Key Points
MN1 promotes self-renewal and inhibits differentiation of CD34+ cord blood cells in vitro.
De novo leukemogenesis is engineered by MN1 and NUP98HOXD13 expression in cord blood cells.
Abstract
Leukemic transformation of human cells is a complex process. Here we show that forced expression of MN1 in primitive human cord blood cells maintained on stromal cells in vitro induces a transient, but not serially transplantable, myeloproliferation in engrafted mice. However, cotransduction of an activated HOX gene (NUP98HOXD13) with MN1 induces a serially transplantable acute myeloid leukemia (AML). Further characterization of the leukemic cells generated from the dually transduced cells showed the activation of stem cell gene expression signatures also found in primary human AML. These findings show a new forward genetic model of human leukemogenesis and further highlight the relevance of homeobox transcription factors in the transformation process.
Introduction
Many classes of genes are now implicated in the pathogenesis of acute myeloid leukemia (AML) including transcription factors, tyrosine kinases, and epigenetic regulators.1 The specific role of many of these genes is, however, not clear. Of particular interest in this regard are the HOX genes and their cofactor MEIS1. These genes have been found to promote the self-renewal of normal hematopoietic stem cells, are frequently overexpressed in primary human AML cells, and are capable of causing AML when introduced into primitive mouse hematopoietic cells.2 To date, however, only mixed lineage leukemia (MLL)-fusion genes have been found to induce AML de novo. Interestingly, however, normal human cord blood (CB) cells transduced with MLL-AF9 (an upstream regulator of HOX genes) consistently gave rise to acute lymphoblastic leukemia in nonobese-diabetic-scid mice, whereas the same cells generated AML in nonobese-diabetic-scid transgenic mice expressing multiple human growth factors, underscoring the importance of instructive signals from the microenvironment in MLL-transformed leukemias.3
Increased expression of MN1 has been identified as a poor-risk prognostic marker in cytogenetically normal AML patients.4 Conversely, loss of MN1 expression impaired the proliferation and clonogenic activity of cells from a human leukemia cell line.5 We previously identified HOXA9 and NUP98-HOXD13 (ND13) as potent collaborating genes with MN1 in leukemic transformation in mice,6 and we identified MN1 as a cofactor of the HOXA9/MEIS1 transcriptional complex.7
In this report, we investigate the transforming potential of MN1 in human CB cells and present a model of stepwise transformation to human AML.
Study design
See supplemental Data available at the Blood Web site for a full description of materials and methods.
Viral vectors
MN1 was expressed from pCCl-c-MNDUSMN1pgkGFP, NUP98HOXD13 (ND13) from pCCl-c-MNDUSFND13pgkGFP and pCCl-c-MNDUSFND13pgkYFP lentiviral vectors (supplemental Figure 1A).8 Umbilical CB cells were obtained at the time of cesarean delivery of healthy, full-term infants, with consent according to procedures approved by the Research Ethics Board of the University of British Columbia.
Mice
NOD.Cg-Prkdcscid Il2rγtm1Wj1/SzJ NOD/SCID-IL-2Rγc-null, NSG, (originally obtained from Jackson Labs) and NSG mice transgenic for human interleukin-3 (IL-3), granulocyte-macrophage-colony-stimulating factor and Steel factor (NSG-3GS)9 were bred in the animal resource center at the British Columbia Cancer Research Centre. All mouse experimental procedures were performed in accordance with Canadian Council on Animal Care guidelines with approval from the University of British Columbia.
Results and discussion
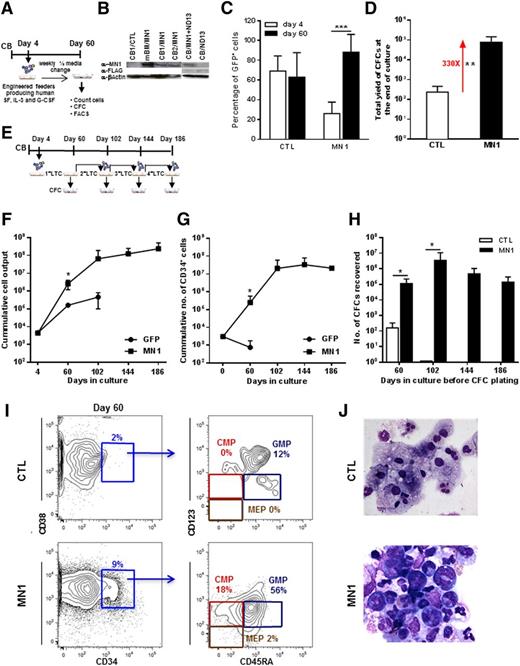
To investigate the effect of increased MN1 expression in primitive normal human hematopoietic cells, we exposed paired populations of CD34+ CB cells to human full-length MN1 (MN1) or control GFP (CTL) viruses prepared from either γ-retroviral or lentiviral vectors (4 and 10 experiments, respectively, Figure 1A and supplemental Figure 1A). Transduction efficiencies of 26 ± 12% for MN1 and 69 ± 15% for CTL virus (n = 14 and 9, respectively) and RNA/protein expression of MN1 were documented (Figure 1B and supplemental Figure 1B-C). MN1-transduced cells produced more GFP+ cells (Figure 1C) and colonies than CTL cells when cultured in stromal feeder-containing long-term cultures (LTCs) (Figure 1D). Growth-stimulating effects of MN1 on primitive hematopoietic cells were also evident by an increased frequency (fivefold) of long-term culture-initiating (LTC-IC) cells, increased yield of CFCs per LTC-IC (∼threefold) (supplemental Figure 1D-F); a greatly increased output of total cells, CD34+ cells, and CFCs on extended serial LTCs up to 186 days (Figure 1E-H); and enriched stem and progenitor phenotypes assessed by FACS and morphology, whereas CTL cells primarily consisted of neutrophils (CD33+CD15+) and monocytes/macrophages (CD33+CD11b+CD14+) (see Figure 1I-J for representative FACS blots and morphology; see supplemental Figure 1I for summary of FACS analyses). Remarkably, cells with the phenotype of CMPs, GMPs, and MEPs and morphologies of blast and primitive myeloid cells were still present after 20 weeks in the cultures of MN1-transduced cells (supplemental Figure 1G-H).
Constitutive expression of MN1 immortalizes human CB cells in vitro. (A) Outline of stromal feeder-containing LTC-IC assay or LTC with human CB cells. (B) Western blot showing protein expression of MN1 in MN1-transduced mouse BM and CB cells and FLAG protein (FLAG-ND13) in ND13-transduced CB cells. β-actin was probed on the same blot as loading control. (C) Proportion of GFP+ cells 2 days after the end of transduction and after culture on feeder cells for 56 days (at the time of CFC plating) for CTL- and MN1-transduced CB cells (CTL, n = 9; MN1, n = 14). (D) Total yield of colonies from primary LTC assay of CTL- or MN1-transduced CB cells (CTL, n = 9; MN1, n = 14). (E) Outline of serial stromal feeder-containing LTC assay. After each LTC, cells were placed in CFC media and a proportion of them were replated on fresh-feeder layers for the next round of LTC assay. (F) Cumulative total cell output from primary to quaternary LTC assays (CTL, n = 2; MN1, 2nd n = 6, 3rd n = 4, 4th n = 2). (G) Cumulative CD34+ cell output from primary to quaternary LTC assays (CTL, 1st n = 5; MN1, 1st n = 10, 2nd n = 5, 3rd n = 2, 4th n = 1). (H) Number of CFCs recovered from primary to quaternary LTC assays (CTL, first n = 2, second n = 2; MN1, first n = 6, second n = 6, third n = 4, fourth n = 2). (I) Proportion of hematopoietic progenitor cells in LTCs of CTL- and MN1-transduced CB cells (day 60 on feeder cells, a representative fluorescence-activated cell sorter [FACS} blot is shown). (J) Morphology of CTL- and MN1-transduced CB cells at the end of the 60-day culture period. Micrographs were acquired using an Axioplan2 Zeiss microscope and images were captured using OpenLab software (Perkin Elmer); original magnification ×400 for CTL and ×630 for MN1. Data represent mean ± standard deviation. *P < .05, **P < .01, ***P < .001. CMP, common myeloid progenitor CD34+CD38+CD123loCD45RA−; GMP, granulocyte-macrophage progenitor CD34+CD38+CD123loCD45RA+; MEP, megakaryocyte-erythrocyte progenitor CD34+CD38+CD123−CD45RA−.
Constitutive expression of MN1 immortalizes human CB cells in vitro. (A) Outline of stromal feeder-containing LTC-IC assay or LTC with human CB cells. (B) Western blot showing protein expression of MN1 in MN1-transduced mouse BM and CB cells and FLAG protein (FLAG-ND13) in ND13-transduced CB cells. β-actin was probed on the same blot as loading control. (C) Proportion of GFP+ cells 2 days after the end of transduction and after culture on feeder cells for 56 days (at the time of CFC plating) for CTL- and MN1-transduced CB cells (CTL, n = 9; MN1, n = 14). (D) Total yield of colonies from primary LTC assay of CTL- or MN1-transduced CB cells (CTL, n = 9; MN1, n = 14). (E) Outline of serial stromal feeder-containing LTC assay. After each LTC, cells were placed in CFC media and a proportion of them were replated on fresh-feeder layers for the next round of LTC assay. (F) Cumulative total cell output from primary to quaternary LTC assays (CTL, n = 2; MN1, 2nd n = 6, 3rd n = 4, 4th n = 2). (G) Cumulative CD34+ cell output from primary to quaternary LTC assays (CTL, 1st n = 5; MN1, 1st n = 10, 2nd n = 5, 3rd n = 2, 4th n = 1). (H) Number of CFCs recovered from primary to quaternary LTC assays (CTL, first n = 2, second n = 2; MN1, first n = 6, second n = 6, third n = 4, fourth n = 2). (I) Proportion of hematopoietic progenitor cells in LTCs of CTL- and MN1-transduced CB cells (day 60 on feeder cells, a representative fluorescence-activated cell sorter [FACS} blot is shown). (J) Morphology of CTL- and MN1-transduced CB cells at the end of the 60-day culture period. Micrographs were acquired using an Axioplan2 Zeiss microscope and images were captured using OpenLab software (Perkin Elmer); original magnification ×400 for CTL and ×630 for MN1. Data represent mean ± standard deviation. *P < .05, **P < .01, ***P < .001. CMP, common myeloid progenitor CD34+CD38+CD123loCD45RA−; GMP, granulocyte-macrophage progenitor CD34+CD38+CD123loCD45RA+; MEP, megakaryocyte-erythrocyte progenitor CD34+CD38+CD123−CD45RA−.
Cytotoxicity testing with all-trans retinoic acid showed that the IC50 for MN1-transduced cells in vitro was 100-fold higher than for the CTL cells (30 μM vs 0.3 μM, supplemental Figure 1J; P = .02). In summary, transduction of CD34+ CB cells with MN1 enhanced their proliferation and self-renewal, altered and delayed their differentiation at the CMP and GMP stage, and conferred resistance to all-trans retinoic acid.
When MN1-transduced CB cells were transplanted in NSG-3GS mice, a bias toward myeloid engraftment in bone marrow (BM) was observed (supplemental Figure 2A-B,D). MN1 cells were CD34-CD33+CD11b+CD15+123+117+ (data not shown) and MN1 mice displayed left-shifted myelopoiesis (supplemental Figure 2C right panel). Engraftment of transduced cells in peripheral blood, BM, and spleen was lower for MN1 than for CTL cells (supplemental Figure 2D) because of a lower initial transduction efficiency (24 ± 1% vs 49 ± 6%) and lower numbers of transplanted cells (median 1.3 × 105 vs 2 × 105). Median survival was shorter in MN1 mice (82 days) than in CTL mice (117 days) (P = .06, supplemental Figure 2E). Three of 4 MN1 mice showed increased BM blast counts of 5% to 7%, whereas CTL mice exhibited <1%. CTL mice died with signs of graft-versus-host disease, such as hypocellular BM and T-cell infiltration of the spleen.10-12 Based on myeloid phenotype, blast count, and inability of disease transplantability, we concluded that MN1 alone induces a transient myeloproliferation, but not AML.
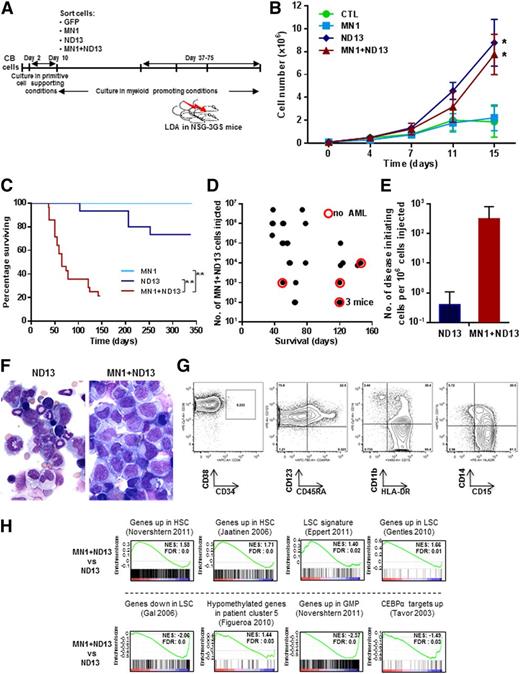
In an effort to stimulate full leukemic transformation, we coexpressed MN1 and ND13 in CB cells and sorted and expanded these cells in vitro under stroma-free conditions (Figure 2A). Both ND13+ and MN1+ND13+ CB cells showed enhanced growth in the first 2 weeks compared with MN1- or CTL-transduced cells (Figure 2B). Cells from these 1- to 3-month-old cultures were then injected intrafemorally into NSG-3GS mice (1 × 102 to 5 × 106 cells per mouse). Strikingly, mice injected with MN1+ND13+ cells developed a rapidly fatal AML with a median survival of 65 days (in ND13 or MN1 mice, median survival was 148 or 338 days, respectively, Figure 2C). The development of AML and survival correlated with the number of MN1+ND13+ cells injected (Figure 2D). Most of these mice (22 of 28) displayed splenomegaly (spleen weight of 142 ± 97 mg, n = 16). In contrast, only 4 of 10 mice transplanted with ND13+ cells that had initially engrafted (from a total of 16) showed persistence of immature myeloid cells (41 ± 3% CD45+CD33+ cells, n = 4) in the BM at the time of sacrifice and no splenomegaly (spleen weight of 38 ± 15 mg, n = 4). Five of 6 recipients of MN1-transduced cells showed engraftment initially in the injected femur, but these had all disappeared by 2 months after transplantation, suggesting that MN1 cells are dependent on stroma support to sustain stem-like cells in vitro. Assessment of mice that were administered limiting dilution transplants showed that the frequency of disease-initiating cells was almost 1000-fold higher for MN1+ND13+ cells as compared with ND13+ cells (3200 vs 2.6 × 106, Figure 2E), demonstrating the leukemic synergy that can be obtained when MN1 and ND13 are coexpressed in CB cells. MN1+ND13+ cells were CD33+CD34−CD38+CD123+HLA-DR+CD15+, corresponding to aberrant progenitor cells that do not express CD34 with partial expression of CD14 and CD11b and variable expression of granulocytic markers (Figure 2F left panel and 2G). Serial transplantation of AML cells from 3 primary mice into secondary recipients showed engraftment with GFP+YFP+ positive (MN1+ND13+) human myeloid cells in the BM (33% to 87% CD45+CD33+cells 64 days after injection) in 4 of 15 secondary recipients. Successful engraftment was correlated to the number of injected cells, showing that 1.2 million cells sufficed to induce secondary AML, whereas transplants of 250 000 cells did not engraft.
MN1 and NUP98HOXD13 cooperate to induce leukemogenesis from human CB cells in vivo. (A) Outline of the experimental design to evaluate the in vitro leukemic stem cell expansion potential of transduced CD34+ CB cells in suspension culture. (B) In vitro proliferation of transduced and sorted cells cultured in myeloid-promoting conditions (Steel factor, FL, IL-3, IL-6, and TPO, each 10 ng/mL) (starting between 33, 38, and 50 days after transduction; mean ± standard error of the mean, CTL n = 4, for MN1, ND13, MN1+ND13 n = 6). *P < .05 for the comparison CTL vs ND13, CTL vs MN1+ND13, MN1 vs ND13, and MN1 vs MN1+ND13 on day 15. (C) Survival of NSG-3GS mice receiving transplants of CB cells cultured for 1 to 3 months in vitro (MN1, n = 6; ND13, n = 16; MN1+ND13, n = 28). **P < .01. (D) Correlation between diagnosis of AML and survival and the number of MN1+ND13+ CB cells transplanted in NSG-3GS mice (mice from survival curve in Figure 2C). Each dot represents a mouse. Red circle indicates the absence of AML at the time of sacrifice. (E) Number of disease initiating cells per 106 injected cells in ND13 and MN1+ND13-transduced CB cells transplanted in NSG-3GS mice after 1 to 3 months of culture. (F) Morphology of BM cells of ND13 (103 days posttransplant, left panel) and MN1+ND13 (38 days posttransplant, right panel) mice at sacrifice. Wright-Giemsa–stained marrow cytospins from representative mice. Micrographs were acquired using an Axioplan2 Zeiss microscope and images were captured using OpenLab software (Perkin Elmer); original magnification ×400. (G) Immunophenotype of GFP+YFP+ BM cells from a MN1+ND13 mouse with AML at sacrifice. Representative FACS blots are shown. (H) Gene set enrichment analysis of indicated signatures comparing MN1+ND13+ and ND13+ CB cells cultured for 60 days in myeloid promoting conditions. FDR, false discovery rate; NES, normalized enrichment score.
MN1 and NUP98HOXD13 cooperate to induce leukemogenesis from human CB cells in vivo. (A) Outline of the experimental design to evaluate the in vitro leukemic stem cell expansion potential of transduced CD34+ CB cells in suspension culture. (B) In vitro proliferation of transduced and sorted cells cultured in myeloid-promoting conditions (Steel factor, FL, IL-3, IL-6, and TPO, each 10 ng/mL) (starting between 33, 38, and 50 days after transduction; mean ± standard error of the mean, CTL n = 4, for MN1, ND13, MN1+ND13 n = 6). *P < .05 for the comparison CTL vs ND13, CTL vs MN1+ND13, MN1 vs ND13, and MN1 vs MN1+ND13 on day 15. (C) Survival of NSG-3GS mice receiving transplants of CB cells cultured for 1 to 3 months in vitro (MN1, n = 6; ND13, n = 16; MN1+ND13, n = 28). **P < .01. (D) Correlation between diagnosis of AML and survival and the number of MN1+ND13+ CB cells transplanted in NSG-3GS mice (mice from survival curve in Figure 2C). Each dot represents a mouse. Red circle indicates the absence of AML at the time of sacrifice. (E) Number of disease initiating cells per 106 injected cells in ND13 and MN1+ND13-transduced CB cells transplanted in NSG-3GS mice after 1 to 3 months of culture. (F) Morphology of BM cells of ND13 (103 days posttransplant, left panel) and MN1+ND13 (38 days posttransplant, right panel) mice at sacrifice. Wright-Giemsa–stained marrow cytospins from representative mice. Micrographs were acquired using an Axioplan2 Zeiss microscope and images were captured using OpenLab software (Perkin Elmer); original magnification ×400. (G) Immunophenotype of GFP+YFP+ BM cells from a MN1+ND13 mouse with AML at sacrifice. Representative FACS blots are shown. (H) Gene set enrichment analysis of indicated signatures comparing MN1+ND13+ and ND13+ CB cells cultured for 60 days in myeloid promoting conditions. FDR, false discovery rate; NES, normalized enrichment score.
Given the marked increase in leukemogenic potential of MN1+ND13+ cells compared with ND13+ cells, we performed gene expression profiling on these cells. Gene set enrichment analysis showed that human-hematopoietic stem cell13,14 and patient-derived LSC15-17 gene signatures were enriched in MN1+ND13+ cells consistent with their high leukemic potential. Furthermore, genes found to be hypomethylated in a subpopulation of AML patients18 were also found to be enriched in the gene signature of MN1+ND13+ cells. In contrast, gene signatures specific to GMP13 and CEBPA target genes19 were downregulated in MN1+ND13+ cells when compared with ND13+ cells. Taken together, these data suggest that the leukemic potential of MN1+ND13+ cells is at least in part mediated through genetic pathways that govern AML pathogenesis and stem cell self-renewal.
Here we report a new model for human AML de novo from normal human hematopoietic cells. Key to this process is the forced increased expression of 2 genes of known importance to spontaneously arising AML in humans: MN1 and the HOX fusion gene ND13. Additionally important was the continuous long-term exposure of the cells to human cytokines in NSG-3GS mice, because serially transplantable AML was not observed in NSG mice (data not shown). These results strongly reinforce the importance of HOX gene activation for leukemic transformation in human cells. MN1 alone proved to have a potent stimulatory effect on the in vitro expansion of transduced CB cells and could induce an abnormal myeloproliferation from them in vivo, but was insufficient to induce full leukemic transformation in vivo. With the addition, however, of deregulated HOX gene expression, here modeled by the introduced expression of ND13, full transformation to transplantable AML in vivo is reproducibly achieved. This model now allows both mechanistic and pharmacologic studies in vitro and in vivo and will advance our understanding of myeloid transformation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Carola Effertz and Margaret Hale for plasmid construction, Glen Edin for lentivirus production, and Christopher May for immunoblotting. The authors also thank Stephane LeBihan, Anne Haegert, and Anna Lapuk of The Laboratory for Advanced Genome Analysis at the Vancouver Prostate Centre, Vancouver, Canada, for helping with microarray services and statistical analysis.
This work was supported by the Animal Resource Center and Flow Cytometry Core Facility of the British Columbia Cancer Agency Research Centre; by a program project grant funded by Terry Fox Foundation, Canada; by a disease team research award by Canadian Stem Cell Consortium, Canada; by the Cancer Research Society, Canada, grants 109003, 110284, 110292, and 111267 from Deutsche Krebshilfe; by the Deutsche-José-Carreras Leukämie-Stiftung e.V grant DJCLS R13/14; and by Deutsche Forschungsgemeinschaft grant HE 5240/5-1.
Authorship
Contribution: S.I. and M.H. conceived the project, designed and performed experiments, analyzed and interpreted data, and wrote the manuscript; M.G. contributed to flow cytometric analysis; G.L.N. contributed to flow cytometric and gene expression analysis; P.X. and T.B. performed experiments and analyzed and interpreted data; P.A.B., A.P.W., G.P., and Y.M. analyzed morphological slides and interpreted data; L.C., P.R., and G.W.R. helped perform experiments; C.J.E. interpreted data and wrote the manuscript; and R.K.H. conceived the project, designed research, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: M.H. holds the patent for MN1 as a predictive marker in acute myeloid leukemia patients. G.W.R. was affiliated at the time of this work with the Terry Fox Laboratory, British Columbia Cancer Agency. The remaining authors declare no competing financial interests.
The current affiliation for G.W.R. is Oncology iMED, AstraZeneca R&D, Boston, MA.
Correspondence: R. Keith Humphries, Terry Fox Laboratory, British Columbia Cancer Agency, 675 West 10th Ave, Vancouver, BC, V5Z 1L3 Canada; e-mail: khumphri@bccrc.ca.
References
Author notes
S.I. and M.H. contributed equally.
M.G., P.A.B., and G.L.N. contributed equally.