoform expressed. (E) RT-PCR analysis at baseline (lane 1), during remission (lane 2), and at the time of relapse (lane 3).
Ik6 expression is associated in vivo with resistance to imatinib and dasatinib
The mechanism of aberrant overexpression of the non–DNA-binding Ik6 isoform has been previously demonstrated to be restricted to certain forms of leukemia,27 such as blast crisis of CML20 or acute lymphoblastic/myeloid leukemia,28,–30 suggesting a role in pathogenesis of leukemia; however, its role in leukemia resistance has not yet been demonstrated. In this study, to provide new evidence for a possible link between the expression of Ik6 and the resistance to TKIs in Ph+ ALL patients, we examined the expression pattern of Ikaros isoforms at different checkpoints during treatment with TKIs: at baseline, during remission, and at the time of hematologic relapse. On the same samples and at the same time points, we performed an RQ-PCR analysis to quantify the BCR-ABL transcript levels. We divided the patients into 3 groups according to the Ikaros isoforms expressed (DNA-binding isoforms (A), coexpression of both DNA and non–DNA-binding isoforms (B), non–DNA-binding isoforms, and (C) the phase of disease (diagnosis, hematologic/cytogenetic remission and hematologic/cytogenetic relapse; Figure 3; Table 2). Forty-one patients were evaluated for molecular analysis of BCR-ABL and Ikaros at diagnosis. Expression of Ik6 was detected in 36 patients (88%); however, only 14 patients (34%) expressed Ik6 alone versus 12% of patients who expressed only Ik1, Ik2, and Ik4 isoforms (P = not significant), suggesting that the expression of Ik6 alone was not correlated with diagnosis. Furthermore, at diagnosis we did not observe any significant difference in the BCR-ABL transcript levels among the 3 patient groups (P = .399). During remission after tyrosine kinase inhibitor therapy, in all patients we detected only DNA-binding isoforms with a median BCR-ABL/ABL ×100 value of 0.001 (range, 0.07-0.0001). It is important to note that we detected the Ik6 isoform in no patients, reinforcing the idea that Ik6 expression is correlated with the amount of BCR-ABL transcript. Therefore, in patients with relapse and high levels of BCR-ABL transcript, Ik6 became the major isoform expressed both alone (67%) and in association with other isoforms (24%) versus 9% of patients expressing predominantly Ik1, Ik2, and Ik4 isoforms. We did not observe any difference in the expression of Ik6 at the time of relapse between patients who were resistant to imatinib (10 of 16, 63%) and patients who were resistant to dasatinib (4 of 8, 50%). Overall, these results demonstrate that the expression of the Ik6 isoform is associated with resistance to both imatinib and dasatinib.
Correlation between the Ik isoforms (DNA-binding isoform [A], Ik1-Ik2; coexpression of both DNA-binding and non–DNA-binding isoforms [B]; dominant non–DNA-binding isoform Ik6 [C]), the status of disease and the BCR-ABL transcript levels, expressed as a ratio (BCR-ABL/ABL) and monitored by RQ-PCR
| Disease phase . | A . | B . | C . | |||
|---|---|---|---|---|---|---|
| No. (%) of patients . | Ratio, % . | No. (%) of patients . | Ratio, % . | No. (%) of patients . | Ratio, % . | |
| Diagnosis | 5 (12) | 33.41 | 22 (54) | 51.40 | 14 (34) | 69.01 |
| Remission | 27 (100) | 0.001 | ||||
| Relapse | 2 (9) | 52.66 | 5 (24) | 23.33 | 14 (67) | 58.75 |
| Disease phase . | A . | B . | C . | |||
|---|---|---|---|---|---|---|
| No. (%) of patients . | Ratio, % . | No. (%) of patients . | Ratio, % . | No. (%) of patients . | Ratio, % . | |
| Diagnosis | 5 (12) | 33.41 | 22 (54) | 51.40 | 14 (34) | 69.01 |
| Remission | 27 (100) | 0.001 | ||||
| Relapse | 2 (9) | 52.66 | 5 (24) | 23.33 | 14 (67) | 58.75 |
Ik isoforms are expressed as n; ratios are expressed as BCR-ABL/ABL (%; median).
TKIs induced apoptosis in full-length Ikaros-expressing cells but not in Ik6 expressing-cells
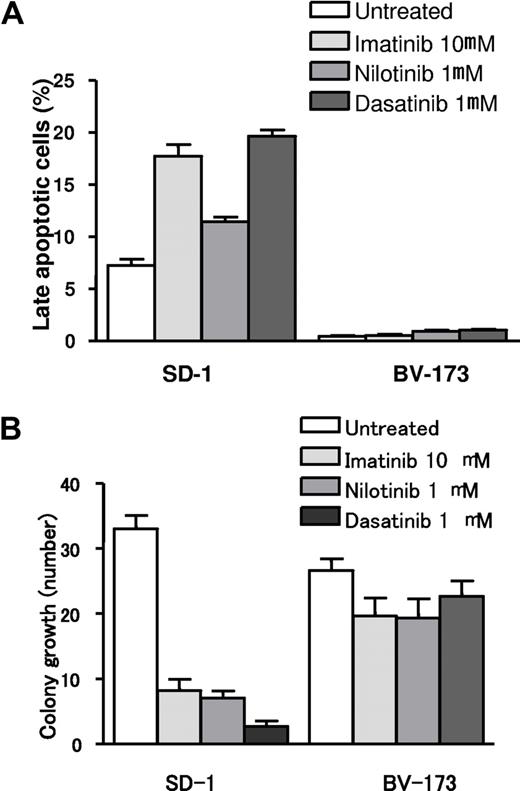
To assess whether in vitro the selection of cells with higher expression of short Ikaros isoforms affects the sensitivity to TKIs in Ph+ ALL, we treated full-length Ikaros-expressing cells (SD-1) and Ik6-expressing cells (BV-173) with different TKIs (imatinib, nilotinib, and dasatinib). As shown in Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) and Figure 4A, in SD-1 cells we observed a mean increase in apoptotic cells (18% ± 2%, 11% ± 1%, and 20% ± 1% after incubation with imatinib, nilotinib, and dasatinib, respectively). By contrast, in Ik6-expressing cells, TKIs did not increase the number of apoptotic cells compared with the control samples. These results were confirmed by colony growth assay (Figure 4B). The in vitro incubations of SD-1 cells with imatinib, nilotinib, and dasatinib resulted in a marked inhibition of colony growth (mean number of colonies was 32 ± 4 for the untreated cells and 8 ± 3, 7 ± 2, and 3 ± 1.5 after incubation with imatinib, nilotinib, and dasatinib, respectively). In contrast, no significant inhibition of colony growth was detected after incubation of Ik6-expressing cells with TKIs (mean number of colonies was 26 ± 3 for the untreated cells and 18 ± 4, 20 ± 4, and 22 ± 5 with imatinib, nilotinib, and dasatinib, respectively).
Apoptosis and colony growth assays in full-length Ikaros- and Ik6-expressing cells after treatment with TKIs. (A) Apoptotic rates of untreated and TKI (imatinib, nilotinib, and dasatinib)-treated cells. Treatment with TKIs increased the apoptotic rate in Ikaros full-length expressing cells (SD-1), whereas no differences were identified in Ik6 expressing cells (BV-173) between untreated and TKI-treated cells. (B) Mean value of colony growth from SD-1 and BV-173 cell lines in control cells (untreated) and after incubation with imatinib, nilotinib, and dasatinib.
Apoptosis and colony growth assays in full-length Ikaros- and Ik6-expressing cells after treatment with TKIs. (A) Apoptotic rates of untreated and TKI (imatinib, nilotinib, and dasatinib)-treated cells. Treatment with TKIs increased the apoptotic rate in Ikaros full-length expressing cells (SD-1), whereas no differences were identified in Ik6 expressing cells (BV-173) between untreated and TKI-treated cells. (B) Mean value of colony growth from SD-1 and BV-173 cell lines in control cells (untreated) and after incubation with imatinib, nilotinib, and dasatinib.
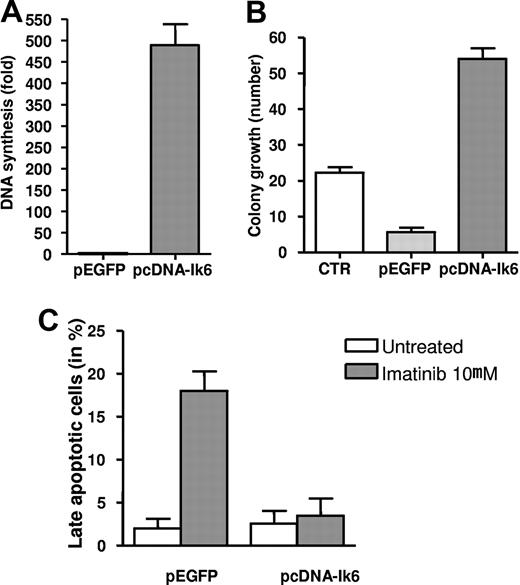
Transfection of Ik6 in an Imatinib-sensitive Ik6-negative Ph+ ALL cell line decreases sensitivity to TKIs
To mechanistically demonstrate whether in vitro the overexpression of Ik6 impairs response to TKIs and contributes to resistance, we transfected an imatinib-sensitive Ik6-negative Ph+ ALL cell line (SUP-B15) with the complete Ik6 DNA coding sequence. First, we assessed the proliferation activity of the cells transfected with Ik6 (pcDNA-Ik6) or with the empty vector (pEGFP). Interestingly, we observed a strong increase in DNA synthesis (500-fold increase in the mean value with an SD of 85) in cells transfected with pcDNA-Ik6 (Figure 5A). Similar results were obtained by colony growth assay, as shown in Figure 5B. The expression of the Ik6 isoform in SUP-B15 cells strikingly increased colony growth (mean number of colonies was 8 ± 2 and 60 ± 5 for the empty vector and for cells transfected with Ik6, respectively). Moreover, in pcDNA-Ik6 SUP-B15 transfected cells, the number of apoptotic cells did not significantly increase after imatinib incubation compared with the samples transfected with the empty vector (mean values of 3.5% ± 3% vs 18% ± 4%).
Transfection of Ik6 in an imatinib-sensitive Ik6-negative Ph+ ALL cell line. Induction of DNA synthesis (A) and colony growth (B) in pcDNA-Ik6 SUP-B15 transfected cells. Mean value of colony growth from control cells and after incubation with imatinib, nilotinib, and dasatinib. (C) Apoptotic rates evaluated by FACS for the detection of annexin V–positive cells in pcDNA-Ik6 SUP-B15–transfected cells and in cells transfected with the empty vector (pEGFP) after incubation with imatinib.
Transfection of Ik6 in an imatinib-sensitive Ik6-negative Ph+ ALL cell line. Induction of DNA synthesis (A) and colony growth (B) in pcDNA-Ik6 SUP-B15 transfected cells. Mean value of colony growth from control cells and after incubation with imatinib, nilotinib, and dasatinib. (C) Apoptotic rates evaluated by FACS for the detection of annexin V–positive cells in pcDNA-Ik6 SUP-B15–transfected cells and in cells transfected with the empty vector (pEGFP) after incubation with imatinib.
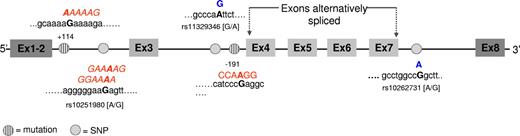
Cis-acting mutations may be responsible for alternative splicing of Ikaros transcript
Amplification and genomic sequence analyses of the exon splice junction regions were performed in search of mutations in the regions spanning the cryptic splice site, as well as the predominant 5′ (donor) or 3′ (acceptor) splice sites in Ph+ ALL patients who expressed the Ik6 isoform. In these regions, we observed several different single nucleotide polymorphisms (SNPs): rs10251980 [A/G] in the exon 2/3 splice junction, rs11329346 [A/G] in the intron 3, and rs10262731 [A/G] in the exon 7/8 splice junction in 50%, 16%, and 36% of analyzed patients, respectively (Figure 6). Two different single nucleotide substitutions, which were not recognized as SNPs, were also observed. The first one was a substitution of a G with an A in the exon 2/3 splice junction at position +114 counting from the first nucleotide of the intron 2, in 80% of Ik6-expressing patients. The other one was always a G/A substitution in the exon 3/4 splice junction at position −191 to the end of intron 3, identified in 30% of patients. Furthermore, we examined expression of the SNP affecting the third base of the triplet codon for a proline (CCC or CCA) in the highly conserved bipartite activation region of exon 8 (A or C at position 1170 numbering from the translation start site of Ik-1, GenBank accession number U40462, NM_006060.331 ). This region is conserved in the various Ikaros splice variants, thereby allowing typing of all Ikaros isoforms. To assess whether SNPs or point mutations in intron or exon sequences could affect alternative splicing to create or abolish splicing enhancers and silencers, we applied a computational RESCUE-ESE/ISE23,24 approach, by which we analyzed the effect of sequence variation entering both the wild-type and the variant sequence into the input window. In our samples, the mutation at position + 114 in intron 2 was identified as a putative ESE creating the AAAAAG motif (Figure 6; the mutated base is in bold type.). ESE alteration events were also observed in the case of the rs10251980 SNP in the same intron where the allelic variant containing an A instead of a G was part of the GGAAAA or GAAAAG ESE motif. In the case of the mutation at the 3′ of intron 3, the RESCUE approach identified the putative exon splicing silencer (ESS) site CCAAGGT.
Results from genome sequence analysis of the splice junction regions. Point mutations and single nucleotide polymorphisms (SNPs) are represented in different colors. The letters written in red represent the ESE/ESS motifs identified using the RESCUE-ESE/ISE computational approach (http://genes.mit.edu/burgelab/rescue-ese/), and the mutated base is in bold type. Nucleotide substitutions that are not predicted to create ESE/ESS motifs are indicated in blue.
Results from genome sequence analysis of the splice junction regions. Point mutations and single nucleotide polymorphisms (SNPs) are represented in different colors. The letters written in red represent the ESE/ESS motifs identified using the RESCUE-ESE/ISE computational approach (http://genes.mit.edu/burgelab/rescue-ese/), and the mutated base is in bold type. Nucleotide substitutions that are not predicted to create ESE/ESS motifs are indicated in blue.
Discussion
The rationale for many studies to investigate whether normal Ikaros expression and function might be altered in human hematologic malignancies is based on the fact that Ikaros functions as a critical regulator of normal lymphocyte development32 and the rapid development of leukemia in mice expressing non–DNA-binding isoforms. An excess of short Ikaros isoforms has been described in leukemic cells obtained from infant and children B and T ALLs,18,19,28,33 in de novo adult B ALL,34 in cells from transformed CML20 and from de novo acute myelomonocytic and monocytic leukemias,30,35 demonstrating that aberrant regulation of splicing is a new mechanism of activation of an oncogene in ALL. Because expression of non–DNA-binding Ikaros isoforms during early lymphopoiesis may dysregulate normal lymphocyte development predisposing lymphocyte precursors to second hits and leukemic transformation and/or progression, we analyzed for the first time the expression pattern of Ikaros isoforms in Ph+ ALL patients who were resistant to imatinib and dasatinib for the purpose of determining whether Ikaros aberrant spliced isoforms correlated with the BCR-ABL transcript levels and whether they were associates with resistance to TKIs.
Using RT-PCR and sequencing analysis, we found a different pattern of spliced Ikaros isoforms in Ph+ ALL patients resistant to imatinib and dasatinib. In 43 of 47 (91%) patients, the Ik6 isoform lacking all 4 N-terminal zinc-fingers responsible for DNA-binding was detected; and in 23 of these patients (49%), it was the predominant isoform. The presented data are consistent with the evidence from previous studies that reported an aberrant expression of spliced isoforms of Ikaros in some subtypes of human leukemia.18,28,–30,33,34,36 Moreover, in a recent study, a comparison of the genome-wide gene expression profiles of normal B-cell subsets and BCR-ABL pre-B lymphoblastic leukemia cells by serial analysis of gene expression (SAGE) showed loss of B-lymphoid identity and aberrant expression of myeloid lineage-specific molecules in leukemia cells.37 In the same report, BCR-ABL was demonstrated to induce the expression of dominant-negative Ik6, which contributes to lineage infidelity observed in BCR-ABL pre-B lymphoblastic leukemia cells. Focusing on Ph+ ALL in this study, we confirmed not only that BCR-ABL may induce the expression of the Ik6 isoform, but we also demonstrated that Ik6 does not depend on the type of BCR-ABL arrangement because it was detected in patients carrying both a p210 and a p190 oncoprotein. Furthermore, the molecular monitoring of minimal residual disease showed for the first time in vivo that the dominant-negative Ik6 expression correlated with the BCR-ABL transcript levels. Patient-derived leukemia cells expressed dominant-negative Ik6 before, but not during response to TKIs, and predominantly at the time of relapse. To mechanistically demonstrate whether in vitro the overexpression of Ik6 impairs the response to TKIs and contributes to resistance, we transfected an imatinib-sensitive Ik6-negative Ph+ ALL cell line (SUP-B15) with the complete Ik6 DNA coding sequence. We confirmed that expression of Ik6 strongly increases proliferation and inhibits apoptosis in TKI sensitive cells establishing a previously unknown link between specific molecular defects that involve the IKZF1 gene and the resistance to TKIs in Ph+ ALL patients.
Expression of aberrant Ikaros isoforms in leukemic cells may result from cis-sequence alterations or from leukemia-associated alterations in trans-acting factors. Although the splice sites themselves are located in the introns, additional sequences that enhance splicing from an exonic location are also known. These ESEs are short oligonucleotide sequences that are often recognized by proteins of the SR (serine-arginine) family. To establish which mechanism may be responsible for the spliced Ikaros isoforms in our patients, amplification and genomic sequence analysis of the exon splice junction regions were performed in search of mutations. We applied for the first time on the IKZF1 gene a computational method, RESCUE-ESE/RESCUE-ISE, to determine which sequences were capable of functioning as splicing enhancers, finding that some mutations in our samples may function as splicing enhancer or silencers. Given the overwhelming number of ESEs in human genes, it is probable that not all of them are the binding targets of the SR proteins; consequently, it is difficult to reliably determine the true incidence of ESE alteration within the set of mutations predicted to alter ESEs. The presence of some predicted ESEs may frequently occur in human genes just by chance and in this case the non–DNA-binding Ikaros isoforms may be the result of changes in trans-acting splicing regulators or to changes at the genomic level. This aspect is under investigation.
In conclusion, our data demonstrate that the posttranscriptional regulation of alternative splicing of IKZF1 pre-mRNA is defective in the majority of Ph+ ALL patients treated with TKIs. Our hypothesis is that the overexpression of Ik6 resulting in B-cell differentiation blocking may contribute to TKI resistance, which allows an extended time frame during which leukemia cells acquire secondary transforming events. During the manuscript revision, Mullighan et al38 described a deletion of IKZF1 involving exons 4 to 7, which are responsible for the generation of the Ik6 transcript variant isoform in Ph+ ALL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by European LeukemiaNet, COFIN 2003 (M.B.), Associazione Italiana contro le Leucemie, Associazione Italiana per la Ricerca sul Cancro, Fondazione Del Monte di Bologna e Ravenna, Fondo per gli Investimenti della Ricerca di Base 2006, and Ateneo 60% grants.
Authorship
Contribution: I.I. designed research, performed molecular analysis, and drafted the manuscript; A.L. performed molecular analysis; D.C., F.M., and F.A. performed molecular analysis, immunofluorescence analysis, and experiments of transfection and drug sensitivity; E.O., S.P., C.P., P.P.P., P.G., S.S., M.A., A.P., G.S., F.P., A.V., S.C., R.F., and G.P. contributed to the development of the study and data collection; G.B. and A.B. performed Western blot analysis; and M.B. and G.M. designed and supervised the study and gave final approval to the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Giovanni Martinelli, Molecular Biology Unit, Institute of Hematology and Medical Oncology Seràgnoli, University of Bologna, Via Massarenti 9, 40138 Bologna, Italy; e-mail: giovanni.martinelli2@unibo.it.