Key Points
The rs6021191 variant in NFATC2 is associated with an increased risk of asparaginase hypersensitivity and is an expression quantitative trait locus associated with expression of NFATC2.
Exome interrogation confirms the importance of the HLA-DRB1*07:01 allele in asparaginase hypersensitivity.
Abstract
Asparaginase is used to treat acute lymphoblastic leukemia (ALL); however, hypersensitivity reactions can lead to suboptimal asparaginase exposure. Our objective was to use a genome-wide approach to identify loci associated with asparaginase hypersensitivity in children with ALL enrolled on St. Jude Children’s Research Hospital (SJCRH) protocols Total XIIIA (n = 154), Total XV (n = 498), and Total XVI (n = 271), or Children’s Oncology Group protocols POG 9906 (n = 222) and AALL0232 (n = 2163). Germline DNA was genotyped using the Affymetrix 500K, Affymetrix 6.0, or the Illumina Exome BeadChip array. In multivariate logistic regression, the intronic rs6021191 variant in nuclear factor of activated T cells 2 (NFATC2) had the strongest association with hypersensitivity (P = 4.1 × 10−8; odds ratio [OR] = 3.11). RNA-seq data available from 65 SJCRH ALL tumor samples and 52 Yoruba HapMap samples showed that samples carrying the rs6021191 variant had higher NFATC2 expression compared with noncarriers (P = 1.1 × 10−3 and 0.03, respectively). The top ranked nonsynonymous polymorphism was rs17885382 in HLA-DRB1 (P = 3.2 × 10−6; OR = 1.63), which is in near complete linkage disequilibrium with the HLA-DRB1*07:01 allele we previously observed in a candidate gene study. The strongest risk factors for asparaginase allergy are variants within genes regulating the immune response.
Introduction
Current acute lymphoblastic leukemia (ALL) and lymphoma therapies depend heavily upon the use of glucocorticoids and asparaginase, among other drugs. Asparaginase is a heterologous enzyme that can influence the exposure of other drugs, including glucocorticoids.1 Moreover, allergy to asparaginase can compromise the effectiveness of both asparaginase and dexamethasone by decreasing plasma exposure to both agents, and can result in a higher risk of relapse.2 Hypersensitivity reactions to asparaginase during treatment are common, and have been associated with anti-asparaginase immunoglobulin G (IgG) antibodies rather than IgE antibodies.3,4 However, recent reports indicate that patients receiving PEGylated Escherichia coli asparaginase can develop hypersensitivity reactions to the drug without any evidence of detectable antibodies; because IgE may be bound to mast cells and may be present for only a limited time period, it is possible that IgE plays a role in asparaginase-induced reactions but eludes detection.5 Therefore, multiple pathways of asparaginase hypersensitivity are possible and currently they are not well understood.
Few studies have investigated genetic risk factors for asparaginase hypersensitivity. In a study of 485 pediatric ALL patients on St. Jude Children’s Research Hospital (SJCRH) Total XV, the rs4958351 variant in the glutamate receptor gene GRIA1 was associated with asparaginase hypersensitivity.6 Our recent study investigating the role of HLA-DRB1 genes on asparaginase hypersensitivity in 1870 patients of European ancestry identified an association with the HLA-DRB1*07:01 allele.7 However, additional agnostic genome-wide studies that encompass the racial diversity of patients with ALL are required to better understand the risk and the mechanisms of asparaginase hypersensitivity.
The present study is the largest investigation of genetic predispositions to asparaginase hypersensitivity and includes 3308 patients of diverse ancestry. We used a genome-wide association approach, including a focus on exonic variants in addition to noncoding variants, to identify genetic loci associated with asparaginase hypersensitivity.
Materials and methods
Patients
Asparaginase hypersensitivity was assessed in children with ALL enrolled on SJCRH protocols Total XIIIA (n = 154), Total XV (n = 498, ClinicalTrials.gov #NCT00137111), and Total XVI (n = 271, ClinicalTrials.gov #NCT00549848) or Children’s Oncology Group (COG) protocols POG 9906 (n = 222, ClinicalTrials.gov #NCT00005603) and AALL0232 (n = 2163, ClinicalTrials.gov #NCT00075725). Informed consent from the parents or guardians, and consent from the patients as appropriate were obtained according to Institutional Review Board guidelines for genomic research and for treatment. The schedules and doses of asparaginase used during treatment varied by protocol and have been described elsewhere.7 The association between HLA variants and asparaginase allergy among patients of European ancestry enrolled in these studies has been reported previously7 (see supplemental Table 1, available on the Blood Web site), and we have previously reported an analysis of standard noncoding “GWAS” array single nucleotide polymorphisms (SNPs) for allergy for a subset of the Total XV patients6 (supplemental Table 1). Patients enrolled on protocols Total XIIIA, Total XV, and POG 9906 received native E coli asparaginase (26%), whereas patients on Total XVI and AALL0232 received PEGylated E coli asparaginase (74%). Hypersensitivities were graded using the scales described in the National Cancer Institute’s common toxicity criteria version 1.0 for Total XIIIA,8 version 2.0 for POG 9906, version 3.0 for Total XV6 and Total XVI, and version 4.0 for COG AALL0232. Hypersensitivity reactions of grade 2 and above were considered cases. The clinical symptoms of reactions grade 2 and above can include rash, flushing, urticaria, fever ≥38°C, symptomatic bronchospasm, and anaphylaxis.
Genotyping
Germline DNA was genotyped using the Affymetrix Human Mapping 500K Array Set, the Affymetrix Genome-Wide Human SNP Array 6.0, or the Illumina Exome BeadChip array.9 Hardy-Weinberg equilibrium tests were performed using PLINK in patients of European ancestry. SNPs with genotyping call rates <95% and SNPs that were not in Hardy-Weinberg equilibrium (P < .001 for SNPs with a minor allele frequency [MAF] ≥1%) were excluded from the association analysis. The genetic ancestry of patients was estimated using STRUCTURE as previously described,10 with the European-ancestry group defined as having >90% Northern European ancestry (CEU), the Asian-ancestry group defined as having >90% East Asian ancestry (CHB/JBT), the African-ancestry group defined as having >70% West African ancestry (YRI), the Hispanics defined as having Native American ancestry >10% and whose Native American ancestry was greater than their % of African ancestry, and finally, others whose ancestry was outside the above boundaries. Affymetrix genotyping was available for 94% of patients with asparaginase hypersensitivity data (supplemental Table 1). Illumina exome array genotyping was available for 95% of the patients in the combined patient cohort (supplemental Table 1). Overall, 90% of the ALL patients with asparaginase hypersensitivity data had genotyping available from both platforms (supplemental Table 1).
Nuclear factor of activated T cells 2 (NFATC2) gene expression analysis
NFATC2 gene expression was assessed in ALL tumor samples from patients enrolled on SJCRH protocols Total XV (n = 57) and Total XVI (n = 8). RNA-seq was performed as previously described.11,12 Expression levels of NFATC2 were estimated as fragment per kilobase of transcript per million mapped reads (FPKM), and gene FPKM values were computed by summing the transcript FPKM values for each gene as described previously.13,14 NFATC2 gene expression and genotyping was also available for 52 unrelated Yoruba (YRI) HapMap samples (GSE9703).15,16 NFATC2 expression levels in HapMap YRI samples were estimated by adding the hybridization signals of probes within spliced messenger RNA regions of the gene.
Data analysis
Univariate and multivariate logistic regression was used to identify clinical risk factors associated with asparaginase hypersensitivity. The association analysis between SNP genotypes and asparaginase hypersensitivity reactions was performed using logistic regression, adjusting for significant covariates and assuming an additive genetic model.17 Genotype and phenotype association analysis was performed in PLINK.17 The association analysis between NFATC2 gene expression and rs6021191 genotyping were performed using a general linear model with R statistical software (version 2.13.2).
Pathway analysis using ingenuity
Ingenuity Pathway Analysis (IPA) was used to prioritize biological pathways related to asparaginase hypersensitivity reactions within the genome-wide analysis. Genes containing interrogated SNPs with the strongest associations (lowest 0.1% P values, with P ≤ 1.1 × 10−3) to asparaginase hypersensitivity were included in the pathway analysis (334 genes representing 499 SNPs; supplemental Table 2). Fisher’s exact test was used to determine if the genes most associated with asparaginase hypersensitivity were over-represented in each canonical pathway.
Results
Hypersensitivity reaction data to native E coli asparaginase or PEGylated E coli asparaginase were available from a total of 3308 pediatric ALL patients enrolled on 5 different protocols (Table 1). Their median age was 9.3 years (range, 0.17 to 30), there were slightly more males than females (56.2% vs 43.8%; Table 1), and 59.5% of the genotyped patients were of European genetic ancestry (Table 1).
A total of 589 patients had asparaginase hypersensitivity reactions (17.8%; Table 2). By univariate analysis both treatment protocol (P = 2.1 × 10−51; Table 2) and asparaginase formulation (P = 2.1 × 10−50; Table 2) were associated with hypersensitivity. Protocols including native E coli asparaginase had a higher frequency of hypersensitivity than those using PEGylated E coli asparaginase (35.1% vs 11.6%; Table 2). Age <10 years, ancestry, and B-cell ALL were also associated with the risk of hypersensitivity (Table 2). In multivariate analysis including treatment protocol, gender, age group, ancestry, and ALL lineage (Table 2), only protocol (P = 2.3 × 10−46), gender (P = .04), and ALL lineage (P = .003) were associated with asparaginase hypersensitivity. Patients with T-cell ALL were only enrolled on SJCRH protocols; within the standard/high-risk arms of these studies, there were more asparaginase hypersensitivity reactions among patients with B-lineage compared with those with T-cell ALL receiving identical treatment (supplemental Figure 1; P = .001). Asparaginase preparation was confounded with protocol, but it also associated with hypersensitivity in multivariate analysis (P = 7.2 × 10−45) when removing protocol as a covariate from the regression model.
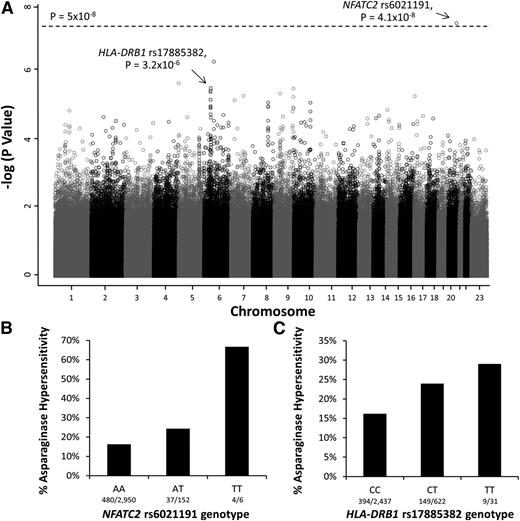
To identify genomic variants associated with asparaginase hypersensitivity, a multivariate logistic regression model adjusted for treatment arm, ancestry (as a categorical covariate), gender, age, and ALL lineage was used. The minor allele of an intronic polymorphism in NFATC2 (rs6021191) was associated with a higher risk of hypersensitivity at the genome-wide significance threshold (P = 4.1 × 10−8, odds ratio [OR] = 3.11; Figure 1A-B). Focusing on nonsynonymous SNPs, the minor allele at rs17885382 in the HLA-DRB1 gene (P = 3.23 × 10−26, OR = 1.63; Figure 1A,C) had the strongest association with asparaginase hypersensitivity.
Manhattan plot of P values for genome-wide SNP association with asparaginase hypersensitivity. (A) The Manhattan plot shows the negative log10 of the P values for association with asparaginase hypersensitivity vs each chromosome on the x-axis. The open circles indicate SNPs genotyped using the Affymetrix 6.0 array, whereas the open squares indicate SNPs genotyped by the Illumina Exome Beadchip array. The dashed line denotes the genome-wide threshold (P = 5 × 10−8). (B) SNPs genotyped using the Affymetrix SNP 6.0 array identified the NFATC2 rs6021191 variant associated with asparaginase hypersensitivity (P = 4.1 × 10−8, OR = 3.11, CI = 2.07-4.66). AA, subjects homozygous for the reference allele (A); TT, subjects homozygous for the variant allele (T); AT, heterozygotes. (C) Illumina Exome array genotyping identified the strongest association with the HLA-DRB1 rs17885382 variant (P = 3.2 × 10−6, OR = 1.63, CI = 1.33-2.00). CC, subjects homozygous for the reference allele (C); TT, subjects homozygous for the variant allele (T); CT, heterozygotes. Associations were determined using a general linear model adjusted for treatment, ALL immunophenotype, gender, age group, and ancestry.
Manhattan plot of P values for genome-wide SNP association with asparaginase hypersensitivity. (A) The Manhattan plot shows the negative log10 of the P values for association with asparaginase hypersensitivity vs each chromosome on the x-axis. The open circles indicate SNPs genotyped using the Affymetrix 6.0 array, whereas the open squares indicate SNPs genotyped by the Illumina Exome Beadchip array. The dashed line denotes the genome-wide threshold (P = 5 × 10−8). (B) SNPs genotyped using the Affymetrix SNP 6.0 array identified the NFATC2 rs6021191 variant associated with asparaginase hypersensitivity (P = 4.1 × 10−8, OR = 3.11, CI = 2.07-4.66). AA, subjects homozygous for the reference allele (A); TT, subjects homozygous for the variant allele (T); AT, heterozygotes. (C) Illumina Exome array genotyping identified the strongest association with the HLA-DRB1 rs17885382 variant (P = 3.2 × 10−6, OR = 1.63, CI = 1.33-2.00). CC, subjects homozygous for the reference allele (C); TT, subjects homozygous for the variant allele (T); CT, heterozygotes. Associations were determined using a general linear model adjusted for treatment, ALL immunophenotype, gender, age group, and ancestry.
The MAF for the NFATC2 rs6021191 variant was highest in patients of non-European ancestry (0.062; supplemental Table 3), and only 0.001 among patients of European ancestry, MAFs that are consistent with those reported in the general population (supplemental Table 3). The rs6021191 variant was associated with hypersensitivity across all non-European ancestries (supplemental Table 3), with the largest effect size seen in those with primarily Asian ancestry (OR = 8.71; supplemental Figure 2A), followed by those with African ancestry (OR = 4.45; supplemental Figure 2A). The association was also present across all protocols (supplemental Figure 2B) and with both asparaginase preparations (supplemental Figure 2C).
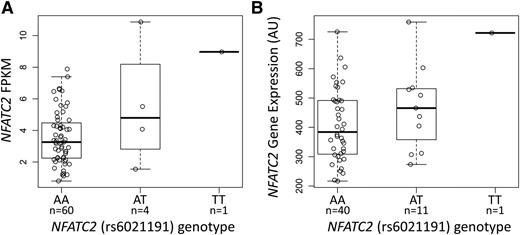
The NFATC2 rs6021191 variant is located in an intron of the gene and it is in a region with weak enhancer activity in HUVEC and K562 cell lines,18 suggesting that the SNP may influence the expression of NFATC2. We found higher NFATC2 gene expression in both ALL tumor samples and in YRI HapMap lymphoblastoid cell line samples carrying the rs6021191 variant allele (P = 1.1 × 10−3 and 0.03, respectively; Figure 2A-B), indicating that the SNP is an expression quantitative trait locus (eQTL).
The NFATC2 rs6021191 variant is an eQTL. RNA-seq data were available from the ALL tumor samples of 65 SJCRH patients and from 52 unrelated YRI HapMap samples to investigate the association between rs6021191 and NFATC2 gene expression. We found higher NFATC2 gene expression in samples with the rs6021191 variant compared with samples without the variant in both (A) ALL tumor samples (P = 1.1 × 10−3) and (B) YRI HapMap samples (P = .03). Allele abbreviations are explained in Figure 1.
The NFATC2 rs6021191 variant is an eQTL. RNA-seq data were available from the ALL tumor samples of 65 SJCRH patients and from 52 unrelated YRI HapMap samples to investigate the association between rs6021191 and NFATC2 gene expression. We found higher NFATC2 gene expression in samples with the rs6021191 variant compared with samples without the variant in both (A) ALL tumor samples (P = 1.1 × 10−3) and (B) YRI HapMap samples (P = .03). Allele abbreviations are explained in Figure 1.
The MAF for the HLA-DRB1 rs17885382 variant was highest in patients of European ancestry (MAF = 0.125; supplemental Table 4) and present in all non-European ancestries at a MAF ≥0.05 (supplemental Table 4). The effect size was most pronounced in patients of European ancestry (P = 5.9 × 10−5, OR = 1.68; supplemental Figure 3A), but elevated in other ancestries, for all treatment protocols, and for both asparaginase preparations (supplemental Figure 3A-C).
The rs17885382 variant is located in exon 2 of the HLA-DRB1 gene (Arg > Gln, 54). Our previous candidate gene analysis of HLA alleles associated with asparaginase allergies in patients of European ancestry identified an association with 10 amino acids in HLA-DRB1, and position 54 (amino acid position 25 when excluding the leader signal sequence) was among those implicated.7 From a total of 1540 HLA-DRB1 alleles with amino acid sequences included in the International ImMunoGene Tics/HLA database,19 only HLA-DRB1*01:13, HLA-DRB1*03:54, HLA-DRB1*13:92, HLA-DRB1*14:26, HLA-DRB1*15:24, and HLA-DRB1*07 encode for a Gln in position 54. Our prior HLA-focused analysis discovered that HLA-DRB1*07:01 was associated with hypersensitivity,7 and it is the only allele among those encoding for Gln at position 54 with a MAF >1%. Consistent with our previous analysis, the HLA-DRB1 rs17885382 variant was in linkage disequilibrium with the HLA-DRB1*07:01 allele (R2 = 0.94); therefore, it is likely that the haplotype is driving the association between rs17885382 and asparaginase hypersensitivity.
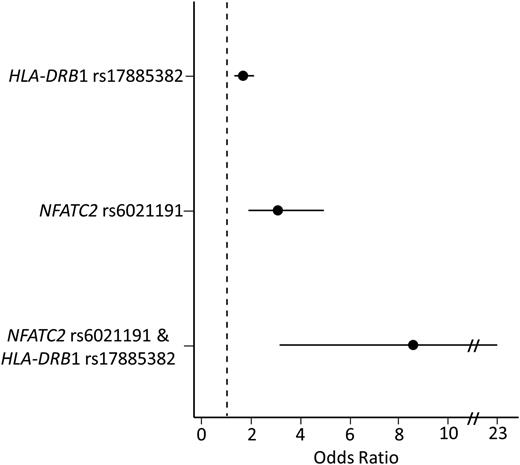
In a multivariate analysis, including both rs6021191 (NFATC2) and rs17885382 (HLA-DRB1) genotype, both variants remained associated with asparaginase hypersensitivity (Prs6021191 = 1.8 × 10−8, Prs17885382 = 3.2 × 10−6), and modest increases in effect size were seen after adjusting for the genotypes (ORrs6021191 = 3.35, ORrs17885382 = 1.66). The frequency of asparaginase hypersensitivity in patients with neither of these variants was 14.8% (321 of 2173). In comparison, 168 of the 738 (22.7%) patients with one variant (at rs6021191 or rs17885382) developed hypersensitivity (Figure 3) (ORrs6021191 = 3.07, Prs6021191 = 5.4 × 10−6; ORrs17885382 = 1.66, Prs17885382 = 3.5 × 10−5). The risk of hypersensitivity was higher still in patients carrying both alleles (9 out of 19 or 47.4%) as compared with patients with no variant alleles (Figure 3) (OR = 8.58, P = 1.7 × 10−5) and to patients carrying a single variant allele (ORrs6021191 = 2.79, Prs6021191 = 0.05; ORrs17885382 = 5.18, Prs17885382 = 1.2 × 10−3). Similar results were found taking the preparation of asparaginase into consideration. Patients receiving native E coli asparaginase carrying both risk alleles had an incidence of allergy of 50% compared with 32% among noncarriers of risk alleles, whereas patients receiving PEGylated E coli asparaginase had an incidence of allergy of 47% compared with 10% among noncarriers. Furthermore, after conditioning on the association for the NFATC2 rs6021191 and HLA-DRB1 rs17885382 variants, no other SNP reached the genome-wide significance threshold.
Patients carrying both the HLA-DRB1 rs17885382 and NFATC2 rs6021191 variant have a higher risk of developing asparaginase hypersensitivity compared with patients with no risk variant. The risk of developing hypersensitivity was determined for patients carrying a single-risk variant (HLA-DRB1 rs17885382 or NFATC2 rs6021191) or for patients carrying both risk variants (NFATC2 rs6021191 and HLA-DRB1 rs17885382). The risk of hypersensitivity was higher in patients carrying a single variant (ORrs6021191 = 3.07, CIrs6021191 = 1.87-4.94, Prs6021191 = 5.4 × 10−6; ORrs17885382 = 1.66, CIrs17885382 = 1.3-2.1, Prs17885382 = 3.5 × 10−5) or both variants (OR = 8.58, CI = 3.14-22.92, P = 1.7 × 10−5) compared with patients with no risk alleles. The associations were determined using a general linear model adjusted for treatment, ALL immunophenotype, gender, age group, and ancestry.
Patients carrying both the HLA-DRB1 rs17885382 and NFATC2 rs6021191 variant have a higher risk of developing asparaginase hypersensitivity compared with patients with no risk variant. The risk of developing hypersensitivity was determined for patients carrying a single-risk variant (HLA-DRB1 rs17885382 or NFATC2 rs6021191) or for patients carrying both risk variants (NFATC2 rs6021191 and HLA-DRB1 rs17885382). The risk of hypersensitivity was higher in patients carrying a single variant (ORrs6021191 = 3.07, CIrs6021191 = 1.87-4.94, Prs6021191 = 5.4 × 10−6; ORrs17885382 = 1.66, CIrs17885382 = 1.3-2.1, Prs17885382 = 3.5 × 10−5) or both variants (OR = 8.58, CI = 3.14-22.92, P = 1.7 × 10−5) compared with patients with no risk alleles. The associations were determined using a general linear model adjusted for treatment, ALL immunophenotype, gender, age group, and ancestry.
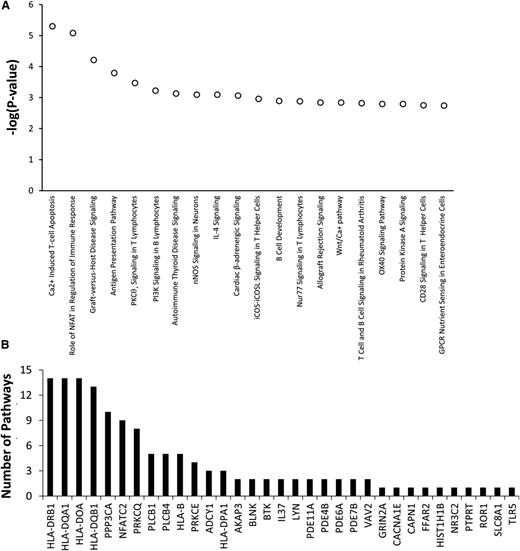
Among the top 20 pathways with P values <3 × 10−3 invoked by the 334 genes having top ranked SNPs associated with asparaginase hypersensitivity, 13 pathways involved T-cell apoptosis, T-cell signaling, T-cell activation, or T-cell disorders (Figure 4A). Genes within the top 20 pathways included PPP3CA, PRKCQ, and PRKCE, which are involved in T-cell function (Figure 4B). Therefore, the pathway analysis suggests that genetic variants within genes involved in T-cell function may play a role in the development of asparaginase hypersensitivity.
Genes involved in T-cell function may contribute to the risk of developing asparaginase hypersensitivity reactions. IPA was used to determine if genes most associated with asparaginase hypersensitivity were overrepresented in specific biological pathways. A total of 334 genes invoked by 499 SNPs associated with asparaginase hypersensitivity annotated to 234 distinct IPA pathways. (A) The top 20 pathways identified, all with P < 3 × 10−3, were enriched for genes involved in T-cell apoptosis, T-cell signaling, T-cell activation, or T-cell disorders. (B) A total of 33 genes containing SNPs associated with hypersensitivity were included within the top 20 canonical pathways (shown on x-axis). Many of these genes were included in multiple pathways. The number of pathways (out of 20) in which each of these 33 genes is involved is plotted on the y-axis. For example, HLA-DRB1 is involved in 14 out of the 20 top canonical pathways. GPCR, G-protein coupled receptor; nNOS, neuronal NOS; IL, interleukin; iCOS, inducible T-cell costimulator and its ligand, iCOSL.
Genes involved in T-cell function may contribute to the risk of developing asparaginase hypersensitivity reactions. IPA was used to determine if genes most associated with asparaginase hypersensitivity were overrepresented in specific biological pathways. A total of 334 genes invoked by 499 SNPs associated with asparaginase hypersensitivity annotated to 234 distinct IPA pathways. (A) The top 20 pathways identified, all with P < 3 × 10−3, were enriched for genes involved in T-cell apoptosis, T-cell signaling, T-cell activation, or T-cell disorders. (B) A total of 33 genes containing SNPs associated with hypersensitivity were included within the top 20 canonical pathways (shown on x-axis). Many of these genes were included in multiple pathways. The number of pathways (out of 20) in which each of these 33 genes is involved is plotted on the y-axis. For example, HLA-DRB1 is involved in 14 out of the 20 top canonical pathways. GPCR, G-protein coupled receptor; nNOS, neuronal NOS; IL, interleukin; iCOS, inducible T-cell costimulator and its ligand, iCOSL.
Discussion
Asparaginase is a mainstay in the treatment of ALL, but immune responses to asparaginase can decrease the systemic exposure to asparaginase and result in a higher risk of relapse.2 In this study, we used a genome-wide association approach to identify genetic loci associated with asparaginase hypersensitivity within a racially diverse set of pediatric patients with ALL. We found that the rs6021191 variant is associated with a higher risk of asparaginase hypersensitivity at the genome-wide significance level (Figure 1A-B), and the variant is associated with higher NFATC2 messenger RNA expression (Figure 2A-B). The NFATC2 variant was rare in patients of European ancestry (supplemental Table 3); nevertheless, we were able to identify the association between the NFATC2 variant and asparaginase hypersensitivity because a multiethnic cohort was available for our investigation. Other variants in NFATC2 have also been related to immune-mediated phenotypes, such as narcolepsy,20 self-reported allergies (cat, pollen, and dust-mite allergy),21 and thiazolidinedione-induced edema.22 Interestingly, both self-reported allergies and narcolepsy also have associations with HLA genetic variants.21,23
The NFATC2 gene encodes a cytoplasmic component of the NFAT transcription factor family. Upon T-cell receptor stimulation, cytoplasmic NFATC2 is dephosphorylated and translocated to the nucleus where it participates in gene regulation.24 The role of NFATC2 on the risk of drug-induced allergy is unknown, yet studies have shown that NFATC2 can influence the development and function of regulatory T cells and can have either a negative25 or positive26 regulatory influence on the immune response, depending on the antigen. Nevertheless, several studies using NFAT inhibitors for different immune-related disease models support that inhibition of the NFAT pathway can attenuate an immune response.27-32 Moreover, among individuals with Down syndrome, the inhibition of the NFAT pathway (partly via overexpression of DYRK1A and DSCR1 due to trisomy 21)33,34 is hypothesized to be related to the cardiac, neurologic, and immunologic defects associated with Down syndrome,34 and individuals with Down syndrome may have attenuated antibody responses to several vaccine antigens.35 Based on this role of the NFAT pathway in Down syndrome, and on our finding of a higher incidence of asparaginase hypersensitivity among those carrying the NFATC2 variant, we hypothesized that Down syndrome patients might have a lower incidence of asparaginase hypersensitivity. Indeed, we observed a lower incidence of asparaginase hypersensitivity among ALL patients with Down syndrome than in those without Down syndrome (6% vs 17%, P multivariate = 8.4 × 10−3; supplemental Figure 4), and this effect was found among protocols using either preparation of asparaginase, with 8% (4 of 48) of patients with Down syndrome having allergic reactions to PEGylated E coli asparaginase compared to 12% of patients without Down syndrome, and 0% (0 of 22) of patients with Down syndrome having allergic reactions to native E coli asparaginase compared with 37% among patients without Down syndrome. These results further support the notion that variability in NFATC2 plays a role in the risk of allergy to asparaginase, and that inhibition of NFATC2 might mitigate sensitization to asparaginase.
Our study included not only standard “GWAS” arrays (mostly noncoding SNPs), but also the exome array for interrogation of coding variants. The top hit for a coding variant was in the HLA-DRB1 gene and the SNP was in linkage disequilibrium with the HLA-DRB1*07:01 allele.7 Therefore, the top coding variant that we identified using the exome array was the same variant identified by our candidate gene study. However, the patients included in our candidate gene study were solely of European ancestry (supplemental Table 1), and the current study extends our previous report identifying HLA-DBRB1*07:01 as a risk allele for asparaginase hypersensitivity by showing that HLA-DRB1*07:01 is also a risk allele in non-European patients (supplemental Figure 3). Furthermore, we show that the association between the HLA-DRB1 rs17885382 variant is independent from the NFATC2 rs6021191 variant and that patients carrying both alleles have a higher risk of developing asparaginase hypersensitivity compared with patients with no risk alleles or with a single risk allele (Figure 3). Interestingly, the GRIA1 rs4958351 variant that we previously found to associate with asparaginase hypersensitivity6 was also associated with the risk of asparaginase hypersensitivity in our combined patient cohort (P = .03; OR = 1.2). However, the risk of allergy was stronger in patients receiving native E coli asparaginase (OR = 1.55) than in patients receiving PEGylated E coli asparaginase (OR = 1.02).
In conclusion, we have identified that the NFATC2 rs6021191 variant is an eQTL that is associated with the risk of developing asparaginase hypersensitivity. This is the first study to identify an association between NFATC2 and a drug-induced hypersensitivity. Moreover, the results from our single SNP and pathway analyses suggest that T cells play an important role in the development of asparaginase hypersensitivity, likely due to their role in B-cell activation and differentiation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by grants GM 92666, CA 21765, CA 142665, CA 36401, RCA 156449, and U10 CA98543, COG Statistical Center (U10 CA98413), and COG Specimen Banking (U24 CA114766) from the National Insitutes of Health and by the American Lebanese Syrian Associated Charities.
Authorship
Contribution: C.A.F. and M.V.R. designed the project, analyzed and interpreted the data, and drafted the manuscript; C.A.F., C.S., and W.Y. performed statistical analysis; and C.S., W.Y., C.G.M., C.Q., E.L.,W.P.B., C.L., L.B.R., T.C., S.E.K., M.L.L., E.A.R., N.J.W. S.P.H., W.L.C., S.J., C.-H.P., W.E.E., and M.D. contributed to the writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mary V. Relling, St. Jude Children’s Research Hospital, 262 Danny Thomas Place, Mail Stop 313, Memphis, TN 38105; e-mail: mary.relling@stjude.org.