Abstract
Recent major advances in understanding the molecular basis of acute myeloid leukemia (AML) provide a double-edged sword. Although defining the topology and key features of the molecular landscape are fundamental to development of novel treatment approaches and provide opportunities for greater individualization of therapy, confirmation of the genetic complexity presents a huge challenge to successful translation into routine clinical practice. It is now clear that many genes are recurrently mutated in AML; moreover, individual leukemias harbor multiple mutations and are potentially composed of subclones with differing mutational composition, rendering each patient’s AML genetically unique. In order to make sense of the overwhelming mutational data and capitalize on this clinically, it is important to identify (1) critical AML-defining molecular abnormalities that distinguish biological disease entities; (2) mutations, typically arising in subclones, that may influence prognosis but are unlikely to be ideal therapeutic targets; (3) mutations associated with preleukemic clones; and (4) mutations that have been robustly shown to confer independent prognostic information or are therapeutically relevant. The reward of identifying AML-defining molecular lesions present in all leukemic populations (including subclones) has been exemplified by acute promyelocytic leukemia, where successful targeting of the underlying PML-RARα oncoprotein has eliminated the need for chemotherapy for disease cure. Despite the molecular heterogeneity and recognizing that treatment options for other forms of AML are limited, this review will consider the scope for using novel molecular information to improve diagnosis, identify subsets of patients eligible for targeted therapies, refine outcome prediction, and track treatment response.
Cytogenetic characterization of AML
Forty years ago, pioneering work led by Janet Rowley established that acute myeloid leukemia (AML) is a genetic disease, with the discovery of somatic chromosomal abnormalities including balanced translocations (eg, t[8;21] and t[15;17]) in the leukemic cells of some patients.1,2 These studies paved the way to the identification of the genes disrupted at the chromosomal breakpoints of leukemia-associated translocations and inversions, providing major insights into normal hematopoiesis and uncovering common mechanistic themes in the pathogenesis of AML (for review, see Grimwade and Mrózek3 ). The balanced chromosomal rearrangements were found to give rise to in-frame chimeric fusion genes and recurrently target genes encoding hematopoietic transcription factors (eg, RARA, RUNX1, or CBFβ subunits of the core binding factor [CBF] complex), epigenetic regulators (eg, MLL [KMT2A], NSD1, CREBBP [KAT3A]), and components of the nuclear pore complex (NUP98, NUP214).
The generation of chimeric fusion genes because of translocation or inversion events occurring in a permissive hematopoietic stem/progenitor cell is considered to be the critical initiating step in the pathogenesis of a significant proportion of AML arising in children and younger adults (Figure 1).4 ,,,,,,,,,,,,,,,,,,,,,,, -29 Successive World Health Organization (WHO) classifications have recognized a number of balanced chromosomal rearrangements and their resulting chimeric fusion genes, which are designated as “recurrent genetic abnormalities” and deemed sufficient to make a diagnosis of AML irrespective of bone marrow blast count.30 Cytogenetic analyses of large cohorts of AML patients have shown that translocations/inversions underlie disease pathogenesis in ∼50% and 30% of AML arising in children and younger adults, respectively, whereas only a minority of AMLs presenting in older adults have balanced rearrangements.3 These studies highlight that the outcome for AML patients differs markedly according to the cytogenetic abnormality.3
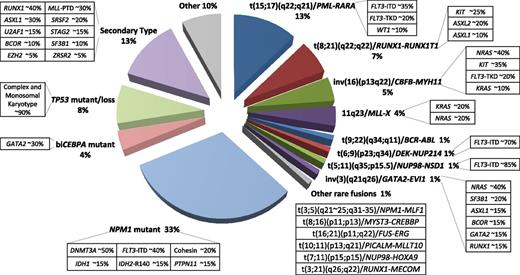
Distribution of cytogenetically and molecularly defined subsets of AML presenting in younger adults. Based on analysis of large cohorts of patients and patterns of mutual exclusivity between cytogenetic and molecular genetic features, the majority of AML cases can be segregated into a number of biologically and prognostically distinct subgroups. In approximately a third of cases, AML is characterized by the presence of balanced chromosomal rearrangements,4 which lead to the generation of chimeric oncoproteins, considered to be initiating events in disease pathogenesis. These chromosomal abnormalities are mutually exclusive of mutations in the nucleophosmin gene (NPM1) and biallelic CEBPA (biCEBPA) mutations, which are recognized as recurrent AML-defining genetic abnormalities and typically associated with a normal karyotype. Recent studies have established a close correlation between complex karyotype/monosomal karyotype and underlying mutation in the TP53 gene,5 which defines a biological subgroup with very poor prognosis. Recent studies have distinguished a mutational profile involving alterations to a panel of genes including those encoding ASXL1 and spliceosome components associated with secondary AML arising on a background of myelodysplasia (MDS).6 For each cytogenetically and genetically defined subset of AML denoted in the pie chart, frequent associated cooperating mutations are shown in the respective boxes. Mutational frequencies are derived from integration of data from previous studies.4
Distribution of cytogenetically and molecularly defined subsets of AML presenting in younger adults. Based on analysis of large cohorts of patients and patterns of mutual exclusivity between cytogenetic and molecular genetic features, the majority of AML cases can be segregated into a number of biologically and prognostically distinct subgroups. In approximately a third of cases, AML is characterized by the presence of balanced chromosomal rearrangements,4 which lead to the generation of chimeric oncoproteins, considered to be initiating events in disease pathogenesis. These chromosomal abnormalities are mutually exclusive of mutations in the nucleophosmin gene (NPM1) and biallelic CEBPA (biCEBPA) mutations, which are recognized as recurrent AML-defining genetic abnormalities and typically associated with a normal karyotype. Recent studies have established a close correlation between complex karyotype/monosomal karyotype and underlying mutation in the TP53 gene,5 which defines a biological subgroup with very poor prognosis. Recent studies have distinguished a mutational profile involving alterations to a panel of genes including those encoding ASXL1 and spliceosome components associated with secondary AML arising on a background of myelodysplasia (MDS).6 For each cytogenetically and genetically defined subset of AML denoted in the pie chart, frequent associated cooperating mutations are shown in the respective boxes. Mutational frequencies are derived from integration of data from previous studies.4
Although cytogenetics provides powerful independent prognostic information and continues to provide the framework for risk stratification in children and younger adults, it has a number of limitations. Apart from technical failures, by definition cytogenetics cannot identify cryptic gene fusions, for example NUP98-NSD1, CBFA2T3-GLIS2, and MNX1-ETV6, which predict a poor outcome in pediatric AML.3,31,32 Moreover, ∼5% of cases of PML-RARA+ acute promyelocytic leukemia (APL) lack the classic t(15;17), with the fusion gene formed as a result of insertion events or more complex rearrangements.33 These patients respond to molecularly targeted therapies (ie, all-trans retinoic acid [ATRA] and arsenic trioxide [ATO]) and share the favorable prognosis of those with the t(15;17). Similarly cryptic rearrangements of CBF or MLL fusion genes have been reported.3 Such cases are important to identify because, based purely on conventional cytogenetics, they could be assigned to the wrong risk group and the patients treated inappropriately. Cytogenetic analysis is also limited in its resolution. This can be particularly problematic where breakpoints occur in close proximity; for example, at least 5 different genes that can potentially recombine with the MLL locus fall within the 19p13.1∼13.3 region.34 Apart from the considerable heterogeneity within cytogenetic risk groups, there may also be marked variation in outcomes between patients with the same primary chromosomal abnormality that cannot be explained by the pattern of additional chromosomal changes. Moreover, cytogenetics provides no insights into molecular mechanisms underlying AMLs with numerical or other structural changes, or importantly, those with normal karyotype (CN-AML), which account for ∼40% of adult AML and are highly heterogeneous in terms of clinical outcome.3 However, over the course of the past 2 decades, major advances have been made to start to address these questions as the mutational landscape of AML has been mapped.
Defining the mutational spectrum of AML
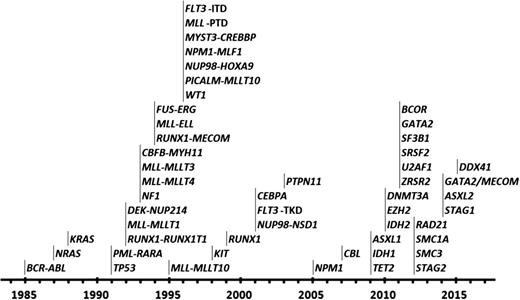
With advances in technology from chromosome banding, fluorescence in situ hybridization/chromosomal painting, array comparative genomic hybridization, genomic breakpoint cloning, and Sanger sequencing of candidate genes through to development of single nucleotide polymorphism profiling, whole genome sequencing (WGS), whole exome sequencing (WES), and RNA sequencing, there have been incremental improvements in understanding the genetic basis of AML (Figures 1 and 2; Table 1). These have provided important insights into the molecular abnormalities underlying AML with normal cytogenetics and those with chromosomal losses or gains that were previously poorly understood.
Progress in defining the molecular landscape of AML. Timing of the identification of leukemic fusion genes and mutations underlying the pathogenesis of AML.
Progress in defining the molecular landscape of AML. Timing of the identification of leukemic fusion genes and mutations underlying the pathogenesis of AML.
Prevalence, association, and prognosis of mutations detected in AML presenting in younger adults (16-60 y)
| Category/mutant gene . | % . | Associated mutations/ chromosomal abnormalities . | Prognostic implications . |
|---|---|---|---|
| DNA methylation | |||
| DNMT3A | 20 | NPM1, FLT3-ITD, NK | Adverse |
| DNA demethylation | |||
| TET2 | 8 | ASXL1, NK | Poorer in IR-AML |
| IDH1 | 7 | NPM1, NK | Poorer in FLT3-ITD-neg AML |
| IDH2-R140 | 7 | NPM1, NK | Favorable |
| IDH2-R172 | 2 | NK | Adverse |
| WT1 | 9 | FLT3-ITD | Poorer in NK-AML |
| Activated signaling | |||
| FLT3-ITD | 27 | NPM1, DNMT3A, NK, t(15;17)/ PML-RARA, t(6;9)/DEK-NUP214, t(5;11)/NUP98-NSD1 | Poorer in IR-AML |
| FLT3-TKD | 7 | inv(16)/CBFB-MYH11, | Variable according to study |
| t(15;17)/PML-RARA | |||
| NRAS | 11 | inv(16)/CBFB-MYH11, | NS |
| t(3;5)/NPM1-MLF1, 11q23/MLL-X | |||
| KRAS | 5 | inv(16)/CBFB-MYH11, 11q23/MLL-X | NS |
| PTPN11 | 5 | NPM1 | ND |
| NF1* | 4 | MK, −17/17q | ND |
| KIT | 4 | t(8;21)/RUNX1/RUNX1T1, inv(16)/CBFB-MYH11 | Poorer outcome in CBF AML |
| CBL | 1 | TBC | |
| Myeloid transcription factors | |||
| RUNX1 | 5 | MLL-PTD, ASXL1, IDH2, NK, +13 | Adverse |
| biCEBPA | 4 | GATA2, NK | Favorable |
| Tumor suppressor/ multifactorial | |||
| TP53* | 8 | Complex, MK, −5/-5q, −7/-7q, −17/17p | Adverse |
| NPM1 | 33 | DNMT3A, IDH1, IDH2-R140, | Favorable in absence of FLT3-ITD and mutant DNMT3A |
| FLT3-ITD, PTPN11, cohesin, NK | |||
| Chromatin regulation | |||
| ASXL1 | 5 | RUNX1, IDH2-R140, | Poorer in IR-AML |
| t(8;21)/RUNX1-RUNX1T1, +8 | |||
| MLL-PTD | 5 | +11, NK, RUNX1, FLT3 | Adverse |
| PHF6 | 3 | RUNX1 | TBC |
| ASXL2 | 2 | t(8;21)/RUNX1-RUNX1T1 | ND |
| BCOR | 1 | NK, DNMT3A | TBC |
| EZH2 | 1 | ND | |
| Spliceosome | |||
| SRSF2 | 2 | +13, ASXL1, RUNX1, IDH1/2 | ND |
| SF3B1 | 3 | RUNX1, inv(3)/GATA2-EVI1 | |
| U2AF1 | 2 | ||
| ZRSR2 | <1 | ||
| Cohesin | |||
| RAD21 | 6-9 | NPM1 | NS |
| SMC1A | |||
| SMC3 | |||
| STAG1 | |||
| STAG2 |
| Category/mutant gene . | % . | Associated mutations/ chromosomal abnormalities . | Prognostic implications . |
|---|---|---|---|
| DNA methylation | |||
| DNMT3A | 20 | NPM1, FLT3-ITD, NK | Adverse |
| DNA demethylation | |||
| TET2 | 8 | ASXL1, NK | Poorer in IR-AML |
| IDH1 | 7 | NPM1, NK | Poorer in FLT3-ITD-neg AML |
| IDH2-R140 | 7 | NPM1, NK | Favorable |
| IDH2-R172 | 2 | NK | Adverse |
| WT1 | 9 | FLT3-ITD | Poorer in NK-AML |
| Activated signaling | |||
| FLT3-ITD | 27 | NPM1, DNMT3A, NK, t(15;17)/ PML-RARA, t(6;9)/DEK-NUP214, t(5;11)/NUP98-NSD1 | Poorer in IR-AML |
| FLT3-TKD | 7 | inv(16)/CBFB-MYH11, | Variable according to study |
| t(15;17)/PML-RARA | |||
| NRAS | 11 | inv(16)/CBFB-MYH11, | NS |
| t(3;5)/NPM1-MLF1, 11q23/MLL-X | |||
| KRAS | 5 | inv(16)/CBFB-MYH11, 11q23/MLL-X | NS |
| PTPN11 | 5 | NPM1 | ND |
| NF1* | 4 | MK, −17/17q | ND |
| KIT | 4 | t(8;21)/RUNX1/RUNX1T1, inv(16)/CBFB-MYH11 | Poorer outcome in CBF AML |
| CBL | 1 | TBC | |
| Myeloid transcription factors | |||
| RUNX1 | 5 | MLL-PTD, ASXL1, IDH2, NK, +13 | Adverse |
| biCEBPA | 4 | GATA2, NK | Favorable |
| Tumor suppressor/ multifactorial | |||
| TP53* | 8 | Complex, MK, −5/-5q, −7/-7q, −17/17p | Adverse |
| NPM1 | 33 | DNMT3A, IDH1, IDH2-R140, | Favorable in absence of FLT3-ITD and mutant DNMT3A |
| FLT3-ITD, PTPN11, cohesin, NK | |||
| Chromatin regulation | |||
| ASXL1 | 5 | RUNX1, IDH2-R140, | Poorer in IR-AML |
| t(8;21)/RUNX1-RUNX1T1, +8 | |||
| MLL-PTD | 5 | +11, NK, RUNX1, FLT3 | Adverse |
| PHF6 | 3 | RUNX1 | TBC |
| ASXL2 | 2 | t(8;21)/RUNX1-RUNX1T1 | ND |
| BCOR | 1 | NK, DNMT3A | TBC |
| EZH2 | 1 | ND | |
| Spliceosome | |||
| SRSF2 | 2 | +13, ASXL1, RUNX1, IDH1/2 | ND |
| SF3B1 | 3 | RUNX1, inv(3)/GATA2-EVI1 | |
| U2AF1 | 2 | ||
| ZRSR2 | <1 | ||
| Cohesin | |||
| RAD21 | 6-9 | NPM1 | NS |
| SMC1A | |||
| SMC3 | |||
| STAG1 | |||
| STAG2 |
Mutations within the same functional category are negatively associated, and positive associations are listed.
bi, biallelic; IR, intermediate risk; ITD, internal tandem duplication; MK, monosomal karyotype; ND, not determined; NK, normal karyotype; NS, not significant; PTD, partial tandem duplication; TBC, to be confirmed; TKD, tyrosine kinase domain.
Includes mutations and gene deletions.
Mutations in signaling pathway components
A major step forward was the discovery that the receptor tyrosine kinase fms-like tyrosine kinase 3 (FLT3), which plays a key role in normal hematopoiesis, is mutated in a third of AML cases (for review, see Levis35 ). These mutations involve in-frame duplications within the juxtamembrane region (FLT3-ITD) and point mutations in the tyrosine kinase domain (FLT3-TKD) in ∼25% and ∼7% of AML, respectively. Both classes of mutation lead to constitutive activation of the receptor, yet surprisingly they carry different prognostic implications. Numerous studies have confirmed FLT3-ITD as an independent predictor of higher relapse rate and poorer overall survival, with cases with high mutant allelic burden (because of acquired uniparental disomy) having a particularly poor prognosis,35 whereas presence of an FLT3-TKD mutation has been associated with better survival.11 How these 2 classes of mutation that involve the same gene are associated with such disparate outcomes is intriguing and may reflect differences in the spectra of associated cooperating mutations, as well as differences in signaling pathways downstream of FLT3 impacting disease biology.
Successive studies established that a number of other genes encoding signaling pathway components including RAS, cKIT, CBL, NF1, and PTPN11 are recurrent mutation targets in AML (Table 1). Although RAS mutations are prognostically neutral (for review, see Mrózek et al36 ), a number of studies have highlighted an association between the cKIT mutation and CBF leukemia (Figure 1), which predicts a poorer outcome (for review, see Mrózek et al37 ). The observation that mutations in signaling proteins frequently accompany chromosomal rearrangements that target hematopoietic transcription factors (eg, PML-RARα, CBFβ-MYH11, RUNX1-RUNX1T1) led to the proposal of a 2-hit model of AML, involving cooperation of mutations conferring a proliferative advantage (designated class I mutations) and those predicted to induce a block in myeloid differentiation (designated class II mutations).38 However, with the capacity to sequence AML genomes, it has become clear that the situation is much more complex, with ∼40% of AML lacking mutations in genes encoding classical signaling pathway components.12
CEBPA, NPM1, and RUNX1 mutations
Major insights into the molecular pathogenesis of cytogenetic standard risk AML, including CN-AML, were provided by discoveries of mutations in the genes encoding CCAAT/enhancer binding protein α (CEBPA) and nucleophosmin (NPM1).39,40 These entities were recognized in the 2008 WHO Classification and define prognostically relevant subsets of AML. CEBPA encodes a myeloid transcription factor; mutations are found in ∼10% of CN-AML, can also occur in the context of abnormal karyotype (eg, del(9q)), and are mutually exclusive of balanced chromosomal rearrangements. Mutations cluster in both the amino- and carboxy-terminal regions, with the former leading to expression of a truncated isoform of CEBPA (p30) and loss of the full-length protein (p42).39 Carboxy-terminal mutations affect regions involved in mediating dimerization and DNA binding. In more than half of patients with CEBPA mutations, both alleles are involved, combining an upstream mutation in 1 allele with a downstream mutation in the other (for review, see Smith et al41 ). Interestingly, in a proportion of patients, CEBPA mutation is inherited through the germ line, with development of AML associated with acquisition of additional mutations, which can include involvement of the other CEBPA allele.41 Murine models have provided important insights into the biology, showing how loss of p42 expression (mimicking biallelic N-terminal CEBPA mutations) or compound heterozygous mutations affecting amino- and carboxy-terminal regions in combination affect hematopoiesis and give rise to AML.42,43 Although early studies reported that CEBPA mutation predicts a relatively favorable outcome, it subsequently became clear that this effect relates to the subgroup with biallelic mutations. These AMLs are associated with GATA2 mutations and typically lack FLT3-ITD.13-17
Mutations involving the NPM1 gene, discovered by Brunangelo Falini and colleagues in 2005,40 represent the commonest AML-defining molecular lesion identified to date, occurring in a third of cases, including >50% of those with CN-AML. More than 40 different mutations have been described (type A, B, and D mutations collectively account for ∼90%), which involve the C-terminal region of the protein and lead to loss of tryptophan residues and generation of a nuclear export signal causing delocalization of nucleophosmin from the nucleoli to the cytoplasm. Murine modeling has shown that NPM1 mutation (designated NPM1c) can enhance self-renewal of hematopoietic progenitors, associated with expanded myelopoiesis and lead to development of AML,44 with FLT3-ITD dramatically reducing latency and increasing penetrance of the leukemic phenotype.45 Analyses of large clinical trial cohorts have consistently shown that young adults with AML harboring an NPM1 mutation in the absence of FLT3-ITD have a relatively favorable prognosis (for review, see Kühnl and Grimwade46 ), and coexisting FLT3-ITD and/or mutation in DNA methyltransferase 3A (DNMT3A, see “Epigenetic modifier mutations” section) predicts an increased risk of relapse and poorer outcome.18
RUNX1 is a relatively common somatic mutation target in cytogenetic standard risk and CN-AML (for review, see Lam and Zhang47 ). Germ-line mutations of RUNX1 have also been described, which lead to familial platelet disorder and predispose to AML. In adult AML, RUNX1 mutations are distributed across the gene and can be biallelic. They tend to be mutually exclusive of balanced translocations and mutations involving NPM1 and CEBPA, being associated with M0 FAB type, trisomy 13, MLL-PTD, IDH2, and ASXL1 mutations (see “Epigenetic modifier mutations” section). RUNX1 is also mutated in ∼10% of MDS cases48 and could therefore indicate a subgroup of AML patients with secondary disease arising on a background of MDS. Large studies of younger adults with AML have consistently reported that RUNX1 mutation is an independent predictor of adverse clinical outcome.19-21
Epigenetic modifier mutations
Application of a range of both genome-wide and candidate gene sequencing approaches has identified a large number of genes encoding epigenetic regulators to be recurrent mutation targets in AML (Table 1; for review, see Wouters and Delwel49 ). Alteration of a number of these genes, including DNMT3A, TET2, ASXL1, CREBBP/KAT3A, and EZH2 is not specific to AML, with disease phenotype likely to be determined by the cell of origin as well as the mutational context, in terms of temporal order and nature of the associated cooperating mutations.
DNMT3A, a methyltransferase that generates de novo DNA methylation via the conversion of cytosine to 5-methylcytosine, is mutated in ∼30% of cytogenetic standard risk AML.18,22 Mutations in DNMT3A, NPM1, and FLT3-ITD commonly cosegregate, suggesting cooperativity in AML pathogenesis (Figures 1, 3, and 4; Table 1)50-52 ; DNMT3A is mutated in half of NPM1c AML, and the majority (80%) of AML cases harboring mutant DNMT3A are also NPM1 mutant. DNMT3A mutations are typically heterozygous, with ∼60% falling in a hotspot encoding arginine at codon 882 (R882) within the catalytic domain. Recent data indicate that R882 mutations act in a dominant negative manner with the mutant protein interfering with the capacity of residual wild-type DNMT3A to form active tetramers leading to reduced enzyme activity and focal hypomethylation at specific cytosine guanine dinucleotides in primary AML cells.53 Molecular mechanisms underlying non-R882 loss-of-function mutations are less well understood; however, both types of mutation have been associated with poorer prognosis in patients with NPM1c and NPM1 wild-type AML.18,22
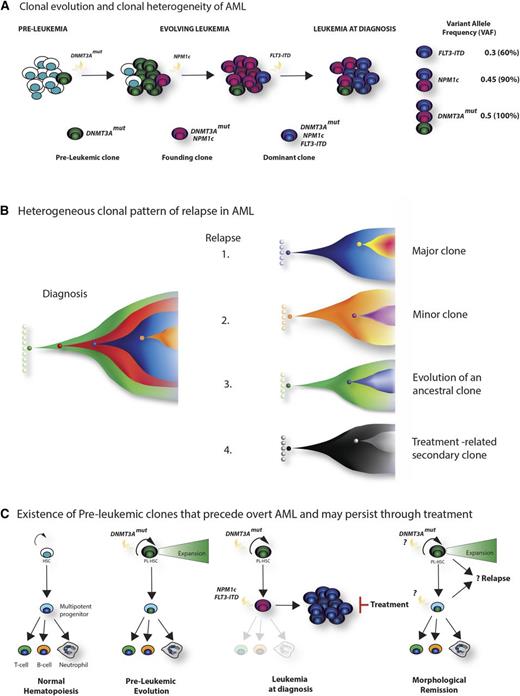
Clonal architecture, patterns of relapse, and the existence of preleukemic stem cells. (A) Clonal evolution and clonal heterogeneity of AML. Evolution of AML in a hypothetical patient whose tumor carries cooperating DNMT3A, NPM1c, and FLT3-ITD mutations. Mutation of DNMT3A is the earliest event, and although facilitating clonal expansion, occurs prior to overt disease development. Subsequently, NPM1c occurs as the disease-defining mutation in the founding clone, with further acquisition of a FLT3-ITD mutation in a hyperproliferative clone during leukemia expansion, which becomes dominant at diagnosis. Quantification of the variant allele frequency (VAF) of each mutation (by VAF, right) allows a demonstration of the temporal acquisition of mutations and the clonal hierarchy of the bulk tumor. For simplicity, a linear evolutionary pattern is shown, although commonly branching evolution can be demonstrated. (B) Heterogeneous clonal pattern of relapse in AML. Potential patterns of relapse from the hypothetical tumor in panel A are shown in the Fish plots: 1, relapse of the dominant clone at diagnosis; 2, relapse of a subclone present at diagnosis; 3, relapse from an ancestrally related clone; 4, “apparent relapse,” where the new tumor is not clonally related to the initial leukemia, such as might happen in therapy-related AML. (C) Existence of preleukemic clones that precede overt AML and may persist through treatment. Recently, the existence of preleukemic stem cells has been demonstrated.50-52 These harbor AML-associated mutations such as DNMT3A, TET2, and IDH2 that permit multipotent differentiation, but also facilitate clonal expansion within the stem and progenitor compartment (panel, second right). Upon the acquisition of further mutations (again NPM1c and FLT3-ITD are shown in the third panel), overt disease develops. However, treatment, although successful in eradicating the AML blasts, does not eradicate the preleukemic stem cells. Evidence suggests that these cells form the reservoir for relapse and resistance (left panel), although further study is required to characterize their biology and prognostic significance.
Clonal architecture, patterns of relapse, and the existence of preleukemic stem cells. (A) Clonal evolution and clonal heterogeneity of AML. Evolution of AML in a hypothetical patient whose tumor carries cooperating DNMT3A, NPM1c, and FLT3-ITD mutations. Mutation of DNMT3A is the earliest event, and although facilitating clonal expansion, occurs prior to overt disease development. Subsequently, NPM1c occurs as the disease-defining mutation in the founding clone, with further acquisition of a FLT3-ITD mutation in a hyperproliferative clone during leukemia expansion, which becomes dominant at diagnosis. Quantification of the variant allele frequency (VAF) of each mutation (by VAF, right) allows a demonstration of the temporal acquisition of mutations and the clonal hierarchy of the bulk tumor. For simplicity, a linear evolutionary pattern is shown, although commonly branching evolution can be demonstrated. (B) Heterogeneous clonal pattern of relapse in AML. Potential patterns of relapse from the hypothetical tumor in panel A are shown in the Fish plots: 1, relapse of the dominant clone at diagnosis; 2, relapse of a subclone present at diagnosis; 3, relapse from an ancestrally related clone; 4, “apparent relapse,” where the new tumor is not clonally related to the initial leukemia, such as might happen in therapy-related AML. (C) Existence of preleukemic clones that precede overt AML and may persist through treatment. Recently, the existence of preleukemic stem cells has been demonstrated.50-52 These harbor AML-associated mutations such as DNMT3A, TET2, and IDH2 that permit multipotent differentiation, but also facilitate clonal expansion within the stem and progenitor compartment (panel, second right). Upon the acquisition of further mutations (again NPM1c and FLT3-ITD are shown in the third panel), overt disease develops. However, treatment, although successful in eradicating the AML blasts, does not eradicate the preleukemic stem cells. Evidence suggests that these cells form the reservoir for relapse and resistance (left panel), although further study is required to characterize their biology and prognostic significance.
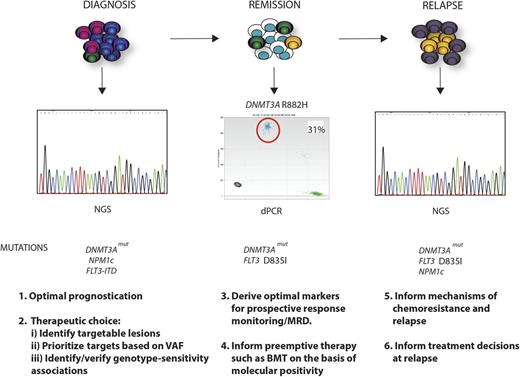
Clinical relevance of the mutational landscape in AML. A longitudinal schematic of various phases of disease are shown for AML. For each time point, possible clinical applications relating to knowledge of the specific mutational complement of the tumor, or the presence of specific mutations, are highlighted. At diagnosis, where in this example DNMT3A, NPM1c, and FLT3-ITD mutations drive disease, WES or panel-based next-generation sequencing (NGS) analysis could optimize prognostication and therapeutic choice, identifying mutations with an existing therapy. Moreover, quantitation of mutation frequency by the VAF could prioritize mutations that exist within every cell as critical targets, and the mutational complement could identify/validate prospective associations between specific mutational genotypes and sensitivity to individual or combined agents. In remission, perhaps using sensitive techniques such as digital polymerase chain reaction (dPCR), identification of the optimal markers for prospective response monitoring could allow early and robust adaptive postinduction therapy such as identifying candidates for stem cell transplant. In the example shown, this would relate to persistence of the DNMT3A mutation, as illustrated by dPCR and the occurrence of an occult FLT3 resistance mutation D835I (yellow clone). At clinical relapse, further WES or panel-based analysis may inform potential mechanisms of chemoresistance and relapse (FLT3 mutation) and in doing so may inform treatment decisions for reinduction and/or further therapy. In this example, the patient has relapsed with a FLT3 mutation (D835I) that would confer therapeutic resistance to some but not all FLT3 inhibitors. The patient has also developed a further subclonal NPM1c mutation in addition to the DNMT3A and FLT3 mutation (purple clone).
Clinical relevance of the mutational landscape in AML. A longitudinal schematic of various phases of disease are shown for AML. For each time point, possible clinical applications relating to knowledge of the specific mutational complement of the tumor, or the presence of specific mutations, are highlighted. At diagnosis, where in this example DNMT3A, NPM1c, and FLT3-ITD mutations drive disease, WES or panel-based next-generation sequencing (NGS) analysis could optimize prognostication and therapeutic choice, identifying mutations with an existing therapy. Moreover, quantitation of mutation frequency by the VAF could prioritize mutations that exist within every cell as critical targets, and the mutational complement could identify/validate prospective associations between specific mutational genotypes and sensitivity to individual or combined agents. In remission, perhaps using sensitive techniques such as digital polymerase chain reaction (dPCR), identification of the optimal markers for prospective response monitoring could allow early and robust adaptive postinduction therapy such as identifying candidates for stem cell transplant. In the example shown, this would relate to persistence of the DNMT3A mutation, as illustrated by dPCR and the occurrence of an occult FLT3 resistance mutation D835I (yellow clone). At clinical relapse, further WES or panel-based analysis may inform potential mechanisms of chemoresistance and relapse (FLT3 mutation) and in doing so may inform treatment decisions for reinduction and/or further therapy. In this example, the patient has relapsed with a FLT3 mutation (D835I) that would confer therapeutic resistance to some but not all FLT3 inhibitors. The patient has also developed a further subclonal NPM1c mutation in addition to the DNMT3A and FLT3 mutation (purple clone).
The potential of mutational profiling to inform downstream functional analysis of AML pathogenesis has been highlighted by recent studies investigating the consequences of mutations involving genes encoding TET2 (ten eleven translocation 2), WT1 (Wilms’ tumor 1), and isocitrate dehydrogenase enzymes IDH1 and IDH2.54-56 Mutations in these genes were found to be mutually exclusive but shared common signatures on DNA methylation profiling. The key downstream target of this pathway is TET2, which is subject to mutation in ∼8% of AML and is the enzyme that regulates the initial step in DNA demethylation through dioxygenase activity and the conversion of 5-methylcytosine to 5-hydroxymethylcytosine.
IDH1 mutations affecting the arginine amino acid at codon 132 (R132) are found in ∼7% of AML, particularly those with normal karyotype and NPM1c (Table 1). IDH2 is mutated in a further ∼9% of AML, with mutations clustering at codons R140 (7%) and R172 (2%). IDH2 provides a further example of a gene where mutations involving different sites have been reported to be prognostically distinct.23,24 The R140 and R172 mutations differ in mutational complement, with R140 associated with NPM1c and better clinical outcome, whereas IDH2 R172 is not found in NPM1c AML and predicts a poor prognosis. The IDH enzymes catalyze the conversion of isocitrate to α-ketoglutarate, generating reduced NAD phosphate. IDH mutations not only reduce α-ketoglutarate formation, but also have neomorphic activity, additionally altering enzyme function to generate the “oncometabolite” 2-hydroxyglutarate, which inhibits the function of TET2 and other dioxygenase enzymes. Recently, WT1 has been identified as an interaction partner of TET2, regulating its binding to DNA.55,56 WT1 mutation (found in ∼9% AML) therefore provides a further mechanism by which TET2 function can be impaired leading to site-specific changes in DNA hydroxymethylation and altered gene expression. These data have highlighted the importance of deregulation of the IDH/WT1/TET2 axis in AML pathogenesis, which may be operative in a third of cases arising in younger adults.
Splicing factor gene mutations
A further major step forward in understanding the molecular landscape of AML, followed the discovery that genes encoding components of the splicing machinery, involved in pre–messenger RNA (mRNA) processing prior to protein translation, are recurrent mutation targets in MDS.57,58 The most common genes involved are SF3B1, U2AF1, SRSF2, and ZRSR2. Splicing factor gene mutations are mutually exclusive; they are found in approximately half of MDS cases, where they are considered to be founder mutations and segregate with distinct subsets of disease (for review, see Cazzola et al48 ). In particular, SF3B1 mutations correlate with presence of ring sideroblasts,58 and SRSF2 mutations are associated with more aggressive disease and chronic myelomonocytic leukemia.57,59 Further insights into the functional consequences of splicing factor gene mutations have been gained through in vivo modeling of the U2AF1 hotspot mutation in a transgenic mouse model, which altered hematopoiesis associated with changes in pre-mRNA splicing of genes that are recurrent mutation targets in MDS and AML including BCOR and KMT2D (MLL2).60 A recent complementary study focusing on SRSF2 showed that the mutation impairs hematopoietic differentiation in vivo. SRSF2 mutation was found to alter the binding specificity of SRSF2 to splice recognition motifs leading to missplicing of targets such as EZH2, thereby explaining the mutual exclusivity of EZH2 and SRSF2 mutations in myeloid malignancy.61 Spliceosome mutations are also found in a proportion of newly diagnosed AML (Figure 1; Table 1), with a recent study involving a well annotated cohort of 194 AML patients concluding that alterations of SRSF2, SF3B1, U2AF1, or ZRSR2 are among a panel of hallmark mutations that can be considered pathognomonic of secondary AML arising on a background of MDS6 (Figure 1).
Mutations in cohesin complex members
Application of WGS and WES led to the identification of genes encoding components of the cohesin complex as recurrent mutation targets in AML.12 The cohesin complex is a ringlike structure composed of SMC1A, SMC3, RAD21, and one of either STAG1 or STAG2, which is involved in sister chromatid exchange during anaphase, regulation of DNA repair, and transcriptional control by the coordination of interactions between promoters and cis-regulatory elements via DNA looping (for review, see Leeke et al62 ). Interestingly, the former function appears irrelevant in AML, with cohesin gene mutations being associated with noncomplex and normal karyotype, and recent murine models implicating altered transcriptional regulation in the development of leukemia.106-109 Examination of a large cohort of younger adults with AML identified mutations in 6% of non-APL patients; mutations were found to be mutually exclusive and commonly associated with NPM1c, with no impact on prognosis.25 Cohesin complex gene mutations are also detected in 10% to 15% of MDS and 20% of secondary AML patients, being associated with mutations involving RUNX1, BCOR, and ASXL1.26
Timing of mutations: early mutations, persistent mutations associated with relapse, and the identification of preleukemic stem cells and clonal hematopoiesis
Next generation sequencing (NGS) studies have also indirectly documented the stepwise acquisition of mutations that fashion AML development, through the demonstration of differences in the relative proportion of co-occurring mutations within the bulk tumor at the time of diagnosis. Greater sequencing depth has allowed these differences to be quantified as the VAF, where the proportion of reads that contain the mutated allele is compared with that of wild-type allele reads, with the relative proportions able to infer clonal architecture (Figure 3A). Such analysis has demonstrated the emergence of new clones carrying novel mutations at different times during the evolution of the leukemia.63,64 At diagnosis, mutations will therefore show a range of VAF, from close to 0.5 (ie, present in 100% of the cells of the tumor) ranging down to much lower frequencies. Mutations with a higher VAF are predicted to occur early during the development of leukemia, whereas mutations present only in a minority of cells are likely to be acquired at later stages of leukemia development (Figure 3A). Higher VAF may also be observed as a result of acquired uniparental disomy, for example in a proportion of AMLs with associated FLT3-ITD and TET2 mutations.65,66 This clonal heterogeneity has also been functionally demonstrated, where longitudinal sequencing studies of diagnostic and relapse material from the same patient have shown heterogeneous patterns of relapse, with relapse occurring from expansion of both major or minor clones present at diagnosis or from novel clones that share an ancestral relationship to the diagnostic clone67,68 (Figure 3B). These studies have therefore begun to document early or initiating mutations present in all cells of the tumor and also those mutations that persist after treatment failure, prioritizing obvious critical molecular drivers of leukemia development and relapse to therapeutically target.
However, the mutational history and evolution of AML appears to be even more complex. Experimental evidence demonstrates that chimeric fusion genes (eg, PML-RARA, RUNX1-RUNX1T1) are insufficient to induce leukemic transformation in their own right; however, they may provide a competitive advantage, generating populations of cells in which secondary mutations may arise and be selected for. Early studies documenting the continued presence of transcripts of the fusion oncogene RUNX1-RUNXT1 following therapy appear to validate this theory.69 In addition, seminal studies conducted by Mel Greaves in twins discordant for acute lymphoblastic leukemia development have documented that preleukemic stem cells carrying the ETV6-RUNX1 fusion were also present in the twin that had not developed leukemia.70 A recent series of elegant studies from the Majeti and Dick laboratories have further extended this work, documenting the presence of AML-associated mutations in the T cells of patients in various phases of the disease.50,50-52 Careful examination of defined hematopoietic stem and progenitor populations and clonal colony studies, using the mutations as clonal markers, allowed the demonstration that a number of mutations, including DNMT3A, TET2, IDH1, IDH2, and ASXL1 occur in hematopoietic stem and progenitor cells that retain the normal characteristics of multipotent differentiation (Figure 3C). These mutations therefore precede the development of overt leukemia, which happens when further mutations such as NPM1c co-occur in later progenitor compartments such as the granulocyte-monocyte progenitor.52 Of clinical importance, some preleukemic mutations (ie, DNMT3A, TET2, and ASXL1) are associated with a poorer outcome18,22,27-29 and, mechanistically, are predicted to alter epigenetic regulation and chromatin function. Using longitudinal studies of allelic burden in xenotransplant experiments, Shlush and colleagues further demonstrated clonal expansion for preleukemic hematopoietic stem cells that carry a DNMT3A mutation,52 although the nature of this proliferative advantage is yet to be elucidated.
The presence of mutations that precede development of overt leukemia likely reflects a cell’s cumulative inability to completely repair the multitude of mutations that occur randomly over time. Recent controversial data have also been presented to suggest that cancer risk in a given tissue can be at least partially explained by the predicted number of stem cell divisions within that tissue.71 Examination of the hematopoietic system has provided supporting evidence for this hypothesis, where WGS sequencing has determined the “background” mutation rate in normal hematopoietic stem cells in 7 individuals along an age spectrum (from cord blood to individuals >70 years), with mutation frequency shown to increase as a function of age.72 Extending this analysis in much larger cohorts of individuals with normal hematologic indices, 4 recent studies have further documented a high and age-related incidence of clonal hematopoiesis, present in nearly 20% of people >90 years, with the suggestion that this percentage is likely a significant underestimate related to the limited number of mutational hotspots assessed and sensitivity of the assays.73-76 Clonality was confirmed by the presence of mutations associated with myeloid malignancies, including DNMT3A, TET2, ASXL1, SF3B1, and SRSF2 and the mutations were associated with clonal hematopoietic expansion, an increased risk of blood cancer development, other adverse outcomes, including ischemic stroke and coronary heart disease, as well as an increased all-cause mortality. Taken all together, a greater knowledge of the clonal architecture in individual tumors and the role of clonal hematopoiesis in the development and prognosis of blood cancers is critical for optimal AML therapy and will need to be factored into the future design and implementation of trials of standard and novel therapies.
Therapeutic targeting of individual AML mutations and the promise of personalized medicine
Our continued use of “7 + 3” as the backbone of AML therapy is a damning indictment of what little progress we have made over the past 3 decades. It also strongly reiterates the need for new, effective therapies. However, genuine optimism currently exists, with the development of a plethora of novel, rational, and in some cases specifically targeted therapies, the majority of whose mechanisms of action differ from standard cytotoxics. These promising agents include, but are not limited to, a battery of novel epigenetic therapies, antiapoptotic agents, selective inhibitors of nuclear export, and immunotherapies including chimeric antibody receptor T-cells and bispecific and other therapeutic antibodies (Table 2). Annotation of the mutational landscape in AML has greatly facilitated the identification and development of some of these agents, where it has documented a number of specific cellular processes discussed previously, including intracellular signaling, epigenetic regulation, transcriptional control, and mRNA splicing to be recurrently perturbed by mutation12,77 and has placed these alterations into individual “complementation groups” reflecting the function of the gene mutated. Although many of these mutations are predicted or have been experimentally demonstrated to be loss-of-function mutations, rendering them less tractable targets, a significant number have been demonstrated to be activating or neomorphic mutations. These include those involving signaling proteins such as FLT3 and C-KIT and mutations in IDH1 and 2.
Examples of molecularly targeted early phase and adaptive later stage trials where mutational information may inform therapeutics
| Therapeutic target . | Potentially sensitive genotypes . | Inhibitor . | Phase . | Trial identifier number . |
|---|---|---|---|---|
| Early phase | ||||
| Highly specific | ||||
| hDOT1L* | MLL rearranged | EPZ-5676 | I | NCT01684510 |
| IDH2 | IDH2 mutated | AG-221 | I | NCT019115498 |
| CDK6 | MLL rearranged | Palbociclib | Ib/IIa | NCT02310243† |
| Broader specificity | ||||
| BET proteins* | MLL rearranged, NPM1c, | OTX015 | I | NCT01713582 |
| other | GSK525762 | I | NCT01943851† | |
| XPO1/CRM-1 | NPM1c, other | KPT-330 (Selinexor) | I | NCT01607892 |
| BCL-2* | IDH2 mutated, other | venetoclax | II | NCT01994837 |
| Risk adapted later phase | ||||
| FLT3* | FLT3 mutated | Crenolanib | II | NCT01522469 |
| Quizartinib | II | NCT01236144 | ||
| Midostaurin | III | NCT00651261 | ||
| C-KIT* | C-KIT mutated | Dasatinib | III | NCT02013648 |
| CD33 | CBF leukemia | Gemtuzumab ozogamicin | III | ISRCTN17161961† |
| (Mylotarg) |
| Therapeutic target . | Potentially sensitive genotypes . | Inhibitor . | Phase . | Trial identifier number . |
|---|---|---|---|---|
| Early phase | ||||
| Highly specific | ||||
| hDOT1L* | MLL rearranged | EPZ-5676 | I | NCT01684510 |
| IDH2 | IDH2 mutated | AG-221 | I | NCT019115498 |
| CDK6 | MLL rearranged | Palbociclib | Ib/IIa | NCT02310243† |
| Broader specificity | ||||
| BET proteins* | MLL rearranged, NPM1c, | OTX015 | I | NCT01713582 |
| other | GSK525762 | I | NCT01943851† | |
| XPO1/CRM-1 | NPM1c, other | KPT-330 (Selinexor) | I | NCT01607892 |
| BCL-2* | IDH2 mutated, other | venetoclax | II | NCT01994837 |
| Risk adapted later phase | ||||
| FLT3* | FLT3 mutated | Crenolanib | II | NCT01522469 |
| Quizartinib | II | NCT01236144 | ||
| Midostaurin | III | NCT00651261 | ||
| C-KIT* | C-KIT mutated | Dasatinib | III | NCT02013648 |
| CD33 | CBF leukemia | Gemtuzumab ozogamicin | III | ISRCTN17161961† |
| (Mylotarg) |
BCL-2, B-cell CLL/lymphoma 2; BET, bromodomain and extraterminal; C-KIT, kit oncogene; CDK6, cyclin dependent kinase 6; hDOT1L, human disruptor of telomere silencing-1-like; IDH2, isocitrate dehydrogenase 2; XPO-1, exportin 1.
Examples are given where multiple trials for targeted agents exist.
Trials involve AML and non-AML patients. Also see text.
Moreover, mechanistic evidence from basic biological studies and observational preclinical work also suggest that nonintuitive associations exist between genotype and sensitivity to specific therapies. Examples of this would include the sensitivity of MLL-rearranged leukemias to BET, hDOT1L, and CDK6 inhibitors78-82 ; of NPM1c leukemias to ATRA+ATO,83,84 BET inhibitors, and selective inhibitors of nuclear export85 ; and of IDH2 mutated leukemias to BCL-2 inhibitors.86 In these associations, no mutation or transcriptional upregulation of the target is evident from molecular studies, however preclinical sensitivity has been demonstrated, with these associations currently being tested in early phase clinical trials. Therefore, a detailed knowledge of the molecular landscape of individual AML patients a priori will help to identify potential targets within the tumor for which an available therapy exists, which might be used in combination with standard therapy. In addition, all therapeutic trials should be documenting a minimal molecular data set to further strengthen intuitive and nonintuitive genotype-sensitivity associations and to identify novel associations, with the aim of further individualizing therapy on the basis of molecular data. Such molecularly adapted trial design has long been adopted in AML and was the prime driver for the therapeutic segregation of the APL subtype.87 Other examples of molecularly driven adaptive design have included the use of FLT3 inhibitors for FLT3-mutated patients,88,89 Gemtuzumab ozogamicin (Mylotarg) for CBF rearranged AML,90 dasatinib for patients with CBF AML to target C-KIT (EudraCT: 2006-006555-12; www.clinicaltrials.gov, #NCT02013648),91,92 and CDK6 inhibitors for MLL-rearranged patients (www.clinicaltrials.gov, #NCT02310243). However, the first of these examples, that of FLT3 inhibition introduces a cautionary note. In general, the results of adding a FLT3 inhibitor to standard therapy for FLT3-ITD and TKD patients have been underwhelming (for reviews, see Konig and Levis88 and Lancet89 ), with no significant improvements in survival noted. Although this may partially be explained by poor target engagement, a situation that may be improved on by the use of more potent inhibitors such as AC220, it also reflects that FLT3-ITD and TKD are subclonal mutations and therefore poor therapeutic targets and perhaps explains why the first survival advantage described with a FLT3 inhibitor was only recently reported (RATIFY study).109 Although they may be dominant at the time of diagnosis, VAF and longitudinal studies demonstrate that they occur later in disease evolution (Figure 3A) and are therefore not present in all cells of the tumor. Furthermore, they are not stable and may be present or absent at relapse. These data demonstrate the importance of targeting early and/or initiating lesions present within all clones for the eradication of disease rather than later proliferative clones, with proof of this hypothesis coming in APL and in Philadelphia-positive acute lymphoblastic leukemia, where the use of ATRA+ATO and ABL-specific tyrosine kinase inhibitors such as Imatinib, respectively, has led to significant and dramatic improvements in overall survival.93-95 Although the identities of these early lesions are becoming more apparent, these data further highlight the importance of documenting the relative allele frequency of mutations to identify the potential “Achilles heel” of each AML in the prioritization of therapeutic targets.
As intimated previously, it is becoming increasingly apparent that the presence of some mutations at diagnosis has more significant clinical implications than the presence of others and it is obvious that further study is needed to refine and optimize combinatorial knowledge of mutation complement for prognostication, which may also be influenced by disease context (eg, patient age, de novo/secondary/therapy-related AML). A key decision in the treatment pathway is whether to undertake allogeneic transplantation in first remission. This is currently guided by cytogenetics and a very limited panel of molecular genetic markers (NPM1, FLT3-ITD, and CEBPA), used to identify patients with a predicted relapse rate exceeding 35% where the benefit of transplant in terms of reduced relapse risk outweighs procedure-related mortality (for review, see Cornelissen et al96 ). A knowledge of the full mutational inventory may also facilitate assessment of response to therapy and provide prospective prognostic guidance, particularly for those patients who are poorly served by the current prognostic scoring systems, such as cytogenetic standard risk patients. A panel of established real-time quantitative PCR assays are already available to track treatment response in the ∼60% of children and younger adults presenting with NPM1c or fusion gene positive AML (for review, see Grimwade and Freeman97 ). It has become clear that (1) persistent high-level PCR positivity or (2) molecular relapse with rising leukemic transcript level after an initial response, both predict impending disease relapse. Moreover, data are now emerging that minimal residual disease assessment provides a more powerful predictor of disease outcome than mutational profile. With greater access to targeted sequencing in the diagnostic work-up of AML, there are opportunities to extend the proportion of patients informative for molecular detection of minimal residual disease. In cases where “hotspot” mutations are identified, digital PCR may be the preferred approach. Whereas, for mutations that are heterogeneous, involve large genes and for simultaneous tracking of multiple mutations, NGS may provide the optimal approach. Recent studies have shown that early assessment of treatment response using NGS can distinguish patients at differing risk of relapse.98,99 A key challenge will be to determine which are the most informative molecular markers that most reliably track leukemic populations irrespective of the recognized clonal heterogeneity, rather than preleukemic clones that can persist in patients in long-term remission following chemotherapy.
Conclusions and future directions
The recent quantum leap in sequencing technology, aided by a dramatic reduction in sequencing costs, has allowed for the generation of previously unparalleled amounts of information annotating mutational landscapes and their evolution across multiple cancers. As befits the first cancer genome sequenced, nowhere is this more apparent than in AML. In this review, we have focused predominantly on the landscape of AML presenting in young adults (<60 years) and have outlined the likely key areas that a detailed knowledge of this landscape will further inform: an improved biological understanding, better prediction of disease outcome, optimization of induction and postremission treatment, and the identification of robust clonal biomarkers for monitoring therapeutic response (Figure 4). The mutational landscape in pediatric AML is less well resolved and requires further attention. Likewise, in older AML where there is an appreciation that the genetic landscape more resembles secondary AML/MDS,6 but this area also merits more detailed investigation.
Although not detracting from the excitement and opportunity that the current situation affords us, we would introduce a note of caution. Like modern-day archeologists confronted with ancient Egyptian hieroglyphs, our current ability to confidently translate the presence of these mutations into clinical gains for individual patients significantly lags behind our ability to detect them. Therefore in practical terms, it is envisaged that cytogenetic analysis will continue to play an important role in patient management for several years ahead, considering that it can be performed in routine laboratories worldwide, provides a cornerstone of the WHO classification, affords powerful independent prognostic information, and is used to guide transplant decisions, as well as predict risk of relapse posttransplant. However, it is clear that additional laboratory information is needed to inform management because in every facet of its biology, including the mutational complement of individual tumors, AML has proved to be an exemplar of heterogeneity. This is highlighted by the commonest subtype of AML, defined by the NPM1c mutation, which exhibits considerable molecular complexity (Figure 5).9,100 We already know that the addition of DNMT3A or FLT3-ITD mutations alter the prognostic significance of NPM1c mutations in patients treated with standard “7 + 3”–like AML therapy. However the prognostic significance of additional mutations or combinations of mutations is currently unknown. In extrapolating this complexity to the multitude of mutations present in AML, bridging this knowledge gap must be our first priority. Therefore, development of risk group classification schemes considering cytogenetic and molecular genetic abnormalities that have been reported to confer independent prognostic information (Table 3),3,4,101 should be considered a work in progress. Thankfully, studies of the necessary size, involving thousands of retrospective patients treated with standard AML therapy, and comparing disease outcome against mutational genotype are already underway.102 These will further refine the contribution of specific combinations of mutations to discrete disease subtypes and to treatment response, allowing improved prognostication. In addition, we can reasonably expect that correlations between genotype and response to the multiple novel AML therapeutics recently identified will also exist. We further advocate that a “minimal genotype data set” is obtained for all AML patients prospectively treated in clinical trials at diagnosis and subsequent relapse, perhaps by panel based NGS, with this data set correlated to diagnostic clinical characteristics, response to therapy and overall outcome. The use of clonal markers from this data set will allow refinement of response monitoring of residual disease in real time, allowing for early adaption and optimal usage of postinduction therapy, including stem cell transplantation.
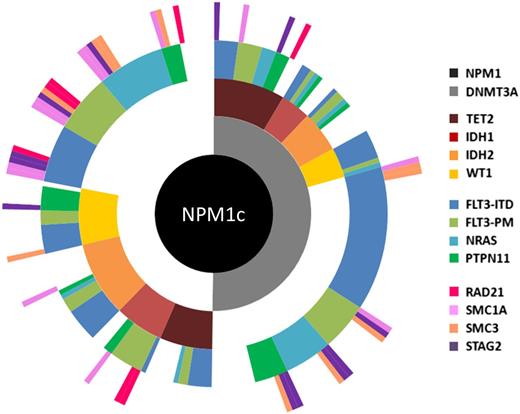
Molecular heterogeneity of AML exemplified by mutational profiling in NPM1c AML. Summary of targeted sequencing data using a published panel100 covering 14 mutational groups conducted in diagnostic samples from a cohort of 223 patients with NPM1c AML.9 Each spoke radiating from the central NPM1c hub represents the mutation pattern of a single patient. Cooperating mutations are grouped into 4 tiers according to function and color coded according to the figure key, and white space indicates no mutation. For example, in the patient displayed at 12 o’clock, mutations were detected in NPM1, DNMT3A, TET2, FLT3 (ITD), and STAG2. Overall, based on mutational combination, patients segregated into >75 different subgroups.
Molecular heterogeneity of AML exemplified by mutational profiling in NPM1c AML. Summary of targeted sequencing data using a published panel100 covering 14 mutational groups conducted in diagnostic samples from a cohort of 223 patients with NPM1c AML.9 Each spoke radiating from the central NPM1c hub represents the mutation pattern of a single patient. Cooperating mutations are grouped into 4 tiers according to function and color coded according to the figure key, and white space indicates no mutation. For example, in the patient displayed at 12 o’clock, mutations were detected in NPM1, DNMT3A, TET2, FLT3 (ITD), and STAG2. Overall, based on mutational combination, patients segregated into >75 different subgroups.
Cytogenetic and molecular genetic abnormalities conferring independent prognostic information in younger adults with AML
| Risk group . | Cytogenetic/molecular genetic abnormality . |
|---|---|
| Favorable | t(15;17)(q22;q21)/PML-RARA |
| t(8;21)(q22;q22)/RUNX1-RUNX1T1 | |
| inv(16)(p13q22)/t(16;16)(p13;q22)/CBFB-MYH11 | |
| NPM1 mutation (in absence of FLT3-ITD or DNMT3A mutation) | |
| Biallelic CEBPA mutation | |
| Intermediate | Cytogenetic/molecular genetic abnormalities not classified as favorable or adverse |
| Adverse | In the absence of favorable risk cytogenetic/molecular genetic abnormalities: |
| abn(3q) [excluding t(3;5)(q21∼25;q31∼35)/NPM1-MLF1], | |
| inv(3)(q21q26)/t(3;3)(q21;q26)/ GATA2/EVI1 | |
| add(5q)/del(5q), −5 | |
| t(5;11)(q35;p15.5)/NUP98-NSD1 | |
| t(6;9)(p23;q34)/DEK-NUP214 | |
| add(7q)/del(7q), −7 | |
| t(11q23) [excluding t(9;11)(p21∼22;q23) and t(11;19)(q23;p13)] | |
| t(9;22)(q34;q11)/BCR-ABL | |
| −17/abn(17p)/TP53 mutation | |
| Complex karyotype (≥4 unrelated abnormalities) | |
| ASXL1 mutation | |
| DNMT3A mutation | |
| FLT3-ITD | |
| MLL-PTD | |
| RUNX1 mutation |
| Risk group . | Cytogenetic/molecular genetic abnormality . |
|---|---|
| Favorable | t(15;17)(q22;q21)/PML-RARA |
| t(8;21)(q22;q22)/RUNX1-RUNX1T1 | |
| inv(16)(p13q22)/t(16;16)(p13;q22)/CBFB-MYH11 | |
| NPM1 mutation (in absence of FLT3-ITD or DNMT3A mutation) | |
| Biallelic CEBPA mutation | |
| Intermediate | Cytogenetic/molecular genetic abnormalities not classified as favorable or adverse |
| Adverse | In the absence of favorable risk cytogenetic/molecular genetic abnormalities: |
| abn(3q) [excluding t(3;5)(q21∼25;q31∼35)/NPM1-MLF1], | |
| inv(3)(q21q26)/t(3;3)(q21;q26)/ GATA2/EVI1 | |
| add(5q)/del(5q), −5 | |
| t(5;11)(q35;p15.5)/NUP98-NSD1 | |
| t(6;9)(p23;q34)/DEK-NUP214 | |
| add(7q)/del(7q), −7 | |
| t(11q23) [excluding t(9;11)(p21∼22;q23) and t(11;19)(q23;p13)] | |
| t(9;22)(q34;q11)/BCR-ABL | |
| −17/abn(17p)/TP53 mutation | |
| Complex karyotype (≥4 unrelated abnormalities) | |
| ASXL1 mutation | |
| DNMT3A mutation | |
| FLT3-ITD | |
| MLL-PTD | |
| RUNX1 mutation |
The cytogenetic classification is based on analysis of 5876 patients treated in successive United Kingdom national AML trials for younger adults.4 The cytogenetic adverse risk category captured virtually all patients (98%) with a monosomal karyotype, which characterizes a group of patients with dismal prognosis.101 Overall, of the 955 patients in the United Kingdom study with adverse risk disease based on cytogenetic criteria, 35% had a monosomal karyotype.4 There has been inconsistency in the definition of complex karyotype between studies.3 In the United Kingdom study, the most informative level of complexity was investigated to distinguish patients with poor prognosis AML with abnormal karyotype, lacking one of the cytogenetic changes that in their own right would have assigned a patient to the favorable or adverse risk group respectively.4 This analysis, which was restricted to patients with standard risk cytogenetic abnormalities, established that >3 unrelated abnormalities was the most informative cutoff for definition of “complex karyotype” to distinguish those with poor prognosis who should be assigned to the adverse risk group.4
Sequencing studies have confirmed and greatly extended the role of aberrant cellular processes such as epigenetic regulation and intracellular signaling in the pathogenesis of AML.12 Dysfunction of these processes can also be captured on a genome-wide scale via epigenetic and proteomic analyses.54,103 Moving forward, correlation of mutational data with these and other “-omic” data sets may further refine our understanding of AML biology, improve outcome prediction and treatment choice.104 In addition, large scale ex vivo functional studies, assessing the response of individual tumors to compound libraries has predicted for in vivo efficacy in selected cases of AML at relapse.105 Although it is not currently practical or financially viable to perform all of these research approaches in every AML patient on a longitudinal basis, coordination and correlation of the previously discussed global analyses may allow the design of directed studies. These would assess for the presence of specific mutations, epigenetic patterns at particular loci, specific signaling abnormalities, and/or ex vivo sensitivities to agents selected based on genotype or other previously discussed characteristic. In the future, such combined-modality, directed studies may provide the Rosetta stone that allows our aspirations of specific therapy, personalized against the mutational complement and biological characteristics of individual tumors, to become a reality. It is hoped that such therapy will accelerate real progress in AML outcomes that have stagnated over the past 30 years.
Acknowledgments
This work was supported by a grant from the National Institute for Health Research (NIHR) under its Programme Grants for Applied Research Programme (RP-PG-0108-10093) (D.G. and A.I.). D.G. also acknowledges support from Leukaemia & Lymphoma Research of Great Britain (now Bloodwise), the NIHR Biomedical Research Centre based at Guy’s and St. Thomas’ National Health Service (NHS) Foundation Trust and King’s College London, the Guy’s and St. Thomas’ Charity, and the Minimal Residual Disease Workpackage (WP12) of the European LeukemiaNet. Work in the laboratory of B.J.P.H. is funded by the Wellcome trust, the Medical Research Council, Bloodwise, the Kay Kendall Research Fund, Worldwide Cancer Research, the Leukemia and Lymphoma Society of America, and the European Research Council. B.J.P.H. acknowledges support from Wellcome trust Strategic Award 100140 and the NIHR Cambridge Biomedical Research Centre. B.J.P.H. would like to thank Faisal Basheer for his kind help with the preparation of Figure 3B.
The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health.
Authorship
Contribution: D.G., A.I., and B.J.P.H. wrote the manuscript and approved the final version.
Conflict-of-interest disclosure: B.J.P.H. is an investigator on #NCT01943851. D.G. and B.J.P.H. are members of the UK National Cancer Research Institute AML Working Group, which is conducting ISRCTN55675535, ISRCTN31682779, and ISRCTN40571019.
Correspondence: David Grimwade, Cancer Genetics Laboratory, Department of Medical & Molecular Genetics, 8th Floor, Tower Wing, Guy’s Hospital, London SE1 9RT, United Kingdom; e-mail: david.grimwade@kcl.ac.uk; and Brian Huntly, Department of Haematology, Cambridge Institute for Medical Research, University of Cambridge, Hills Rd, Cambridge, Cambridgeshire CB2 0XY, United Kingdom; e-mail: bjph2@cam.ac.uk.