Key Points
Oncogenic KRAS causes upregulation of components of the alt-NHEJ pathway, thereby promoting genomic instability.
Inhibition of the alt-NHEJ pathway selectively sensitizes KRAS-mutant leukemic cells to cytotoxic agents.
Abstract
Activating KRAS mutations are detected in a substantial number of hematologic malignancies. In a murine T-cell acute lymphoblastic leukemia (T-ALL) model, we previously showed that expression of oncogenic Kras induced a premalignant state accompanied with an arrest in T-cell differentiation and acquisition of somatic Notch1 mutations. These findings prompted us to investigate whether the expression of oncogenic KRAS directly affects DNA damage repair. Applying divergent, but complementary, genetic approaches, we demonstrate that the expression of KRAS mutants is associated with increased expression of DNA ligase 3α, poly(ADP-ribose) polymerase 1 (PARP1), and X-ray repair cross-complementing protein 1 (XRCC1), all essential components of the error-prone, alternative nonhomologous end-joining (alt-NHEJ) pathway. Functional studies revealed delayed repair kinetics, increased misrepair of DNA double-strand breaks, and the preferential use of microhomologous DNA sequences for end joining. Similar effects were observed in primary murine T-ALL blasts. We further show that KRAS-mutated cells, but not KRAS wild-type cells, rely on the alt-NHEJ repair pathway on genotoxic stress. RNA interference–mediated knockdown of DNA ligase 3α abolished resistance to apoptotic cell death in KRAS-mutated cells. Our data indicate that targeting components of the alt-NHEJ pathway sensitizes KRAS-mutated leukemic cells to standard chemotherapeutics and represents a promising approach for inducing synthetic lethal vulnerability in cells harboring otherwise nondruggable KRAS mutations.
Introduction
RAS proteins belong to the super family of small GTPases. Under physiological settings, these signal molecules are activated by growth factor receptors and integrate external information downstream to several distinct signaling pathways. This in turn involves RAS proteins in the regulation of a variety of important cellular functions such as proliferation, differentiation, and survival.1 The KRAS oncogene is mutated in a significant proportion of epithelial cancers including colon, lung, and pancreatic tumors. In hematologic malignancies, KRAS mutations are found in acute myeloid leukemia (AML), myelodysplastic syndrome, juvenile/chronic myelomonocytic leukemia, and T-cell acute lymphoblastic leukemia (T-ALL).2-5 These mutations cause resistance to GTPase-activating proteins, decrease the intrinsic RAS-GTPase activity, and allow RAS to remain in its active state.1,6 In the process of cellular transformation, KRAS mutations are considered as an early genetic event. For example, in patient samples, KRAS mutations are already detected in colorectal adenomas or pancreatic ductal hyperplasias,7,8 whereas terminal transformation and metastasis are thought to require additional genetic alterations. Furthermore, in murine tumor models, expression of oncogenic Kras induces preneoplastic epithelial lesions in the lung and gastrointestinal tract, whereas additional hits are necessary for frank malignancy.9 In hematologic malignancies, we and others have demonstrated that conditional expression of oncogenic Kras in hematopoietic progenitor cells causes an arrest at the DN2/3 stage during T-cell differentiation followed by the development of an aggressive T-cell lymphoblastic leukemia/lymphoma with long disease latency.10 Interestingly, 50% of analyzed leukemia samples harbored Notch1 mutations, likely acquired during the KrasG12D-mediated block in differentiation. These additional mutations may be acquired by replicative stress or increased reactive oxygen species production. Alternatively, oncogenic RAS may directly affect DNA repair pathways, thereby causing misrepair and, due to concomitant resistance to apoptotic cell death, accumulation of genomic changes.
In mammalian cells, a sophisticated DNA repair system has developed during evolution. To repair DNA damage, cells use different DNA repair pathways depending on the DNA lesion. DNA double-strand breaks (DSBs) induced by a wide variety of environmental carcinogens and anticancer drugs are most problematic as they activate cell death pathways and are also subject to misrepair.11,12 DSBs are repaired by 2 distinct repair mechanisms: namely homologous recombination (HR) and nonhomologous end joining (NHEJ). HR is a highly accurate repair pathway in which the complementary strand of the sister chromatid serves as a template. Hence, the use of this repair pathway is restricted to the S/G2 phase of the cell cycle. DSB repair prior to DNA replication occurs exclusively by the NHEJ pathway, as the homologous DNA template is lacking. Repair by NHEJ is initiated by the Ku70/Ku86 complex, which binds to free DNA ends resulting from DSB formation. Subsequently, the active DNA-dependent protein kinase holoenzyme is formed by recruitment of the DNA-protein kinase catalytic subunit that bridges both DNA ends.13,14 DNA ligase 4 in conjunction with X-ray repair cross-complementing protein 4 (XRCC4) performs the final ligation reaction. In many cases, DSB 5′P and 3′-OH ends are not directly ligatable and need to be processed. This processing is performed be the different nucleases and results in the resection of few nucleotides back to regions displaying DNA sequence microhomologies and subsequently small DNA deletions (<20 bp).15-18
Recent studies provide evidence for the existence of an alternative, Lig4/XRCC4-independent NHEJ pathway (alt-NHEJ).19,20 Several studies have shown that a major player in this pathway is DNA ligase 3α (Lig3α).21 Nuclear Lig3α is stabilized by XRCC1. Consequently, cells deficient for XRCC1 have reduced levels of Lig3α and thus are functionally deficient for the corresponding ligation step. Poly(ADP-ribose) polymerase 1 (PARP1) seems to be another player of the alt-NHEJ pathway, which directly competes with Ku proteins for DNA end binding, thereby causing the switch from the canonical NHEJ (C-NHEJ) to the alt-NHEJ pathway.22 Alt-NHEJ is characterized by delayed repair kinetics, larger DNA deletions, chromosomal translocations, and the preferential use of DNA sequence microhomologies for repair.23 These properties demonstrate a direct relationship between the alt-NHEJ, genomic instability, and cancerogenesis.23-25 Interestingly, increased activity of alt-NHEJ proteins in DSB repair has been demonstrated in BCR-ABL–positive chronic myeloid leukemia cells.26 Rassool and colleagues observed increased expression of Lig3α. In contrast, Lig4 and the endonuclease Artemis were down-regulated in BCR-ABL–positive cells. Short interfering RNA-mediated knockdown of Lig3α caused an increase in DSBs and misrepair, likely due to decreased end-joining efficacy. Moreover, internal tandem duplications of fms-like tyrosine kinase 3 (FLT3-ITD)–positive leukemic cell lines and primary murine bone marrow cells express increased levels of Lig3α and decreased levels of Ku proteins.27 End joining of spontaneously occurring DSBs was accomplished predominantly by using DNA microhomologies and resulted in an increased frequency of DNA misrepair. Interestingly, BCR-ABL and FLT3-ITD are substantial activators of the KRAS signaling pathway. We therefore speculate that oncogenic KRAS is directly involved in the regulation of error-prone DSB repair, causing a mutator phenotype. In this study, we provide evidence that oncogenic KRAS provokes enhanced activity of the alt-NHEJ pathway and that KRAS-mutated cell lines and primary murine cells rely on this pathway to repair DSBs induced by cytotoxic anticancer drugs.
Materials and methods
Cell lines and tissue culture
Cell lines (CCRF-CEM, CCRF-HSB2, NB-4, Mono-Mac-6, Nomo-1, SKM-1, THP-1, and U-937) were obtained from Amerian Type Culture Collection and maintained in fully supplemented RPMI 1640 media (Gibco). The packaging cell line 293FT (Life Technologies) was grown in fully supplemented Dulbecco's modified Eagle medium (Gibco).
Viral supernatants and transduction
Lentiviral particles were produced by cotransfection of 293FT cells with the psPAX2, pMD2.G, and indicated lentiviral expression vectors. Transfections were carried out using TransIT (Mirus) as per the manufacturer’s instructions. Transduction was carried out in the presence of 5 μg/mL polybrene. Following transduction, cells were selected with 1.0 μg/mL puromycin (Sigma-Aldrich) or 10 µg/mL blasticidine (Applichem).
Immunostaining of cells
Cells were fixed with 4% paraformaldehyde and permeabilized with phosphate-buffered saline supplemented with 3% bovine serum albumin and 0.1% Triton X-100. After blocking fragment crystallizable receptors with a CD16/32 antibody (BD Biosciences), cells were stained with a γH2AX-specific antibody (Cell Signaling), washed twice, and stained with goat anti-rabbit antibody F(ab′)2 fragment conjugated with AlexaFluor488 (Molecular Probes). Nuclei were counterstained with 4,6 diamidino-2-phenylindole (DAPI; Life Technologies). Evaluation of γH2AX-positive cells was performed using a confocal laser-scanning microscope (Zeiss).
Single-cell gel electrophoresis
DSBs were quantified by the neutral single-cell gel electrophoresis assay (comet assay) as previously described.28 DNA migration was analyzed with the image analysis system of Kinetic Imaging Ltd. (Komet 4.0.2; Optilas Systemes). The mean tail moment of 100 cells per sample was determined.
Mouse strains
Lox-stop-lox B6.129 (LSL)-RasG12D mice were crossed to C57BL/6J for ≥6 generations. To obtain double-transgenic mice, LSL-KrasG12D/+ mice were crossed to C57BL/6J Lck-Cre mice or C57BL/6J MX1-Cre mice. For all experiments, either C57BL/6J Lck-Cre mice or LSL-KrasG12D/+ mice were used as controls. Genotyping and verification of Cre-mediated recombination were performed as previously described.29,30 All animal studies were conducted in compliance with institutional guidelines and were approved by the regulatory authority.
Statistical analysis
Unless otherwise specified, data are presented as mean ± standard deviation (SD). Comparisons between 2 groups were performed using the unpaired Student t test. A P value of <.05 was considered significant. Statistical computations were performed using GraphPad Prism software, version 5.0.
Full methods for flow cytometry, quantitative reverse transcriptase-polymerase chain reaction (RQ-PCR), immunoblotting, repair assays, apoptosis, and proliferations assays are available in supplemental Methods, available on the Blood Web site.
Results
Oncogenic KRAS mediates protection from apoptosis
To address the question of whether oncogenic KRAS has an impact on the repair of DSBs, we used 2 different genetic approaches. First, the T-ALL cell line CCRF-HSB2 and the AML cell line U-937 were lentivirally transduced with KRASG13D. Immunoblot analysis demonstrated increased phosphorylation of the RAS downstream targets extracellular signal-regulated kinase (ERK)1/2 and mitogen-activated protein kinase kinase (MEK)1/2 (supplemental Figure 1A). KRAS protein expression levels were only slightly increased, suggesting almost physiological expression of oncogenic KRAS (supplemental Figure 1A). Using a GTPase pull-down assay, KRASG13D-expressing cell lines showed increased GTPase activity compared with control cells (supplemental Figure 1B). In the second model, we used a reverse genetic approach and suppressed KRAS expression in Nomo-1 AML cells, harboring an endogenous KRASG13D-mutation.31 We established a doxocycline-inducible system enabling pharmacologic control of microRNA-short hairpin RNA (miR-shRNA) expression targeting KRAS. As shown in supplemental Figure 1C, ERK1/2 phosphorylation was substantially suppressed on doxycycline treatment. In addition, KRAS knockdown completely abolished KRAS activity in the GTPase pull-down assay (supplemental Figure 1D).
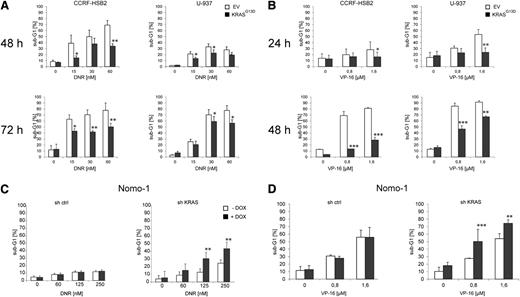
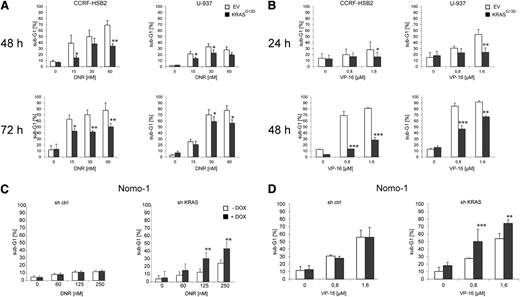
RAS has been shown to exhibit pleiotropic effects within cells, and different studies have demonstrated that deregulated KRAS signaling protects cancer cells from apoptotic cell death.32 Indeed, mutant KRASG13D-expressing CCRF-HSB2 and U-937 cells were protected against apoptosis induced by daunorubicin, etoposide (VP-16), and cytarabine (Figure 1A-B; data not shown). In contrast, the KRASG13D-dependent cell line Nomo-1 was sensitized to daunorubicin- or VP-16–induced apoptosis on KRAS knockdown (Figure 1C-D). Oncogene-mediated replicative stress has been shown to cause DNA damage and to affect DNA damage repair.33 In our model, the expression of KRASG13D in fully transformed leukemic cell lines had no impact on proliferation and cell cycle dynamics before and after treatment with DNA damaging agents as revealed by cell cycle analyses and proliferation assays (supplemental Figures 2A-D and 3A-B).
Oncogenic KRAS signaling mediates drug resistance and inhibits apoptosis. CCRF-HSB2 and U-937 cells stably transduced with either pLenti6.2-empty vector (EV) or pLenti6.2-KRASG13D (KRASG13D) were treated with (A) daunorubicin or (B) VP-16 as indicated. The fraction of apoptotic cells was determined by quantifying the sub-G1 fraction using flow cytometry. Data represent the mean values ± SD of 3 independent experiments (*P < .05, **P < .01, ***P < .001, Student t test). (C) Nomo-1 cells stably transduced with lentiviral vectors expressing a doxycycline-inducible nonsilencing miR-shRNA (sh ctrl) or a miR-shRNA targeting KRAS (sh KRAS) were treated with daunorubicin in the presence (black bars) or absence (white bars) of doxycycline. After 72 hours, the percentage of sub-G1 cells corresponding to apoptotic cells was determined by flow cytometry. Data represent the mean values ± SD of 3 independent experiments (**P < .01, Student t test). (D) Nomo-1 cells stably transduced with doxycycline-inducible sh ctrl or sh KRAS were treated with VP-16 in the presence (black bars) or absence (white bars) of doxycycline. After 48 hours, the percentage of sub-G1 cells corresponding to apoptotic cells was determined by flow cytometry. Data represent the mean values ± SD of 3 independent experiments (**P < .01, ***P < .001, Student t test).
Oncogenic KRAS signaling mediates drug resistance and inhibits apoptosis. CCRF-HSB2 and U-937 cells stably transduced with either pLenti6.2-empty vector (EV) or pLenti6.2-KRASG13D (KRASG13D) were treated with (A) daunorubicin or (B) VP-16 as indicated. The fraction of apoptotic cells was determined by quantifying the sub-G1 fraction using flow cytometry. Data represent the mean values ± SD of 3 independent experiments (*P < .05, **P < .01, ***P < .001, Student t test). (C) Nomo-1 cells stably transduced with lentiviral vectors expressing a doxycycline-inducible nonsilencing miR-shRNA (sh ctrl) or a miR-shRNA targeting KRAS (sh KRAS) were treated with daunorubicin in the presence (black bars) or absence (white bars) of doxycycline. After 72 hours, the percentage of sub-G1 cells corresponding to apoptotic cells was determined by flow cytometry. Data represent the mean values ± SD of 3 independent experiments (**P < .01, Student t test). (D) Nomo-1 cells stably transduced with doxycycline-inducible sh ctrl or sh KRAS were treated with VP-16 in the presence (black bars) or absence (white bars) of doxycycline. After 48 hours, the percentage of sub-G1 cells corresponding to apoptotic cells was determined by flow cytometry. Data represent the mean values ± SD of 3 independent experiments (**P < .01, ***P < .001, Student t test).
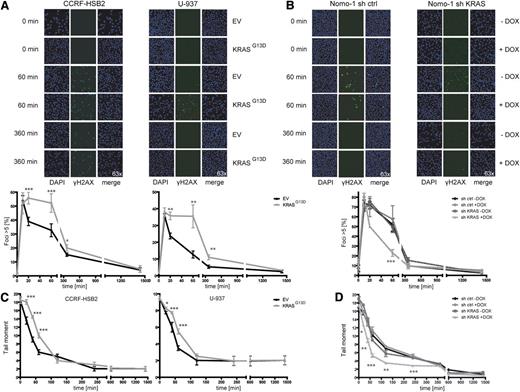
Oncogenic KRAS expression results in delayed DSB repair
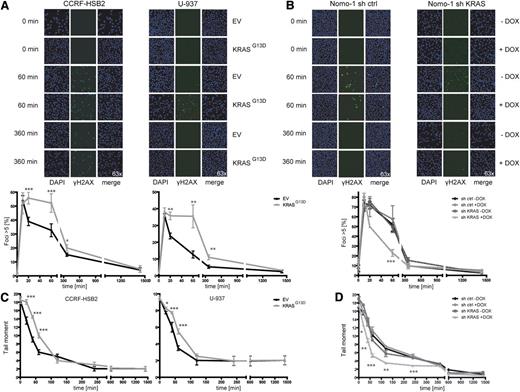
To characterize DNA damage repair in the context of oncogenic KRAS expression, we examined the repair of DSBs induced by exogenous agents and monitored the induction of γH2AX foci, a marker of DSBs.34 Cells were irradiated with 10 Gy, and the number of γH2AX foci–positive cells was determined at different time points after treatment. Whereas similar numbers of γH2AX foci were observed in KRAS wild-type and KRAS mutant cells at early time points, a significant higher number of γH2AX-positive cells persisted for up to 360 minutes in KRASG13D cells compared with controls (Figure 2A). The number of γH2AX-positive cells declined over time and was equal to control cells after 1.440 minutes. Vice versa, knockdown of KRAS in KRAS mutant Nomo-1 cells resulted in a significant lower number of γH2AX-positive cells starting at the 20-minute time point and approximated the control curves after 360 minutes (Figure 2B). Similar effects were seen after treatment with daunorubicin (supplemental Figure 4A-B).
KRASG13D-positive cells show delayed repair kinetics of DSBs. (A) (Left) CCRF-HSB2 cells and (right) U-937 cells transduced with either EV or pLenti6.2-KRASG13D were irradiated with 10 Gy. Cells were stained with anti-γH2AX-Alexa488 and DAPI at the indicated time points. Graphs show the percentage of cells with >5 γH2AX foci (200 cells per experiment were analyzed). Shown are the mean values ± SD of 3 independent experiments (*P < .05, **P < .01, ***P < .001, Student t test). (B) Nomo-1 cells transduced with either (left) nonsilencing miR-shRNA (sh ctrl) or (right) KRAS-miR-shRNA (sh KRAS) were irradiated with 10 Gy. Cells were stained with anti–γH2AX-Alexa488 and DAPI at the indicated time points. Graphs show the percentage of cells with >5 γH2AX foci. Data represent mean values ± SD of 3 independent experiments (***P < .001, Student t test). (C) (Left) CCRF-HSB2, (right) U-937, and (D) Nomo-1 cells (± doxycyline treatment) were irradiated with 10 Gy and repair kinetics of induced DSBs were determined by the neutral comet assay at different time points following irradiation. Data represent the mean of ≥3 independent experiments ± SD (*P < .05, **P < .01, ***P < .001, Student t test).
KRASG13D-positive cells show delayed repair kinetics of DSBs. (A) (Left) CCRF-HSB2 cells and (right) U-937 cells transduced with either EV or pLenti6.2-KRASG13D were irradiated with 10 Gy. Cells were stained with anti-γH2AX-Alexa488 and DAPI at the indicated time points. Graphs show the percentage of cells with >5 γH2AX foci (200 cells per experiment were analyzed). Shown are the mean values ± SD of 3 independent experiments (*P < .05, **P < .01, ***P < .001, Student t test). (B) Nomo-1 cells transduced with either (left) nonsilencing miR-shRNA (sh ctrl) or (right) KRAS-miR-shRNA (sh KRAS) were irradiated with 10 Gy. Cells were stained with anti–γH2AX-Alexa488 and DAPI at the indicated time points. Graphs show the percentage of cells with >5 γH2AX foci. Data represent mean values ± SD of 3 independent experiments (***P < .001, Student t test). (C) (Left) CCRF-HSB2, (right) U-937, and (D) Nomo-1 cells (± doxycyline treatment) were irradiated with 10 Gy and repair kinetics of induced DSBs were determined by the neutral comet assay at different time points following irradiation. Data represent the mean of ≥3 independent experiments ± SD (*P < .05, **P < .01, ***P < .001, Student t test).
To explore whether the observed difference in γH2AX-positive cells was due to augmented DNA damage or slowed repair kinetics, we performed neutral comet assays to investigate the level of DSBs at different time points following irradiation. Whereas tail moments were similar immediately after irradiation, we observed a pronounced delay in the clearance of DSBs in KRASG13D cells compared with EV controls at 20, 40, and 60 minutes (Figure 2C). In contrast, doxycycline-induced knockdown of oncogenic KRAS resulted in accelerated repair kinetics of DSBs (Figure 2D). When cells were treated with daunorubicin, a drug commonly used in leukemia therapy, expression of KRASG13D again was associated with a significant delay in the clearance of DSBs or vice versa, with rapid clearance on KRAS knockdown (supplemental Figure 4C-D). Similar results were obtained following treatment with t-BOOH as genotoxin (data not shown).
Expression of oncogenic KRAS is associated with misrepair of DSBs
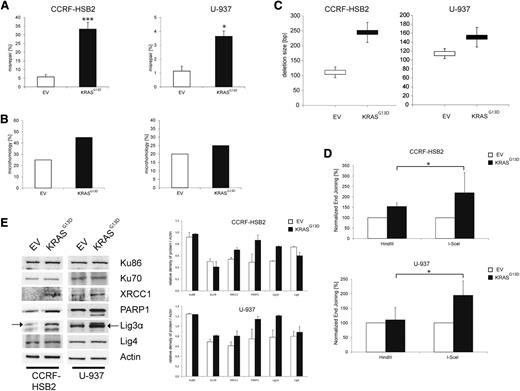
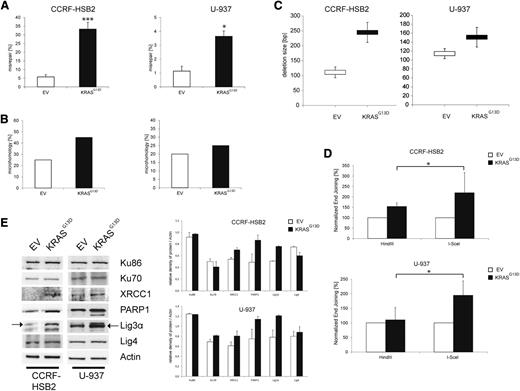
To further characterize DSB repair caused by oncogenic KRAS, we performed plasmid reactivation assays. Linearized pUC19 plasmids harboring a DSB within the LacZα gene were incubated with nuclear extracts derived from cells expressing KRASG13D or EV for 24 hours. After recovery of plasmid DNA and transformation of Escherichia coli, repair efficacy was calculated by using the total number of colonies. Misrepair was estimated by the number of white colonies (circular plasmid, but no functional LacZα; incorrectly repaired) relative to the total number of colonies. KRASG13D-expressing cells demonstrated a significant increase in misrepair (Figure 3A). To characterize the nature of these repair errors, we examined the DNA region surrounding the DSB of incorrectly repaired plasmids by sequence analysis. Interestingly, a higher percentage of plasmids incubated with nuclear extracts from KRASG13D-expressing cells was repaired by the use of microhomologies (Figure 3B; supplemental Figure 5). In addition, microhomologies at the breakpoint junctions consisting of >3 bp were only found in plasmids incubated with KRASG13D nuclear extracts. Furthermore, these plasmids showed a tendency toward an increased deletion size (Figure 3C). Of note, we detected insertions in 5% of all analyzed sequences incubated with KRASG13D nuclear extracts, whereas none were found in control plasmids.
Expression of KRASG13D increases the misrepair frequency and deletion size. (A) Misrepair frequency was determined by an in vitro LacZα plasmid reactivation assay. Nuclear extracts were incubated with linearized pUC19 plasmids harboring a DSB in the LacZα gene. Misrepair frequency was calculated by the number of white colonies (misrepaired) related to the total colony numbers. Data represent the mean of ≥3 independent experiments ± SD (*P < .05, ***P = .0002, Student t test). (B) Microhomologies were defined by ≥2 bp. Bar graphs (white, EV; black, KrasG13D) show the percentage of repaired plasmids containing microhomologies at the site of religation. Twenty plasmids randomly selected from 3 experiments were sequenced and analyzed. (C) Analysis of deletion size of randomly selected, misrepaired (white colonies) plasmids incubated with nuclear extracts derived from (upper) CCRF-HSB2 or (lower) U-937. Data represent the mean values ± SD of 20 sequenced plasmids. White, EV; black, KRASG13D. (D) (Upper) CCRF-HSB2 cells and (lower) U-937 cells transduced with either EV or pLenti6.2-KRASG13D were transfected with either HindIII- or I-SceI–digested linearized plasmid in combination with pDsRed2-N1 as transfection control. After an incubation period of 72 hours, end-joining efficacy was determined by flow cytometry. Data shown represent mean values ± SD of 3 independent experiments (*P < .05, Student t test). (E) Protein expression of Ku86, Ku70, XRCC1, PARP1, Lig3α, and ligase 4 in cell extracts of transduced (left) CCRF-HSB2 and (right) U-937 cells. Shown is 1 representative analysis of 2 independent experiments. Bar graph show protein expression levels relative to actin determined by the means of densitometry of 2 independent experiments.
Expression of KRASG13D increases the misrepair frequency and deletion size. (A) Misrepair frequency was determined by an in vitro LacZα plasmid reactivation assay. Nuclear extracts were incubated with linearized pUC19 plasmids harboring a DSB in the LacZα gene. Misrepair frequency was calculated by the number of white colonies (misrepaired) related to the total colony numbers. Data represent the mean of ≥3 independent experiments ± SD (*P < .05, ***P = .0002, Student t test). (B) Microhomologies were defined by ≥2 bp. Bar graphs (white, EV; black, KrasG13D) show the percentage of repaired plasmids containing microhomologies at the site of religation. Twenty plasmids randomly selected from 3 experiments were sequenced and analyzed. (C) Analysis of deletion size of randomly selected, misrepaired (white colonies) plasmids incubated with nuclear extracts derived from (upper) CCRF-HSB2 or (lower) U-937. Data represent the mean values ± SD of 20 sequenced plasmids. White, EV; black, KRASG13D. (D) (Upper) CCRF-HSB2 cells and (lower) U-937 cells transduced with either EV or pLenti6.2-KRASG13D were transfected with either HindIII- or I-SceI–digested linearized plasmid in combination with pDsRed2-N1 as transfection control. After an incubation period of 72 hours, end-joining efficacy was determined by flow cytometry. Data shown represent mean values ± SD of 3 independent experiments (*P < .05, Student t test). (E) Protein expression of Ku86, Ku70, XRCC1, PARP1, Lig3α, and ligase 4 in cell extracts of transduced (left) CCRF-HSB2 and (right) U-937 cells. Shown is 1 representative analysis of 2 independent experiments. Bar graph show protein expression levels relative to actin determined by the means of densitometry of 2 independent experiments.
To further confirm our results we made use of a plasmid-based in vivo end-joining assay.35 The plasmid was either digested with HindIII or I-SceI, generating either compatible or incompatible DSB ends, respectively. Incompatible ends need processing via the NHEJ repair pathway before ligation. In line with our previous results, we observed a twofold increase in the efficiency of NHEJ end joining in cells expressing KRASG13D compared with controls (Figure 3D; supplemental Figure 6).
Proteins of the alt-NHEJ pathway are enhanced in KRASmut-expressing cells
The observed delay in repair kinetics, the increase in error-prone DNA repair and deletion size, and the use of microhomologies prompted us to investigate the NHEJ DNA repair pathways in more detail. To examine the steady-state level of key proteins of the C-NHEJ and alt-NHEJ pathways, we performed immunoblot analyses. Whereas expression levels of Ku70, Ku86, and Lig4, components of the classical pathway, were not altered, we observed a substantial increase in protein expression of components of the alt-NHEJ pathway such as Lig3α, PARP1, and XRCC1 in KRASG13D-positive cells (Figure 3E). Furthermore, we found increased messenger RNA (mRNA) levels of PARP1, Lig3α, and XRCC1 in cells expressing KRASG13D (supplemental Figure 7). These data suggest that expression of oncogenic KRAS causes a shift in DSB repair from the C-NHEJ pathway to a preferred use of the more error-prone alt-NHEJ pathway.
KRASG13D-dependent deregulation of the alt-NHEJ requires mitogen-activated protein kinase signaling
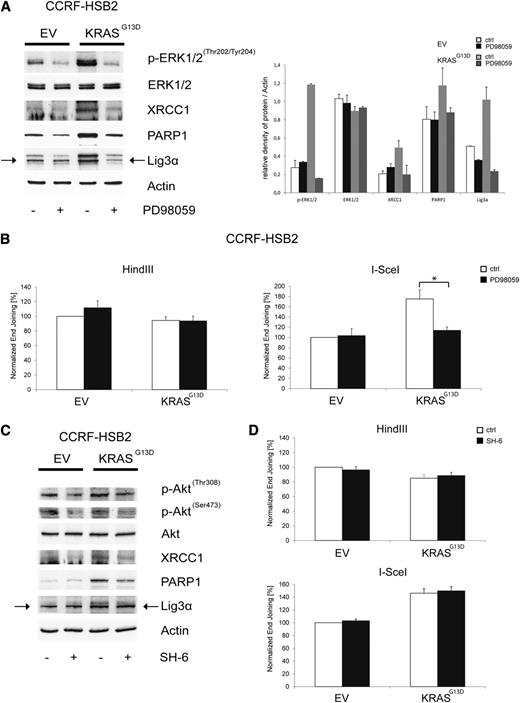
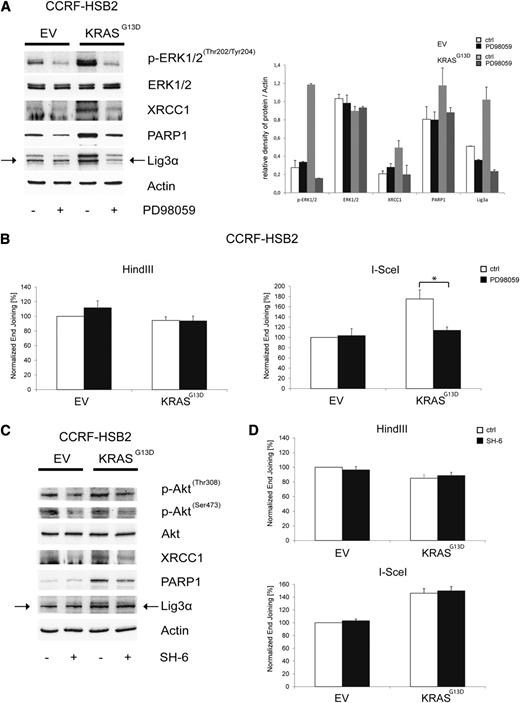
We next asked whether mitogen-activated protein kinase (MAPK) or phosphatidylinositol 3-kinase–protein kinase B (AKT) signaling, both major effector pathways in KRAS mutant cells, are involved in the observed dysregulation of DSB repair. EV and KRASG13D-transduced CCRF-HSB2 cells were treated with PD98059, a pharmacologic MEK inhibitor acting upstream of ERK. Treatment with PD98059 clearly suppressed KRASG13D-induced upregulation of Lig3α, PARP1, and XRCC1 in KRASG13D-positive cells (Figure 4A). In line with these data, we found a significant decrease in the efficiency of NHEJ in KRAS mutant cells treated with PD98059 (Figure 4B). No effects on NHEJ activity in control cells or on HR in both cell lines was observed on treatment with PD98059. In contrast, treatment with the AKT-inhibitor SH6 caused only a minor decrease in XRCC1 and PARP1 protein expression, and no functional effect on NHEJ activity was observed in KRAS mutant cells. These data indicate that NHEJ activity is primarily regulated via the MAPK pathway in KRAS mutant cells (Figure 4C-D).
Pharmacologic inhibition of MEK/ERK kinase reverses the KRASG13D-induced deregulation of the alt-NHEJ. (A) Protein expression of p-ERK1/2Thr202/Tyr204, ERK1/2, XRCC1, PARP1, Lig3α, and actin in cell extracts of transduced CCRF-HSB2 cells treated with the selective MEK inhibitor PD98059 (200 µM) for 24 hours. Shown is 1 representative analysis of 2 independent experiments. Bar graph shows protein expression levels relative to actin determined by the means of densitometry of 2 independent experiments. (B) CCRF-HSB2 cells transduced with either EV or pLenti6.2-KRASG13D were treated with the selective MEK inhibitor PD98059 (200 µM) for 24 hours. After incubation, cells were transfected with either HindIII- or I-SceI–digested linearized plasmid in combination with pDsRed2-N1 as a transfection control. After an incubation period of 24 hours, end-joining efficacy was determined by flow cytometry. Data represent mean values ± SD of 3 independent experiments (*P < .05, Student t test). (C) Protein expression of p-AktThr308, p-AktSer473, Akt, XRCC1, PARP1, Lig3α, and actin in cell extracts of transduced CCRF-HSB2 cells treated with the Akt inhibitor SH-6 (10 µM) for 24 hours. Shown is 1 representative analysis of 2 independent experiments. (D) CCRF-HSB2 cells transduced with either EV or pLenti6.2-KRASG13D were treated with the Akt inhibitor SH-6 (10 µM) for 24 hours. After incubation, cells were transfected with either HindIII- or I-SceI–digested linearized plasmid in combination with pDsRed2-N1 as a transfection control. After an incubation period of 24 hours, end-joining efficacy was determined by flow cytometry. Data represent mean values ± SD of 2 independent experiments.
Pharmacologic inhibition of MEK/ERK kinase reverses the KRASG13D-induced deregulation of the alt-NHEJ. (A) Protein expression of p-ERK1/2Thr202/Tyr204, ERK1/2, XRCC1, PARP1, Lig3α, and actin in cell extracts of transduced CCRF-HSB2 cells treated with the selective MEK inhibitor PD98059 (200 µM) for 24 hours. Shown is 1 representative analysis of 2 independent experiments. Bar graph shows protein expression levels relative to actin determined by the means of densitometry of 2 independent experiments. (B) CCRF-HSB2 cells transduced with either EV or pLenti6.2-KRASG13D were treated with the selective MEK inhibitor PD98059 (200 µM) for 24 hours. After incubation, cells were transfected with either HindIII- or I-SceI–digested linearized plasmid in combination with pDsRed2-N1 as a transfection control. After an incubation period of 24 hours, end-joining efficacy was determined by flow cytometry. Data represent mean values ± SD of 3 independent experiments (*P < .05, Student t test). (C) Protein expression of p-AktThr308, p-AktSer473, Akt, XRCC1, PARP1, Lig3α, and actin in cell extracts of transduced CCRF-HSB2 cells treated with the Akt inhibitor SH-6 (10 µM) for 24 hours. Shown is 1 representative analysis of 2 independent experiments. (D) CCRF-HSB2 cells transduced with either EV or pLenti6.2-KRASG13D were treated with the Akt inhibitor SH-6 (10 µM) for 24 hours. After incubation, cells were transfected with either HindIII- or I-SceI–digested linearized plasmid in combination with pDsRed2-N1 as a transfection control. After an incubation period of 24 hours, end-joining efficacy was determined by flow cytometry. Data represent mean values ± SD of 2 independent experiments.
Inhibition of the alt-NHEJ pathway sensitizes cells toward apoptosis
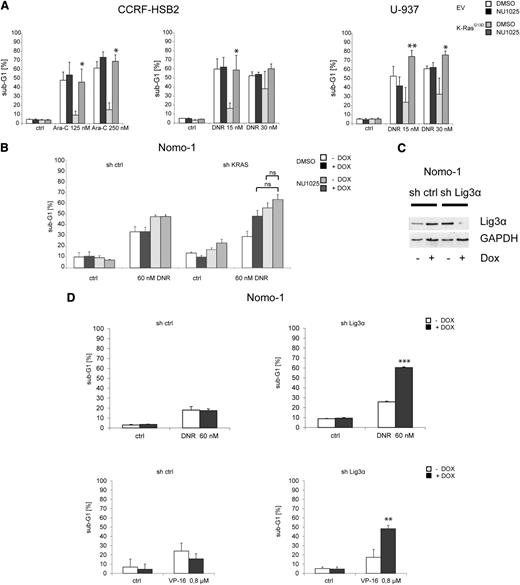
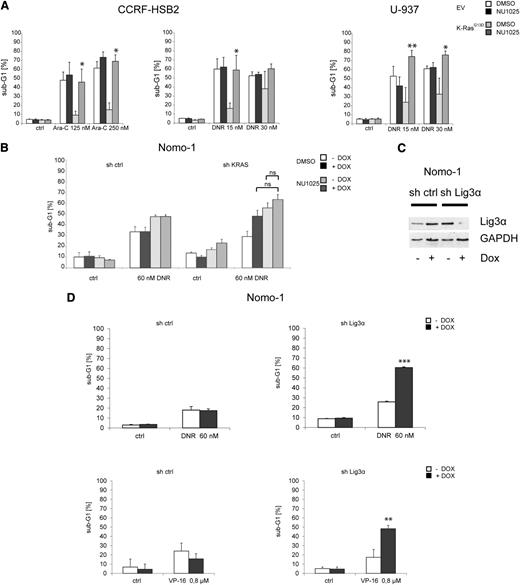
We next explored whether KRASG13D-positive leukemic cells are dependent on alt-NHEJ–mediated DSB repair in the context of genotoxic stress. CCRF-HSB2 and U-937 cells expressing or lacking KRASG13D were treated with the pan-PARP inhibitor NU1025 in combination with chemotherapy (Figure 5A). Again we observed a significant inhibition of apoptotic cell death in KRASG13D-expressing cells on treatment with cytarabine or daunorubicin (Figure 5A). However, this effect was completely reversed when KRASG13D-positive cells were treated in combination with NU1025, whereas the addition of NU1025 had no further effect on cell survival in control cells (Figure 5A). Further, treatment with NU1025 in combination with daunorubicin increased apoptotic cell death in Nomo-1 cells harboring an endogenous KRASG13D mutation (Figure 5B). In contrast, on KRAS knockdown, PARP inhibition only slightly increased apoptosis in combination with daunorubicin (Figure 5B). These data suggest that targeting components of the alt-NHEJ pathway selectively sensitizes KRASmut cells toward genotoxic stress and is able to overcome resistance to apoptosis mediated by oncogenic KRAS.
Targeting proteins of the alt-NHEJ pathway sensitizes cells toward cytotoxic agents. (A) CCRF-HSB2 cells were treated with either (left) cytarabine or (center) daunorubicin in combination with the PARP inhibitor NU1025 (10 mM) or vehicle (dimethylsulfoxide). (Right) U-937 cells were treated with daunorubicin in combination with NU1025 (10 mM) or vehicle (dimethylsulfoxide). After 48 hours, the percentage of sub-G1 cells was determined by flow cytometry. Data represent the mean values ± SD of 3 independent experiments (*P < .05, Student t test). (B) Nomo-1 cells infected with nonsilencing miR-shRNA or KRAS miR-shRNA were treated with daunorubicin (60 nM) in combination with NU1025 (10 mM) in the presence or absence of doxycycline. (C) Nomo-1 cells either expressing tetracycline-controlled transcriptional activation (tet-on–inducible) miR-shRNA targeting Lig3α or nonsilencing miR-shRNA were treated with 1 µg/mL doxycyclin. Immunoblot analysis was performed using an anti-Lig3α antibody. To control equal loading, the blot was stripped and reprobed with anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (D) miR-shRNA expressing Nomo-1 cells were treated with either (upper) daunorubicin (DNR) or (lower) VP-16. After 72 (DNR) or 48 hours (VP-16), the percentage of sub-G1 cells was determined by flow cytometry. Data represent the mean values of 3 independent experiments. Error bars correspond to SD (**P < .01, ***P < .001, Student t test).
Targeting proteins of the alt-NHEJ pathway sensitizes cells toward cytotoxic agents. (A) CCRF-HSB2 cells were treated with either (left) cytarabine or (center) daunorubicin in combination with the PARP inhibitor NU1025 (10 mM) or vehicle (dimethylsulfoxide). (Right) U-937 cells were treated with daunorubicin in combination with NU1025 (10 mM) or vehicle (dimethylsulfoxide). After 48 hours, the percentage of sub-G1 cells was determined by flow cytometry. Data represent the mean values ± SD of 3 independent experiments (*P < .05, Student t test). (B) Nomo-1 cells infected with nonsilencing miR-shRNA or KRAS miR-shRNA were treated with daunorubicin (60 nM) in combination with NU1025 (10 mM) in the presence or absence of doxycycline. (C) Nomo-1 cells either expressing tetracycline-controlled transcriptional activation (tet-on–inducible) miR-shRNA targeting Lig3α or nonsilencing miR-shRNA were treated with 1 µg/mL doxycyclin. Immunoblot analysis was performed using an anti-Lig3α antibody. To control equal loading, the blot was stripped and reprobed with anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (D) miR-shRNA expressing Nomo-1 cells were treated with either (upper) daunorubicin (DNR) or (lower) VP-16. After 72 (DNR) or 48 hours (VP-16), the percentage of sub-G1 cells was determined by flow cytometry. Data represent the mean values of 3 independent experiments. Error bars correspond to SD (**P < .01, ***P < .001, Student t test).
PARP1 has several effects on different DNA repair pathways and is involved in a variety of distinct cellular processes.36 To further validate the role of the alt-NHEJ pathway in the context of oncogenic KRAS, we transduced Nomo-1 cells with lentiviral vectors expressing a tet-on–inducible miR-shRNA directed against Lig3α. Treatment with doxycycline resulted in almost complete suppression of Lig3α expression (Figure 5C). Cells were then treated with either daunorubicin, VP-16, or cytarabine. As shown in Figure 5D and supplemental Figure 8A, knockdown of Lig3α sensitized Nomo-1 cells toward apoptotic cell death on treatment with chemotherapeutic drugs.
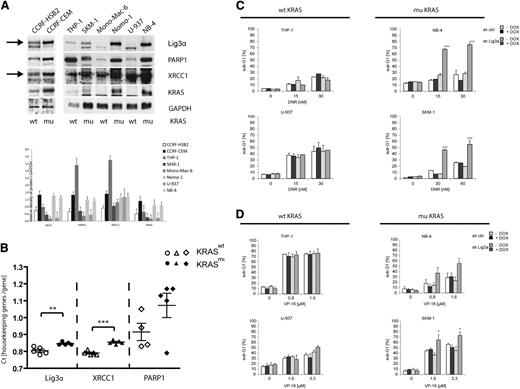
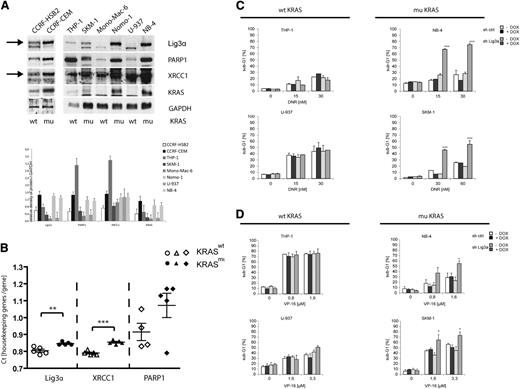
To avoid supraphysiological expression of oncogenic KRAS, we further wanted to confirm our findings using different leukemic cell lines that are well characterized regarding their RAS status. In addition to Nomo-1 cells, KRAS mutations were identified in CCRF-CEM, SKM-1, and NB-4 cells, whereas CCRF-HSB2, THP-1, Mono-Mac-6, and U-937 cells do not harbor KRAS mutations. We first examined the protein expression levels of the alt-NHEJ components Lig3α, PARP1, and XRCC1. As depicted in Figure 6A, all KRAS mutant cell lines showed increased expression of Lig3α compared with KRAS wild-type cells. Although not as homogenous as demonstrated for Lig3α, we also found increased expression levels for PARP1 and XRCC1 in KRAS-mutated cell lines (Figure 6A). The highest protein expression levels for PARP1 and XRCC1 were detected in THP-1 cells. Interestingly, THP-1 cells harbor an endogenous NRAS mutation. We next analyzed mRNA expression levels of PARP1, Lig3α, and XRCC1 in primary AML patient samples. Patient characteristics are listed in supplemental Table 1. In line with our previous findings, we observed increased expression levels in KRAS mutant compared with wild-type patients (Figure 6B).
Endogenous mutated KRAS causes increased expression levels of alt-NHEJ proteins in hematopoietic cell lines and primary AML patient cells. (A) Protein expression and quantitative densitometry of Lig3α, PARP1, XRCC1, and KRAS in cell extracts derived from KRAS-mutated (CCRF-CEM, SKM-1, Nomo-1, NB-4) and KRAS wild-type leukemic cell lines (CCRF-HSB2, THP-1, Mono-Mac-6, U-937). To control equal loading, the blot was stripped and reprobed with an anti-GAPDH antibody. Bar graphs indicate expression levels relative to GAPDH determined by the means of densitometry of 3 independent experiments. (B) RQ-PCR analysis of Lig3α, XRCC1, and PARP1 in primary AML patient samples. Shown are the count values (Ct values) of each gene in relation to actin mRNA and HPRT mRNA. White, KRAS wild-type AML patients; black, KRAS mutant AML patients. Samples were read in duplicates. (**P < .01, ***P < .001, Student t test). Cell lines with (left) wild-type KRAS and (right) mutated KRAS were lentivirally transduced with either tet-on–inducible nonsilencing miR-shRNA or miR-shRNA targeting Lig3α. Knockdown of Lig3α was induced on treatment with doxycycline (black and dark gray bars). Cells were then treated with (C) daunorubicin (72 hours) or (D) VP-16 (48 hours), and the percentage of sub-G1 cells was determined by flow cytometry. Data shown represent mean values ± SD of 3 independent experiments (*P < .05, ***P < .001, Student t test).
Endogenous mutated KRAS causes increased expression levels of alt-NHEJ proteins in hematopoietic cell lines and primary AML patient cells. (A) Protein expression and quantitative densitometry of Lig3α, PARP1, XRCC1, and KRAS in cell extracts derived from KRAS-mutated (CCRF-CEM, SKM-1, Nomo-1, NB-4) and KRAS wild-type leukemic cell lines (CCRF-HSB2, THP-1, Mono-Mac-6, U-937). To control equal loading, the blot was stripped and reprobed with an anti-GAPDH antibody. Bar graphs indicate expression levels relative to GAPDH determined by the means of densitometry of 3 independent experiments. (B) RQ-PCR analysis of Lig3α, XRCC1, and PARP1 in primary AML patient samples. Shown are the count values (Ct values) of each gene in relation to actin mRNA and HPRT mRNA. White, KRAS wild-type AML patients; black, KRAS mutant AML patients. Samples were read in duplicates. (**P < .01, ***P < .001, Student t test). Cell lines with (left) wild-type KRAS and (right) mutated KRAS were lentivirally transduced with either tet-on–inducible nonsilencing miR-shRNA or miR-shRNA targeting Lig3α. Knockdown of Lig3α was induced on treatment with doxycycline (black and dark gray bars). Cells were then treated with (C) daunorubicin (72 hours) or (D) VP-16 (48 hours), and the percentage of sub-G1 cells was determined by flow cytometry. Data shown represent mean values ± SD of 3 independent experiments (*P < .05, ***P < .001, Student t test).
To further investigate the role of oncogenic KRAS and the alt-NHEJ pathway in KRAS mutant or wild-type cells, we generated cell lines expressing an inducible miR-shRNA directed against KRAS or Lig3α. Treatment with doxycycline for 72 hours resulted in effective knockdown of KRAS of Lig3α expression in all cell lines (supplemental Figure 8B). As described for Nomo-1 cells, knockdown of KRAS in NB-4 and SKM-1 cells resulted in increased apoptosis on treatment with daunorubicin or VP-16 (supplemental Figure 8C). In addition and in line with our previous data, knockdown of Lig3α caused a significant increase in apoptotic cell death in KRAS mutant cells on daunorubicin or VP-16 treatment (Figure 6C-D). In contrast, suppression of Lig3α had no additional effect on KRAS wild-type cell lines. Of note, THP-1 cells, which showed high expression levels of essential components of the alt-NHEJ pathway, were not sensitized by Lig3α knockdown. These data indicate that mutant KRAS increases expression levels of components of the alt-NHEJ pathway and preferentially recruits the alt-NHEJ pathway for DSB repair. Inhibition of Lig3α or PARP specifically sensitizes KRAS-mutated cells toward genotoxic anticancer drugs.
Thymocytes derived from KrasG12D knock-in mice demonstrate DSB repair via alt-NHEJ
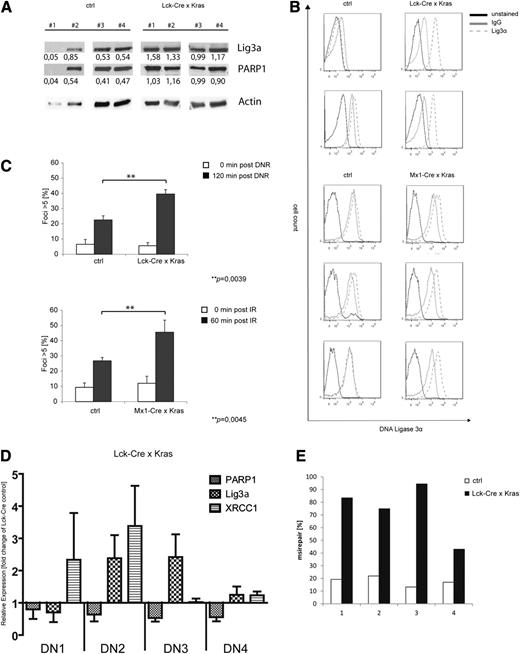
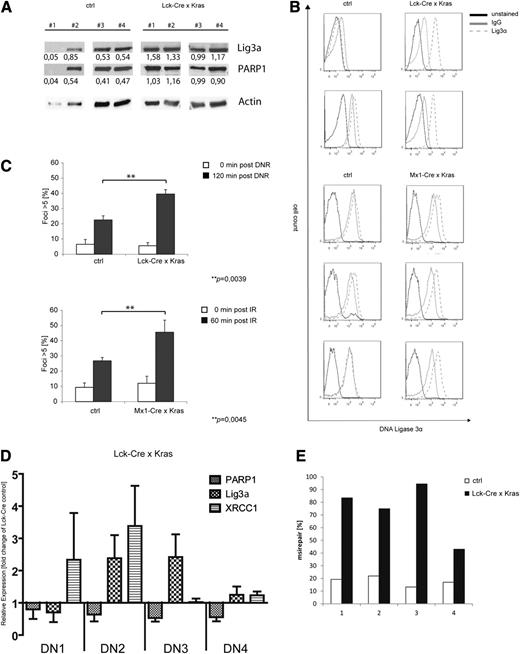
Recently, we demonstrated that conditional expression of KrasG12D in thymocytes caused a block in differentiation followed by the development of a late-onset, aggressive T-lymphoblastic leukemia/lymphoma.10 As 50% of leukemic mice acquired Notch1 mutations, we speculated that the shift toward the alt-NHEJ pathway observed in our in vitro models might also be relevant in vivo. To address this question, we used Lck-Cre/KrasG12D mice, which express oncogenic Kras from its endogenous promoter restricted to immature thymocytes and develop T-cell lymphoma/leukemia with a median latency of 180 days.29 We first analyzed expression levels of Lig3α and PARP1 in leukemic Lck-Cre/KrasG12D mice or healthy littermate controls. As the majority of both populations are CD4/CD8 double positive (data not shown), we used whole thymus extracts for these experiments. Immunoblot analysis showed increased expression of Lig3α and PARP1 in Lck-Cre/KrasG12D mice (1.27 vs 0.49 and 1.02 vs 0.37 for Lig3α and PARP1, respectively; Figure 7A). These findings were confirmed by intracellular staining of Lig3α and fluorescence-activated cell sorter analysis in Lck-Cre/KrasG12D thymus or cells derived from Mx1-Cre/KrasG12D T-cell thymomas (Figure 7B). To analyze DSB repair on genotoxic stress, thymocytes of either KrasG12D or control animals were treated with daunorubicin (Figure 7C, upper) or were irradiated (Figure 7C, lower), and the number of γH2AX foci–positive cells was determined. We detected a significant increase of cells with more than 5 foci in KrasG12D animals compared with littermate controls. Again, the observed increase in γH2AX foci at 60 and 120 minutes was thought to be due to delayed repair kinetics.
Primary murine KrasG12D-expressing T-ALL and preleukemic cells show increased levels of DNA misrepair. (A-C) Single-cell suspensions were prepared from the thymus of leukemic KrasG12D-expressing mice or control animals (KRAS wild type). (A) Immunoblot analysis using anti-Lig3α and PARP1 antibodies as indicated. To control equal loading, the blot was stripped and reprobed with an anti-actin antibody. Numbers indicate expression levels relative to actin determined by the means of densitometry. (B) Expression of intracellular Lig3α in primary thymocytes and fluorescence-activated cell sorter analysis (red, unstained; blue, immunoglobulin G-isotype control; yellow, anti-Lig3α-Alexa488). (C) Cells were treated with either (upper) daunorubicin (125 nM) or (lower) were irradiated (10 Gy). Cells were stained with an anti-γH2AX antibody and counterstained with DAPI. Shown is the percentage of cells with >5 foci relative to the total number of cells. Data shown represent mean values ± SD of 3 independent experiments. (**P = .0039, upper; **P = .0045, lower, Student t test). (D) RQ-PCR analysis of XRCC1, PARP1, and Lig3α in sorted preleukemic thymocyte subsets of double-negative (DN) populations (DN1, DN2, DN3, DN4) from Lck-Cre × KrasG12D mice (n = 6). (E) Nuclear extracts were prepared from preleukemic CD4/CD8 thymocytes (Lck-Cre × KrasG12D) or littermate control animals (Lck-Cre). Shown is the frequency of misrepaired colonies of 4 independent experiments.
Primary murine KrasG12D-expressing T-ALL and preleukemic cells show increased levels of DNA misrepair. (A-C) Single-cell suspensions were prepared from the thymus of leukemic KrasG12D-expressing mice or control animals (KRAS wild type). (A) Immunoblot analysis using anti-Lig3α and PARP1 antibodies as indicated. To control equal loading, the blot was stripped and reprobed with an anti-actin antibody. Numbers indicate expression levels relative to actin determined by the means of densitometry. (B) Expression of intracellular Lig3α in primary thymocytes and fluorescence-activated cell sorter analysis (red, unstained; blue, immunoglobulin G-isotype control; yellow, anti-Lig3α-Alexa488). (C) Cells were treated with either (upper) daunorubicin (125 nM) or (lower) were irradiated (10 Gy). Cells were stained with an anti-γH2AX antibody and counterstained with DAPI. Shown is the percentage of cells with >5 foci relative to the total number of cells. Data shown represent mean values ± SD of 3 independent experiments. (**P = .0039, upper; **P = .0045, lower, Student t test). (D) RQ-PCR analysis of XRCC1, PARP1, and Lig3α in sorted preleukemic thymocyte subsets of double-negative (DN) populations (DN1, DN2, DN3, DN4) from Lck-Cre × KrasG12D mice (n = 6). (E) Nuclear extracts were prepared from preleukemic CD4/CD8 thymocytes (Lck-Cre × KrasG12D) or littermate control animals (Lck-Cre). Shown is the frequency of misrepaired colonies of 4 independent experiments.
To investigate the effect of oncogenic Kras on DSB repair in preleukemic thymocytes, we isolated CD4/CD8 DN populations expressing CD44+/CD25−(DN1), CD44+/CD25+ (DN2), CD44−/CD25+ (DN3), or CD44−/CD25− (DN4) and performed RQ-PCR analysis. These subpopulations represent early stages of T-cell development and likely only differ in the expression of mutated or wild-type Kras. As shown in Figure 7D, we found increased mRNA expression levels of XRCC1 in KrasG12D-DN1/DN2 cells and of Lig3α in KrasG12D-DN2/DN3 compared with age-matched, normal littermate controls. PARP1 mRNA levels remained largely unchanged. This difference compared with our previous data in transformed cells might be attributed to the preleukemic state of cells analyzed here. To study whether oncogenic Kras alters DSB repair efficacy, we performed an ex vivo plasmid reactivation assay using nuclear extracts derived from CD4−/CD8− thymocytes of preleukemic Lck-Cre/KrasG12D mice or healthy littermate controls. Consistent with our previous results, expression of KrasG12D in primary thymocytes was associated with a substantial increase in misrepair frequency (Figure 7E), suggesting an important role of oncogenic Kras in early tumor development.
Discussion
Activating KRAS mutations belong to the most common alterations in human malignancies. In epithelial tumors, KRAS mutations are already detected in preneoplastic lesions, suggesting that oncogenic KRAS is involved in the initiation of cellular transformation; however, additional genetic events are required to complete the process of malignancy. In a murine T-ALL model, expression of oncogenic Kras under the control of its endogenous promoter was associated with a significant decrease in the number of total thymocytes and a block in differentiation at the DN2/3 stage. These changes occurred many weeks before the onset of acute leukemia, suggesting a premalignant stage.10 Interestingly, at the time of leukemia development, about half of the mice had either acquired somatic mutations within the Notch1 gene or chromosomal rearrangements.10,37 These findings prompted us to investigate DNA damage repair in several KRAS-mutated leukemic cell lines and in primary T-ALL cells. Expression of oncogenic KRAS is associated with increased expression levels of Lig3α, PARP1, and XRCC1, all essential elements of the alt-NHEJ pathway. Further, functional analyses revealed end joining of induced DSBs by means of DNA microhomologies, delayed repair kinetics, and an increased frequency of DNA misrepair. Ku proteins have been shown to initiate DNA repair by C-NHEJ through binding to DSBs,38 thus preventing alt-NHEJ proteins from binding to broken DNA ends. However, in the context of oncogenic KRAS, Ku70 and Ku86 need to compete with increased PARP1 levels for binding to DSB ends, likely shifting the balance of DSB repair toward the alt-NHEJ pathway. Although the mechanism of KRAS-induced upregulation of PARP1 remains elusive, there is some evidence that oncogenic KRAS regulates XRCC1 expression via the transcription factor E2F1.39 E2F1 mRNA is directly regulated by RAS signaling or indirectly via release from the tumor suppressor protein retinoblastoma on RAS-regulated phosphorylation of retinoblastoma. Nuclear XRCC1 forms a complex with Lig3α and stabilizes the repair enzyme followed by nuclear accumulation.40 In contrast to FLT3-ITD– or BCR-ABL–expressing cells, we did not observe downregulation of core components of the C-NHEJ pathway, eg, Ku70, Ku86, XRCC4, or LigIV. Beside RAS/RAF/MAPK/ERK, FLT3-ITD and BCR-ABL signal through several additional downstream pathways, including phosphatidylinositol 3-kinase/AKT or signal transducer and activator of transcription 5. Constitutive activation of these alternative pathways may account for the reduced expression of proteins of the C-NHEJ pathway.
We also demonstrate that on treatment with genotoxic anticancer drugs, KRAS-mutated cells exhibited delayed repair kinetics and increased misrepair of DSBs. Interestingly, this phenotype was reversible on knockdown of KRAS in Nomo-1 cells, again suggesting an important role for mutated KRAS in selecting which DNA repair pathway will be used. As Nomo-1 cells are heterozygous for the KRASG13D mutation, we speculate that the mutant KRAS allele is dominant with respect to the described differential DNA repair pathway selection.2 These findings are in line with observations in FLT3-ITD– or BCR-ABL–expressing cells. First, treatment of FLT3-mutated cell lines with the specific FLT3 tyrosine kinase inhibitor CEP-701 caused a significant decrease in spontaneous DSB misrepair accompanied by downregulation of Lig3α.27 Second, knockdown of Lig3α in BCR-ABL–positive cells resulted in an increase in unrepaired DSBs, likely due to concomitant downregulation of components of the C-NHEJ pathway with diminished end-joining activity.26
We further investigated whether targeting components of the alt-NHEJ pathway in KRAS-mutated cells might represent a therapeutic strategy. Pharmacologic targeting of PARP1 successfully sensitized oncogenic KRAS expressing cells to apoptotic cell death on treatment with cytotoxic agents. In addition, knockdown of Lig3α in several KRAS-mutated cell lines partially reversed KRAS-induced resistance to apoptosis. This effect was not as significant as compared with NU1025 treatment; however, besides its role in the alt-NHEJ pathway, PARP is also involved in the regulation of several other biological processes.41 Of note, the reversal of genotoxic drug resistance on inhibition of the alt-NHEJ pathway seems to be specific for KRAS-mutated cells, as no effect was observed in KRAS wild-type cells. Our data indicate that expression of oncogenic KRAS induces a mutator phenotype and confers dependence on components of the alt-NHEJ pathway to repair DSBs on genotoxic stress. Targeting these components is suitable for modulating drug resistance. This strategy is supported by recent findings in artificially selected, therapy-resistant cell lines expressing high levels of PARP1 and Lig3α.42,43 These cell lines were more sensitive to inhibition of either PARP1 or Lig3α alone or in combination with anti-estrogen therapy compared with control cells.
A potential therapeutic effect becomes even more relevant as it is extremely challenging to inhibit KRAS mutations directly. Constitutive activation of KRAS results from a loss of the intrinsic GTPase activity. Therefore, KRAS mutations represent loss-of-function mutations and effective targeting would require restoring enzymatic activity. To bypass this caveat, several groups applied the concept of synthetic lethality to KRAS-mutated cells. By the use of global RNA interference screens, knockdown of distinct target genes such as STK33, TBK1, and PLK1 enabled a selective killing of KRAS-mutated cells.31,44,45 It is important to note that a single protein can have a variety of different functions and specific small molecules likely only inhibit a few of them. For example, only depletion of the entire STK33 protein through HSP90 inhibition, but not selective inhibition of STK33’s enzymatic activity was able to reproduce the impressive effects of RNA interference-mediated knockdown, suggesting different functions of STK33 are involved in the synthetic lethal interaction with oncogenic KRAS.46,47 Here we present a different strategy named synthetic lethal vulnerability. Whereas inhibition of the alt-NHEJ pathway has no effect in KRAS mutant cells, it is able to sensitize malignant cells to genotoxic agents. This approach combines the use of well-known effective chemotherapeutics with small molecule inhibitors in the context of a defined genetic context. Of note, many PARP inhibitors are currently tested in combination with standard chemotherapy in different malignancies.48
In conclusion, expression of oncogenic KRAS shifts the balance of DSB repair toward the highly error-prone alt-NHEJ pathway. Pharmacologic inhibition of central components of this pathway represents a promising strategy to reverse KRASmut-mediated resistance to chemotherapeutics.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Claudia Scholl, Vera Gorbunova, and Claudia Haferlach for providing reagents and samples, and Steffen Lorenz (Confocal Laser Scanning Microscopy, Research Immunology (FZI), Johannes Gutenberg University, Mainz, Germany) for support with Confocal Laser Scanning Microscopy. The authors also thank Teodora Nikolova and members of the Kindler laboratory for helpful discussions and technical assistance.
This work was supported by Deutsche Krebshilfe grant 108821.
Authorship
Contribution: P.S.H. designed and performed experiments, analyzed data, and wrote the paper; B.E., D.S., and L.B. performed research; B.K., W.P.R., and M.T. provided reagents and critically read the manuscript; and T.K. designed experiments, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas Kindler, University Medical Center Mainz, Langenbeckstrasse 1, 55131 Mainz, Germany; e-mail: thomas.kindler@unimedizin-mainz.de.