Abstract
Acute myeloid leukemia (AML) is the most common type of acute leukemia in adults. AML is a heterogeneous malignancy characterized by distinct genetic abnormalities. Recent discoveries have highlighted an additional important role of dysregulated epigenetic mechanisms in the pathogenesis of the disease. In contrast to genetic changes, epigenetic modifications are frequently reversible, which provides opportunities for targeted treatment using specific inhibitors. In this review, we will provide an overview of the current state of epigenetics and epigenetic therapy in AML and will describe perspectives on how to identify promising new approaches for epigenetic targeted treatment.
Introduction
Acute myeloid leukemia (AML) is the most common type of acute leukemia in adults.1 Despite current treatment protocols involving intensive chemotherapy and the judicious use of stem cell transplantation, AML is still fatal in approximately half of younger patients and in about 80% of patients over the age of 60 as a result of primary refractoriness, relapse, or treatment-related mortality.2 Consequently, there is a need for more specific and less toxic drugs that are rationally designed to target leukemia-specific abnormalities.
The term epigenetics refers to changes in gene expression that are inheritable by cell division but not caused by changes in the DNA sequence itself.3 Examples include DNA modifications, such as cytosine methylation, modifications of histone proteins, or RNA-associated gene silencing. Recent discoveries have shed light on the important role of dysregulated epigenetic mechanisms in the pathogenesis of AML.4-6 Several genes playing key roles in epigenetic regulation show recurrent somatic alterations in AML. It is however apparent that epigenetic dysregulation in AML is more widespread than can be explained by recurrent somatic mutations alone.7 For instance, disturbed genome-wide patterns of DNA methylation also exist in AML subtypes not linked to mutations in known epigenetic modifiers.8 In addition to somatic changes in coding regions of genes, alterations in enhancer elements have been reported that perturb normal regulation of gene expression.9
Epigenetic modifications are frequently reversible. Because of this inherent plasticity, epigenetic dysregulation offers potential avenues for targeted treatment using specific inhibitors of histone-modifying proteins or proteins driving or maintaining DNA methylation. In support of this, both experimental evidence as well as clinical observations underline the concept that epigenetic mutations and alterations contribute to a preleukemic state but are usually not sufficient to cause full-blown acute leukemia.10 This would indicate that these defects are already present in early clones and targeting those abnormalities may help to eradicate the disease.
In this review, we give an overview of the current state of epigenetics and epigenetic therapy in AML and will describe perspectives on how to identify promising new approaches for epigenetic-targeted treatment.
Modes of epigenetic (dys)regulation in AML
We first provide an outline of various modes of epigenetic regulation in hematopoietic cells and how these may be disturbed in AML. This has been summarized in Table 1. Several recent reviews have provided excellent descriptions of the various abnormalities in epigenetic-regulating enzymes in hematopoietic malignancies in general and AML in particular, to which the reader is referred for more detail.4-6
DNA methylation
The process of DNA methylation involves the addition of a methyl group to the 5-carbon position of cytosines in CpG dinucleotides, yielding 5-methylcytosine. Aberrant patterns of DNA methylation in malignancies were initially particularly studied in the context of so-called “CpG islands” in gene promoters. Hypermethylation of cytosines in CpG islands is associated with silencing of tumor-suppressor genes.47 It is now increasingly clear that aberrant DNA methylation patterns outside CpG islands may be equally important in leukemogenesis, and that hypomethylation may be as relevant as hypermethylation. Newly identified regions of interest include gene bodies and so-called “CpG shores” (regions at 2 kb at either side of CpG islands). In addition, even (long distant) enhancer regions are frequently regulated by methylation. DNA methylation is established by DNA methyltransferases (DNMTs). Recurrent mutations in DNMT3A, one of the de novo DNA methyltransferases, are found in 6% to 36% of AML patients and are most commonly heterozygous (Table 1). DNMT3A mutations appear to be early events in leukemogenesis. This is evident by studies demonstrating that DNMT3A mutations in AML patients may also be present in T-lymphocytes derived from the same patient.48 These findings indicate that the mutation can arise in a primitive cell that is still able to give rise to both myeloid as well as lymphoid cells. In addition, it was recently demonstrated that DNMT3A mutations can be found in (elderly) individuals without an apparent hematologic malignancy, indicating that these mutations may be involved in clonal hematopoiesis, which in some individuals precedes the development of overt leukemia.10
The mechanism by which mutant DNMT3A contributes to leukemic transformation is not completely clear. It is hypothesized that mutant DNMT3A acts as a dominant negative over wild-type DNMT3A. AML cells with the R882H mutation have been reported to have profound reduction of de novo methyltransferase activity.49 Mice transplanted with Dnmt3a-null hematopoietic stem cells (HSCs) in a competitive transplantation setting do not develop myeloid malignancies.50 However, when the same Dnmt3a-null HSCs are transplanted in irradiated mice, not in a competitive setting, preleukemic disorders are frequently observed.51 Mouse modeling experiments show that the R882H variant, which is the most common DNMT3A mutation, is able to drive the development of myeloproliferative disorders in vivo as well. These data support the hypothesis that mutant DNMT3A acts as a dominant negative.52 Mice that develop full-blown AML generally acquire cooperating activating mutations in signaling factors such as c-Kit.52 This is similar to what is observed in human AML patients with DNMT3A mutations, who frequently carry concomitant FLT3-ITD mutations. Recent work suggests that the presence of DNMT3A R882H promotes chemoresistance, which appears to be associated with impaired DNA damage sensing and impaired chromatin remodeling capacities.53
Specific patterns of genome-wide cytosine methylation are associated with distinct subtypes of AML. Using genome-wide profiling, unique patterns of cytosine methylation are associated with AMLs characterized by specific translocations and fusion genes (eg, RUNX1-RUNX1T1 [AML1-ETO], PML-RARA, mutations in CEBPA, and mutations in NPM1).8 The fact that mutations in genes encoding transcription factors are associated with unique DNA methylation patterns emphasizes the relationship between defective transcriptional regulators and epigenetic alterations. In fact, evidence for a role of oncogenic transcription factors in directing the activity of DNMTs and changing the DNA methylation patterns has been proposed for EVI1,54 PML-RARA55 and RUNX1-RUNX1T1.56
DNA hydroxymethylation
The oxidation of 5-methylcytosine yields 5-hydroxymethylcytosine (5hmC). This process is catalyzed by the Ten-Eleven-Translocation (TET)-enzymes and is dependent on α-ketoglutarate. Mutations in TET2 are observed in 8% to 27% of patients with AML (Table 1). These mutations are associated with reduced levels of 5hmC.57 TET2 mutations confer a poor prognosis in intermediate-risk AML.17 In addition, mutations in genes encoding the enzymes called isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) are frequently observed in AML (Table 1). They appear more prevalent in patients with normal cytogenetics. IDH proteins catalyze the conversion of isocitrate to α-ketoglutarate. Mutations in IDH1 and IDH2 result in the production of an aberrant metabolite, 2-hydroxyglutarate.58 This molecule has been demonstrated to act as a competitive inhibitor of α-ketoglutarate, resulting in the inhibition of TET2. TET2 mutations and IDH1/IDH2 mutations are mutually exclusive in AML.4 In addition, IDH1/IDH2 mutations are associated with global cytosine hypermethylation signatures. Patients with TET2 mutations display overlapping hypermethylation signatures. TET2 mutations are observed in healthy aging individuals, supporting the concept of clonal hematopoiesis that may precede leukemogenesis.10,59 Recently, Levine and colleagues reported that mutations in the WT1 gene are mutually exclusive with mutations in IDH1/IDH2 or TET2.60 Increased global levels of 5hmC were observed upon WT1 overexpression, whereas reduced 5hmC levels were observed when WT1 was silenced. Although the mechanism of 5hmC decrease upon WT silencing or mutation is not understood, the investigators demonstrated that WT1 physically interacts with TET2. Decreased WT1 levels and absence of TET2 resulted in a similar hematopoietic differentiation phenotype. The authors conclude that TET2, IDH1/IDH2, and WT1 mutations define an AML subtype characterized by dysregulated DNA hydroxymethylation.
Murine models to study the mechanism of TET2 and IDH1/2 mutations have recently been developed. Experiments performed in conditional Tet2 knockout mice point to a role for Tet2 in the regulation of self-renewal of hematopoietic stem cells. These mice do not develop leukemia, but the presence of cooperating Flt3-ITD mutations does lead to increased incidence of leukemia.61 Leukemogenesis may be dependent on the downregulation of Gata2 by hypermethylation, an effect that can be overcome by reintroduction of Gata2.61 It is predicted that mice expressing conditional mutant Idh1 or Idh2 alleles in the presence of Flt3-ITD or Nras mutations will develop AML in a comparable manner.
In addition to 5-methylcytosine and 5hmC, two other DNA modifications have been recognized (ie, 5-formylcytosine and 5-carboxylcytosine).62 Similar to 5hmC, both represent oxidative derivates of methylated cytosines and can be the result of the activity of TET enzymes. The biological role of 5hmC and other derivates remains to be established. It has been proposed that they may not merely be intermediates toward CG demethylation, but may have distinct biological roles, for instance in the regulation of gene expression.62
Histone acetylation
Histone acetylation involves the transfer of acetyl groups to lysine residues in histone proteins. The processes of acetylation and deacetylation are governed by histone lysine acetyltransferases (KATs) and histone deacetylases (HDACs), respectively. Acetylation of lysine residues results in open chromatin confirmations, whereas deacetylation results in condensed and closed chromatin.
Sporadic translocations involving KATs have been described in myeloid malignancies, and mutations in HDACs have only rarely been identified (Table 1). However, there is evidence that HDACs may be aberrantly recruited by myeloid oncoproteins, such as EVI1 or PML-RARA.63,64 These oncoproteins frequently interact with scaffold proteins or major complexes, which recruit HDACs. This leads to chromatin remodeling, in particular deacetylation of lysines at histone tails near the transcription factor binding sites. These changes affect transcription of putative target genes.65-67
Acetylated lysine residues in histones can be recognized by reader proteins containing bromodomains, such as the bromodomain and extra terminal (BET) proteins BRD2, BRD3, and BRD4. These proteins contain 2 so-called bromodomains, which bind to acetylated histones. BRD4 is involved in a complex including Mediator and pTEF-b, and through this complex links histone acetylation to transcription.68,69 Although BRD4 and other BET proteins are ubiquitously present at gene promoters and enhancers, inhibition of BET proteins results in disproportionately large changes in expression of particular genes.70-72 Evidence is accumulating that these cell type–specific transcriptional effects of BET inhibition can to a large extent be explained by the association of BRD4 with exceptionally large enhancer elements, called “super-enhancers,” which are involved in lineage-specific gene regulation.73-76 In several types of malignancy, including AML, disease-specific oncogenic super-enhancers, also referred to as “stretched enhancers” or “locus control regions,” can be recognized that drive expression of critical oncogenes, such as MYC.
Histone lysine methylation
Methylation of histone lysine residues can result in mono-, di-, or trimethylation. This is processed by lysine methyltransferases (KMTs). Histone methylation alters the affinity of reader proteins to the methylated histones. Histone methylation can result in activation or repression, the actual effect being dependent on its context.77 Marks associated with activation include methylation of H3K4, H3H36, and H3K79. Marks associated with silenced gene transcription include methylation of H3K9, H3K27, and H4K20. Methylation state (eg, mono- vs trimethylation) may have different functional consequences in the same lysine residue.77
In AML, KMTs that are recurrently involved in translocations or are mutated include mixed-lineage leukemia (MLL) proteins and components of the polycomb repressor complexes (PRCs) (Table 1). KMT2A, more frequently referred to as MLL, is a member of the family of SET domain–containing KMTs. MLL targets H3K4 and leaves marks of transcriptional activation. MLL is involved in translocations in approximately 5% to 10% of AML.29 Partial tandem duplications of MLL are found in 5% to 7% of de novo AML. MLL translocations result in fusion proteins that lack the wild-type SET domain and are frequently replaced by members of genes encoding super elongation complex nuclear proteins, such as AF9, AF10, and ENL. This complex also contains DOT1L, a KMT that targets H3K79. MLL-transformed AML samples show aberrant H3K79 methylation, leading to ongoing expression of target genes, among which the homeobox genes are particularly involved.78
Enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) is an H3K27 methyltransferase that is part of the PRC2 polycomb repressor complex. EZH2 catalyzes di- and trimethylation of H3K27, which results in transcriptional repression. Aberrant gain of action of EZH2 has been associated with transcriptional repression of critical target genes in various types of cancer. In myeloid malignancies, however, EZH2 mutations lead to loss of function (Table 1).79,80 The functional basis for this difference is unclear, but some preliminary observations have been made. In a myelodysplastic syndrome (MDS) mouse model, knockout of Ezh2 promoted the development of MDS by impairing normal HSCs via activating inflammatory cytokine responses, while at the same time preventing the transformation to AML via PRC1-mediated repression of Hoxa9.81 In a study focusing on MLL-AF9 leukemia, EZH2 was not required for leukemogenesis, but was required for tumor progression.82 A recent study highlights another mechanism of interference with EZH2 function. Mutations in the splicing factor gene SRSF2, frequently found in MDS, cause mis-splicing of EZH2, resulting in nonsense-mediated decay.83
Other members of the PRC2 complex include additional sex combs like transcriptional regulator 1 (ASXL1), Jumonji AT-rich interactive domain 2 (JARID2), and SUZ12 polycomb repressive complex 2 subunit (SUZ12), all of which have been implicated in the development of AML (Table 1). Most of the mutations in these genes result in loss of function of the complex. The PRC2-associated protein ASXL1 has an important role in the recruitment of the complex to target loci. Similar to loss-of-function mutations in EZH2, mutations in ASXL1 result in loss of PRC2-mediated H3K27 methylation. ASXL1 mutations may be associated with adverse prognosis, an effect that seems to be correlated to the presence of RUNX1 mutations.36 Mutations in ASXL2 are frequently observed in patients with the RUNX1-RUNX1T1 fusion gene and are mutually exclusive with ASXL1 mutations.42
Similar to ASXL1, JARID2 recruits PRC2 to specific target loci. Deletions of JARID2 have been reported in patients showing leukemic transformation of chronic myeloid malignancies.43 SUZ12 has been reported to be mutated in progression from severe congenital neutropenia to AML.44 Thus, in most if not all mutations found in PRC2 complex members in AML, the ultimate effect is loss of function resulting in loss of H3K27 methylation. These effects are thus suggested to cause increased expression of important target genes.
Another polycomb repressor group gene that has been linked to AML is BMI1. BMI1 is part of the PRC1 polycomb repressor complex. Increased BMI1 expression has been associated with poor prognosis in AML and MDS.84,85
Several AML-related fusion proteins may interfere with normal PRC functioning. The PML-RARA fusion protein can engage in a complex with several PRC2 members, including SUZ12, EZH2, and EED, thereby recruiting them to specific genomic loci.86 Another fusion protein, PZLF-RARA, has been shown to interact with PRC1 components.87
Demethylation of histone proteins is governed by lysine demethylases (KDMs). Two classes of KDMs are known: the amine oxidases, which include lysine-specific demethylase 1 (LSD1/KDM1A), and the α-ketoglutarate–dependent Jumonji domain (JmjC) containing proteins. LSD1 has specificity for H3K4 and H3K9 and has a role both as a transcriptional activator as well as a transcriptional repressor, depending on the cellular context.88 KDM5A (JARID1), a JmjC lysine demethylase, is involved in a fusion protein with NUP98 in ∼10% of children with acute megakaryoblastic leukemia.45,89 KDM6A (UTX) is another JmjC family member and can be altered by inactivating mutations in AML (Table 1).46
Other histone modifications
In addition to the histone modifications described so far, other modifications are known. Methylation of arginine residues in histones is mediated by the family of protein arginine methyltransferases (PRMTs), of which 9 have been identified. Arginine methylation may be associated both with transcriptional activation as well as with silencing.90,91 PRMT5, which symmetrically dimethylates arginine residues and is considered to be a transcriptional repressor, is required for sustaining normal hematopoiesis.92 Dysregulated expression of several PRMTs, including PRMT5, has been observed in malignancies such as leukemia. Tyrosine phosphorylation at histone tails has been reported, but the mechanism by which this may occur and the function of histone phosphorylation is poorly understood.93
Epigenetic targeted treatment of AML
Taking into account the inherent reversibility of epigenetic marks, disordered epigenetic regulation poses the chance to use targeted drugs. The translation of such drugs to standard clinical use is still in an early stage. Compounds in various stages of preclinical and clinical development are summarized in Table 2. We first describe drugs that are currently available and then embark on approaches to identify novel targets.
Drugs targeting disordered patterns of DNA cytosine methylation and hydroxymethylation
Two epigenetic compounds currently approved for clinical use in myeloid malignancies are 5-azacytidine (azacitidine) and 5-aza-2′-deoxycytidine (decitabine). Both are pyrimidine analogs that function as inhibitors of DNA methyltransferases (Table 2). Azacitidine can be converted into decitabine, which is incorporated into DNA. Azacitidine itself is predominantly incorporated into RNA.106 Both drugs have been approved for the treatment of MDS as well as for AML with low blast count. In Europe, decitabine is also approved for the treatment of elderly AML patients with blast counts >30% who are not eligible for intensive treatment. Azacitidine and decitabine are thought to exert their effects by reversing aberrant DNA hypermethylation and thereby restoring expression of critical (tumor-suppressor) genes. There are increasing data to support the efficacy of these hypomethylating compounds in the treatment of patients not eligible for intensive chemotherapy.107-110 There are currently no established biomarkers that predict the response to these drugs, although it would be likely that not all patients are equally susceptible. This hypothesis is supported by the identification of factors that predict for response to azacitidine in MDS and methylation signatures that predict for response to decitabine in chronic myelomonocytic leukemia, respectively.111,112 In MDS, particular gene mutations, such as those in TET2 and DNMT3A, may predict for better responsiveness to treatment with inhibitors of DNMTs.113 Comparable studies need to be carried out to dissect which AML subtypes may benefit from treatment with DNMT inhibitors. One experimental approach would be to test the sensitivity to these drugs in cohorts of AML samples, taking into account subtypes that are particularly defined by unique DNA-methylation signatures. We previously demonstrated that leukemias with well-defined molecular abnormalities can be discriminated based on these epigenetic signatures.8 Moreover, with DNA methylation profiling we could identify AML subgroups that did not harbor any known recurrent genetic defect. It is possible that certain epigenetically defined AML subtypes are more responsive to DNA methyltransferase inhibitors than others. Responsiveness studies may be carried out in vitro using short-term assays testing cellular viability. These experiments could provide an indication of response to the bulk of cells to these agents. Whether the compounds will target the leukemia stem cells needs to be established using in vivo models. Mouse models have been generated for a number of AML subtypes carrying the distinct mutations, which allow investigators to study the effects of hypomethylating agents either alone or in combination with chemotherapy in vivo. One should keep in mind that murine models may not fully represent the actual human situation. Xenotransplant models in which human leukemia cells are transplanted in immunodeficient mice provide an opportunity to study the response of the human leukemias. The combination of these different approaches may, at least in a subset of AML subtypes, provide insight into responsiveness to DNMT inhibitors. Obviously the same strategy can be applied for other newly derived compounds or combinations of agents, either with or without chemotherapy.
Analyzing altered DNA methylation patterns of AML cells may help to uncover critical target genes that upon methylation have become silenced in certain patient groups. Shih and colleagues recently demonstrated how animal models can help to uncover such mechanisms.61 In their study, Tet2-null mice that also carried Flt3-ITD mutations developed AML and expressed a unique methylation signature. Gata2 appeared to be silenced by methylation in those animals. Upon re-expression of Gata2, the investigators reported reprogramming of the tumor cells. It will be of interest to study the effects of DNMT inhibitors in this model and to study reprogramming of these Tet2−/−/Flt3-ITD leukemia cells. No specific therapy for leukemias with TET2 mutations has been developed yet; however, the observed hypermethylation signature in patients as well as in animal models suggests that in particular in this subtype of AML hypomethylating agents may be effective.114 Further studies in xenotransplant models may help to unravel the most optimal combination of drugs, which may also include compounds that target uncontrolled signaling by FLT3-ITD.
As indicated, IDH1/2 mutations are also associated with a distinct methylation signature. IDH1/2 inhibitors have been developed and are already being tested in early clinical trials (Table 2). The outcomes of these studies are pending, but preliminary data are promising.115 It was shown that AGI-6780, a selective inhibitor of the IDH2 R140Q mutation, acts by binding an allosteric site located within the dimer interface and induces differentiation of AML cell lines in vitro.116 Idh1/2-mutant animal models have been developed as well, providing another opportunity to study the effects of the specific agents at a molecular level. There is also experimental evidence that these mutations may be targeted by alternative modes of action, such as BCL2 inhibitors.117 In addition, IDH1 R132H mutations result in a tumor-specific potential neoantigen that may offer a possibility of development of vaccination strategies that induce mutation-specific T-cell responses.118
Drugs targeting disordered patterns of histone acetylation
HDAC inhibitors were among the first epigenetic drugs developed, and several have been tested in clinical trials for malignancies, including AML.119 Responses to HDAC inhibitors as a single agent in AML or MDS appear to be modest.120 More recent work has therefore focused on combining these compounds with other drugs. A phase 2 study testing a combination of vorinostat with idarubicin and cytarabine suggested that the addition of an HDAC inhibitor to standard chemotherapy may lead to improved response rates in patients with newly diagnosed AML and MDS.121 More recently, combinations of DNMT inhibitors and HDAC inhibitors have been tested.122-124 In a phase 2 randomized trial, combination of the HDAC inhibitor valproic acid and low-dose decitabine did not lead to improved outcome in MDS and AML patients.125 Moreover, in another phase 2 study, the HDAC inhibitor entinostat appeared even to be antagonistic when administered concurrently with azacitidine.126
It will be challenging to find the optimal combinations of HDAC inhibitors with other agents. It is also likely that timing of treatment (eg, concurrent vs subsequent administration) may affect responses—an aspect that will in fact be crucial for other epigenetic drugs as well. Answering these questions will require proper in vivo models as discussed earlier. It is also possible that targeting altered histone acetylation or the consequences thereof has to be carried out in a completely different manner. This is supported by recent findings involving the targeting of transcription elongation factors such as bromodomains proteins using small molecules. Bromodomain proteins, including BET proteins, exert their function by binding acetylated lysine residues in histone proteins, such as H3K27. There are to date no known mutations in bromodomain proteins in AML. However, BET inhibitors showed therapeutic effects in various AML mouse models as well as in cell lines and small series of primary samples.9,70,127,128 BET inhibitors may be effective in AML associated with MLL translocations.70 Furthermore, it was recently demonstrated that in AML with chromosome 3q abnormalities, a subtype with an adverse prognosis, the leukemic translocation results in the formation of a novel super-enhancer that drives overexpression of the EVI1 proto-oncogene.9 Indeed, AML t(3;3) or inv(3;3) cell lines appear sensitive to the BET inhibitor JQ1. Not surprisingly, given the large molecular heterogeneity among AML, previous studies have shown that BET inhibitors are not uniformly active against all leukemia cell lines and patient samples.129 BET inhibitors have entered phase 1/2 clinical trials (Table 2).130 Studies in larger series will be needed to identify molecular features linked to response to these drugs and to further clarify the mechanisms underlying the efficacy of BET inhibition in AML and its target genes and proteins. An important question that needs to be answered is how to identify factors that differ between normal super-enhancers, which direct normal cellular functions, and oncogenic super-enhancers, which are unique to particular types of malignancy. Insights into these differences would enable the development of more selective treatment approaches. A recent study describes phthalimide conjugation to the BET inhibitor JQ1 as a modified method to BET inhibition. In this approach, the BET inhibitor is used to target BRD4, whereas the appended phthalimide moiety results in hijacking of the cereblon E3 ubiquitin ligase complex, resulting in targeted BRD4 degradation.131
Drugs targeting disordered patterns of histone methylation
The histone demethylase LSD1 is highly expressed in hematopoietic cells and is specifically required for the terminal differentiation of myeloid cells.132 LSD1 may be overexpressed in AML and appears to show a higher level of expression in less differentiated subtypes of the disease. Recent reports have highlighted a potential application of LSD1 inhibition in AML, and early clinical trials have been initiated (Table 2).133,134 MLL translocations result in fusion proteins that lack the wild-type SET domain and instead frequently recruit another lysine methyltransferase, DOT1L, which targets H3K79. DOT1L can be targeted by small-molecule inhibition.78,135 The first clinical trials involving these compounds have been started and results have yet to follow (Table 2).
Together, single epigenetic drugs such as inhibitors of BET proteins, DOT1L, mutant IDH1/2, or DNMTs may show some significant effects. However, these responses will most likely be partial or transient. Therefore, these novel compounds, derived from functional epigenomics, need to be further studied in representative models in combination with other drugs that target complementary pathways or with chemotherapy. Molecular and epigenetic studies are essential for a better understanding of epigenetic changes that lead to leukemic transformation, and these investigations can lead to novel drugs that target those lesions. We therefore believe that further searching for novel targets and molecules that recognize those elements will be crucial.
How to identify novel targets for epigenetic treatment?
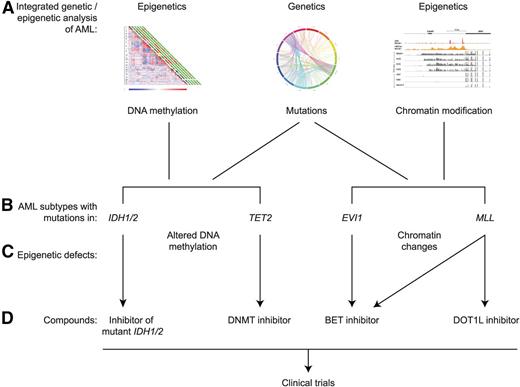
As was pointed out, the application of targeted epigenetic treatment is still in an early phase. Still, there are already multiple examples of AML with unique epigenetic alterations, identified using integrated genetic and epigenetic analysis, that may be matched to appropriate compounds (Figure 1). A number of strategies can be applied to identify novel targets and to match those to appropriate modes of targeted treatment.
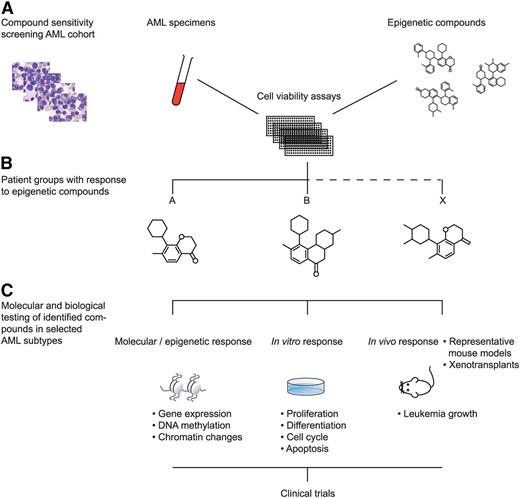
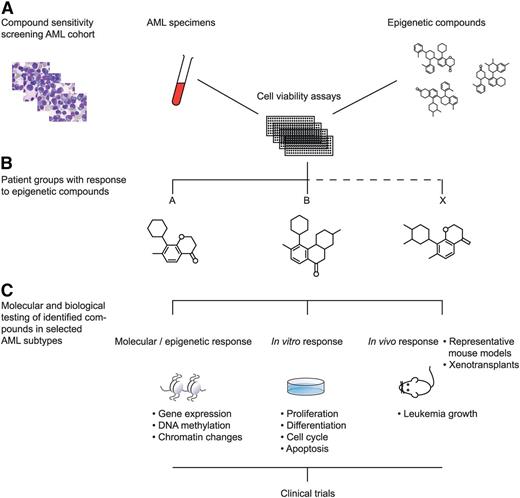
1. A first strategy is to apply high-throughput compound screening approaches. Numerous compounds and small molecules are currently available that interfere with particular known epigenetic processes. Large-scale titration sensitivity experiments of these molecules in representative series of primary AML specimens in vitro, for instance using tritiated thymidine- or MTT assays, will provide compound response signatures of each AML sample. AML patients may be classified based on these response signatures (Figure 2). Because epigenetic changes in response to specific compounds may need several rounds of cell cycle, several time points should be assessed if possible (eg, 3, 7, and 14 days of culture). These experiments should be carried out under well-defined conditions using appropriate combinations of hematopoietic growth stimuli. Based on the outcome of these experiments, specific epigenetic modulators may be predicted to be targetable in certain AML subtypes. Biochemical, molecular, and cell biological studies should be applied to uncover mechanisms of action of the identified compounds. An example of a compound that was identified to target a specific AML subset is the BET inhibitor JQ1. BET inhibitors in general show a higher affinity for super-enhancers compared with normal (small) enhancers. Molecular and biochemical analyses revealed that JQ1 particularly targets the oncogenic super-enhancer that causes overexpression of the EVI1 transforming gene.9 The effects of BET inhibitors on leukemic stem cells of super-enhancer–driven EVI1 AMLs should be tested in mouse models or in xenotransplant models.
A complementary approach involves the use of genetic tools such as shRNA libraries or CRISPR/Cas9 technology targeting epigenetic regulators. These studies may be carried out in vitro as well as in vivo. Using an inducible shRNA library targeting approximately 250 epigenetic modulators in MLL-fusion mouse AML cells, Zuber and colleagues identified Brd4 as a potential target for treatment in vivo.127 These studies pointed to the potential usage of BET inhibitors for this specific AML subtype. As one may anticipate based on the partial response of leukemia cells to particular drugs, a third approach to identify novel targets would be to combine the use of selected compounds (eg, BET inhibitors) with genetic screening approaches such as shRNA.
2. An alternative route would be to start with detailed maps of genetic and epigenetic marks in particular AML subtypes. This way, one could prioritize certain drugs to be tested in distinct subtypes of the disease. For instance, TET2 mutations have been associated with increased levels of cytosine methylation. This may be a rationale for specifically testing DNMT inhibitors in those patients (Figure 1). Indeed, patients with TET2 mutations may be more susceptible to azacitidine than AML patients in general.114 Similarly, one could compile genome-wide maps of super-enhancers by applying ChIP-sequencing using antibodies directed against BRD4 and H3K27. Identifying AML cases with particular super-enhancer profiles could provide leads to test BET inhibitors specifically in subsets of the disease. By combining genetic mapping studies with screening studies using extensive libraries of compounds or genetic libraries, it should be possible to match particular drugs to patient subgroups and to identify biomarkers.
3. A third approach is to use chemical-based engineering techniques to design novel molecules predicted to specifically interfere with certain epigenetic processes. These types of structural approaches heavily rely on chemical methods such as crystallography, frequently in combination with computer-based modeling. This could result in the development of inhibitors with a relatively high affinity for mutated proteins, as has been done for IDH1/2 mutations.116 Alternatively, the development of BET inhibitors and derivates illustrates that also inhibitors of wild-type proteins may be applicable in certain situations.131,136
Examples of integrated genetic and epigenetic analysis of human AML leading to potential epigenetic treatment. The integrated analysis of gene mutations, DNA methylation, and histone modifications (A) has revealed multiple AML subtypes with specific mutations in association with specific epigenetic alterations. Four examples are indicated (B). Mutations in IDH1/2 and TET2 result in aberrant patterns of DNA cytosine methylation and hydroxymethylation (C), whereas rearrangements affecting the EVI and MLL loci result in alterations of chromatin marks (C). For each of these 4 examples, certain epigenetic inhibitors have been designed or are predicted to be active, offering the potential to translate these findings to clinical trials in human patients (D). The various inhibitors are discussed in more detail in the text.
Examples of integrated genetic and epigenetic analysis of human AML leading to potential epigenetic treatment. The integrated analysis of gene mutations, DNA methylation, and histone modifications (A) has revealed multiple AML subtypes with specific mutations in association with specific epigenetic alterations. Four examples are indicated (B). Mutations in IDH1/2 and TET2 result in aberrant patterns of DNA cytosine methylation and hydroxymethylation (C), whereas rearrangements affecting the EVI and MLL loci result in alterations of chromatin marks (C). For each of these 4 examples, certain epigenetic inhibitors have been designed or are predicted to be active, offering the potential to translate these findings to clinical trials in human patients (D). The various inhibitors are discussed in more detail in the text.
Epigenetic modifier screen for human AML. Numerous compounds and small molecules are currently available that interfere with particular known epigenetic processes. Large-scale titration sensitivity experiments of these molecules in primary AML specimens in vitro can provide compound response signatures of each AML sample (A). Using those assays, AML subtypes may be uncovered that uniformly respond to specific epigenetic compounds (B). The molecular, epigenetic, and biological effects to these AML cells may subsequently be studied in vitro and in vivo (C). The outcomes of such experiments should lead to the translation to clinical trials in human patients (D).
Epigenetic modifier screen for human AML. Numerous compounds and small molecules are currently available that interfere with particular known epigenetic processes. Large-scale titration sensitivity experiments of these molecules in primary AML specimens in vitro can provide compound response signatures of each AML sample (A). Using those assays, AML subtypes may be uncovered that uniformly respond to specific epigenetic compounds (B). The molecular, epigenetic, and biological effects to these AML cells may subsequently be studied in vitro and in vivo (C). The outcomes of such experiments should lead to the translation to clinical trials in human patients (D).
Optimal applications of epigenetic therapy
Despite great progress in the understanding of the molecular basis of AML in the past years, treatment of the disease has essentially not shown major changes in the past decades. This review highlights the potential of compounds targeting epigenetic regulators in AML.
An important question is what the role of such compounds should be in clinical practice. There are several challenges that may hamper the clinical introduction of novel targeted therapies in general.137 Some of these challenges include the inherent problems in the translation of preclinical findings to the clinic, the presence of multiple coactive deregulated pathways in the disease, and questions related to the optimal design of clinical trials (how to best select patients to include and which end points to address). In particular, the testing of novel targeted treatment in an isolated fashion may be problematic and may in fact underestimate the effectiveness of these novel compounds. It is reasonable to assume that combination therapy will be key, which may involve either combination with standard chemotherapy or with other targeted therapies that are predicted to target parallel critical pathways.
Together, epigenetic targeted treatment holds great promise. At the same time, there are various challenges that will have to be dealt with. In the first place, many of the types of targeted treatment that will be applied will not be mutation-specific, but will rather target particular epigenetic regulating systems that are also active in nontransformed cells, posing the risk of toxicity. Furthermore, the action of epigenetic marks and regulators is often cell type–specific, the actual result being dependent on particular protein-protein interactions, which also carries risks of off-target toxicity. Along the same lines, many chromatin-modifying enzymes have more diverse functions, including a role in modifying nonhistone substrates. The identification of biomarkers that can predict the susceptibility to certain epigenetic compounds will be essential for a proper choice of epigenetic treatment of the distinct AML subtypes.138 An additional challenge in the treatment of AML that has emerged is the concept of clones and subclones.139,140 Targeted therapy may be successful in eradicating some subclones, but may not be able to eradicate subclones that do not harbor the particular epigenetic abnormality. In this respect, it is promising to note that recent work has indicated that many of the mutations that can be found in clonal hematopoiesis are in epigenetic regulators such as TET2 and DNTM3A. These mutations seem to enhance the self-renewal capacity of hematopoietic progenitor cells and may predispose individuals to the development of leukemia. Targeting such early lesions may eradicate the founding clones. Still, it is most likely that the combination of several types of treatment (eg, epigenetic therapy in combination with conventional chemotherapy) will have the best potential to be successful.
Acknowledgments
The authors thank Egied Simons for help with preparing the figures and Dr Peter Valk for critically reading the manuscript. They apologize to authors whose work could not be cited due to space limitations.
B.J.W. is a recipient of a Fellowship Award by the Dutch Cancer Society (K.W.F.) and a Special Fellowship Award by the Leukemia & Lymphoma Society.
Authorship
Contribution: B.J.W. and R.D. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ruud Delwel, Erasmus Medical Center, Department of Hematology, P.O. Box 2040, 3000 CA Rotterdam, The Netherlands; e-mail: h.delwel@erasmusmc.nl.