Abstract
The transcription factor Meis1 is expressed preferentially in hematopoietic stem cells (HSCs) and overexpressed in certain leukemias. However, the functions of Meis1 in hematopoiesis remain largely unknown. In the present study, we found that Meis1 is required for the maintenance of hematopoiesis under stress and over the long term, whereas steady-state hematopoiesis was sustained in the absence of Meis1 in inducible knock-out mice. BM cells of Meis1-deficient mice showed reduced colony formation and contained significantly fewer numbers of long-term HSCs, which exhibited loss of quiescence. Further, we found that Meis1 deletion led to the accumulation of reactive oxygen species in HSCs and decreased expression of genes implicated in hypoxia response. Finally, reactive oxygen species scavenging by N-acetyl cysteine or stabilization of hypoxia signaling by knockdown of the von-Hippel-Lindau (VHL) protein led to reversal of the effects of Meis1 deletion. The results of the present study demonstrate that Meis1 protects and preserves HSCs by restricting oxidative metabolism.
Introduction
The transcription factor Meis1 belongs to the 3–amino acid loop extension (TALE) family of homeodomain proteins, which also includes Pbx1. Meis1 was originally discovered as a target of activation by retroviral integration in the leukemic mouse strain BXH-2. In these mice, Meis1 was always found coactivated with Hoxa7 or Hoxa9.1 Experimental overexpression of Meis1 and Hoxa9 in hematopoietic cells leads to an aggressive leukemia in mice.2 In normal hematopoiesis, Meis1 expression among the various cell compartments is correlated with the degree of self-renewal, with the levels being highest in hematopoietic stem cells (HSCs) and declining with differentiation.3,4 These results, combined with those from studies in leukemia, suggest a role for Meis1 in HSC self-renewal. However, the functions of Meis1 in established hematopoiesis remain unknown. Meis1−/− mice die by embryonic day 14.5 and the embryos display extensive hemorrhaging, particularly in the CNS. In addition, fetal liver cells obtained from Meis1−/− embryos show reduced myeloid colony formation in vitro and perform poorly in competitive transplantation assays in vivo.5,6 These results show that Meis1 is critical in the development of hematopoiesis. Further, Meis1−/− embryos display defects in capillary formation, suggesting additional roles for Meis1 in mechanisms that regulate the process of angiogenesis, at least during development.
Previous studies showed that Meis1 and Pbx1 can form heterodimeric and heterotrimeric complexes with HOX proteins, augmenting the binding and activation of target genes by HOX proteins such as HOXA9.7,8 Deletion of Pbx1 in the hematopoietic system led to loss of quiescence in HSCs, resulting in hematopoietic failure.9 Given that both Pbx1 and Meis1 are expressed in HSCs, we hypothesized that Meis1 might also be required for the maintenance of the self-renewing HSCs. In the present study, we investigated the role of Meis1 in established hematopoiesis using inducible knock-out mice. We found that Meis1 is required for the maintenance of HSCs by preserving quiescence in these mice. Maintenance of quiescence is critical for preserving self-renewal of long-term stem cells, and various signaling pathways have been shown to modulate quiescence in HSCs, including oxidative stress.10 Recent studies have shown that HSCs reside in hypoxic niches and that the principal regulator of the hypoxia response, Hif-1α, is critical for the maintenance of quiescence in HSCs.11 Further, one of the upstream regulators of this pathway was shown to be Meis1 in a previous study.12 In the present study, we found that Meis1 maintains Hif-1α expression in the BM and restricts the levels of reactive oxygen species (ROS) in HSCs, which in turn sustains self-renewal.
Methods
Generation of Meis1-knockout mice
Exon 8 of the Meis1 gene was targeted in mouse embryonic stem (ES) cells by a standard replacement-type targeting vector constructed by microhomologous recombination in bacteria using a 9.3-kb C57BL/6 mouse genomic fragment retrieved by the bacterial artificial chromosome RP23-306E8.13,14 Electroporation and selection were performed using the v6.4 ES cell line that has been described previously.15 DNAs derived from G418/FIAU-resistant ES clones were screened by diagnostic EcoRV and HindIII restriction enzyme digestions using, respectively, a 5′ and 3′ probe external to the targeting vector sequence. Recombinant clones containing the predicted rearranged band were obtained at a frequency of 1/7. Two independently targeted ES cell clones injected into C57Bl/6 blastocysts generated chimeras that transmitted the mutated allele to the progeny.16 After germline transmission of the targeted ES cell clones, the diagnostic HindIII digest was used for screening because it enabled us to distinguish between the wild-type and all the targeted alleles including those generated after Cre or Flpe recombination. Mutant mice were backcrossed for at least 10 generations onto the C57BL/6 background before use.
Transgenic Meis1-flox/CreER mice were generated by crossing Meis1-flox mice with Rosa26CreER mice.17 Genotypes of the animals were confirmed by PCR on peripheral blood. Mice were injected with tamoxifen (Sigma-Aldrich) IP with a dosing schedule of 1 mg/d for 5 days or a single dose of 4 mg to induce Meis1 deletion. Control mice were given the same schedule. For in vitro deletion, BM cells were incubated with 4-hydroxy tamoxifen (4-OHT; Sigma-Aldrich) at a concentration of 1μM for 48 hours. All mice used in this study were 6-10 weeks of age. All animal experiments used were approved by the institutional animal care and use committee. The congenic CD45.1 BoyJ mice were purchased from The Jackson Laboratory and used as recipients of BM transplantation.
Flow cytometry
BM cell suspensions were obtained by crushing the femurs and tibias in sterile medium. For acquisition studies, these suspensions were stained with surface Abs in staining buffer (PBS supplemented with 0.5% FBS). Peripheral blood was obtained by tail bleeding, treated with Pharm Lyse Lysing Buffer (BD Biosciences), and then stained with surface Abs (eBiosciences) in the staining buffer. For lineage depletion, the mouse lineage depletion kit (Miltenyi Biotec) was used, followed by magnetic separation in an automated cell separator (AutoMACS; Miltenyi Biotec). Data were acquired in FACSCanto III and LSR II flow cytometers (BD Biosciences) and results were analyzed using FACSDiva Version 6.1.2 (BD Biosciences) and FlowJo (TreeStar) software.
Cell-cycle analysis
Mice were injected with tamoxifen and the animals were killed a week later. Lineage-depleted BM cells were incubated with surface marker Abs for 20 minutes on ice, followed by fixation and permeabilization using the Cytofix/Cytoperm kit (BD Biosciences), and then stained with 1 μg/mL of Hoechst 33342 stain (Sigma-Aldrich) and 10 μg/mL of pyronin Y stain (Sigma-Aldrich). The cells were stained for 30 minutes in a reciprocal shaking bath set at 37°C and then analyzed by flow cytometry on an LSR II flow cytometer. For proliferation analysis, tamoxifen-treated mice were injected with 1 mg of bromodeoxyuridine (BrdU flow kit; BD Biosciences) and killed 16 hours later. Lineage-depleted BM cells were first stained with Abs against surface antigens and then with anti-BrdU as per the manufacturer's instructions.
5-FU treatment
The experimental and the control mice were injected with single dose of tamoxifen. Three days after the tamoxifen injection, the mice were injected with 5-fluorouracil (5-FU, 150 mg/kg; APP Pharmaceuticals) every 7 days.
Colony-forming assays
BM cells from the controls and the experimental mice were plated onto methylcellulose-containing medium (M3434; StemCell Technologies). Colonies were scored 7 days after plating. For in vitro deletion, cells were cultured for 48 hours in medium supplemented with cytokines (SCF, G-CSF, and thrombopoietin) in the presence of 4-OHT and then seeded onto methylcellulose medium. N-acetyl-L-cysteine (100 μg/mL; Sigma-Aldrich), cobalt chloride (100μM), or dimethyloxalylglycine (100μM) were added to the 4-OHT–containing medium in certain experiments as described in Results.
Lentiviral shRNA transduction
The lentiviral von-Hippel-Lindau (VHL) and Pbx1 shRNA constructs were purchased from Sigma-Aldrich in the pLKO.1-Puro plasmid. The viral supernatant was generated by transfecting HEK 293T cells as per the manufacturer's instructions. Lineage-negative BM cells were prestimulated by overnight incubation in IMDM supplemented with cytokines as in the previous section. Fresh viral supernatant was then used to transduce the prestimulated cells by spinoculation on retronectin (Fisher Scientific)–coated plates. The transduced cells were selected in puromycin (1 μg/mL) for 72 hours and placed in culture with 4-OHT or ethanol as a vehicle control. After 48 hours, cells were seeded onto methylcellulose medium for assessing the clonogenic potential of the cells. The remaining cells were used for RNA isolation and subsequent gene-expression analysis.
Gene-expression analysis
RNA was extracted from cultured lineage-negative cells using the RNeasy Mini Kit (QIAGEN) and analyzed by Affymetrix GeneChip Version 1.0 ST arrays. Sample processing and normalization of data were according to the manufacturer's instructions. All microarray data have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession no. GSE41133. For RT-PCR, total RNA was converted to cDNA using the Superscript First-strand cDNA Synthesis kit (Invitrogen). The resulting cDNA was then subjected to real-time quantitative PCR with Power SYBRGreen PCR Master Mix using custom designed primers (available on request) or TaqMan PCR Master Mix and commercially available TaqMan primer/probe sets (Applied Biosystems).
ROS analysis
For the detection of ROS, lineage-depleted BM cells were stained with surface Abs and then incubated with 5μM dichlorofluorescein diacetate (Invitrogen) for 20 minutes at 37°C in PBS. The cells were then pelleted and resuspended in staining buffer before acquisition.
Western blotting
Abs used were against Meis1 (Abcam), Pbx1, actin (Cell Signaling), and Hif-1α (kind gift from Dr James Bridges, Cincinnati Children's Hospital Medical Center).18 Nuclear lysates were first isolated using the NE-PER kit (Thermo Scientific) and then separated by SDS-PAGE on a 4%-16% gradient gel (Invitrogen). After transfer to PVDF membranes, blots were blocked with 5% nonfat dry milk and incubated with primary Abs overnight. After washing, blots were treated with appropriate secondary Abs and then developed using a chemiluminescence kit (Thermo Scientific).
Results
Meis1 is required for the maintenance of LT-HSCs
To determine the functional contribution of Meis1 in adult hematopoiesis, we generated inducible Meis1-knockout mice in which the homeodomain of Meis1 (exon 8 in supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) was flanked by loxp sites (henceforth referred to as Meis1-flox). These mice were then bred to Rosa26CreER mice to generate Meis1-flox/CreER (Meis1flox/flox/CreER) mice.17 We achieved efficient and complete recombination of the Meis1 gene on tamoxifen administration to mice (supplemental Figure 2) and also with in vitro treatment of BM cells with 4-OHT. Loss of Meis1 protein was detected by Western blotting (supplemental Figure 2). Loss of Meis1 mRNA was evident in the BM, including in sorted long-term HSCs (LT-HSCs; supplemental Figure 2). In all experiments, Meis1-flox/CreER mice were used to assess the impact of Meis1 deletion; controls consisted of Meis1-flox, Rosa26CreER, or Meis1flox/+/CreER mice.
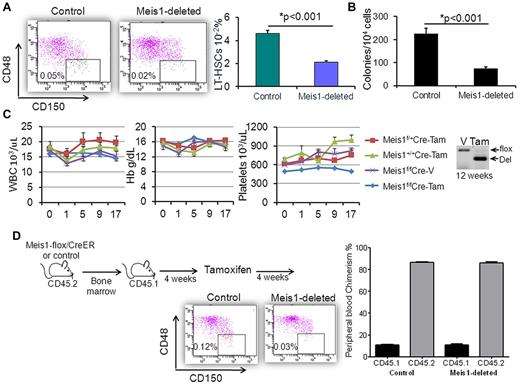
To determine the impact of Meis1 deletion on hematopoiesis, we treated Meis1-flox/CreER and control mice with tamoxifen. After 3 weeks, the BM of the Meis1-deleted mice showed a significant decline in LT-HSCs, defined as Lin−c-kit+sca-1+ (LSK) CD48−CD150+ cells (Figure 1A). In addition, Meis1-deleted BM formed significantly fewer colonies when cultured in semisolid medium (Figure 1B). This effect was evident even when Meis1 was deleted in vitro with 4-OHT, suggesting a cell-intrinsic defect in HSC function in the absence of Meis1. Conversely, the progenitor fractions were not significantly changed with Meis1 deletion (supplemental Figure 3). For up to 17 weeks after treatment, there was little change in the peripheral blood counts of control and Meis1-deleted animals, except for a mild decrease in platelet count with Meis1 deletion (Figure 1C). Analysis of tail blood DNA by PCR showed persistent deletion in these mice, confirming that at least short-term hematopoiesis was sustained in the absence of Meis1. At 19 weeks after Meis1 deletion, tail-blood PCR showed reemergence of the nonrecombined (floxed) allele and additional tamoxifen treatments proved too toxic, resulting in premature death in all groups.
Meis1 deletion leads to loss of LT-HSCs. Meis1-flox/CreER or control mice were treated with tamoxifen as described in the text. (A) Representative flow cytometry plots of CD48 and CD150 staining (gated on LSK fraction) showing significantly fewer LT-HSCs in the BMs of Meis1-deleted mice compared with controls. Bar graph depicts mean ± SEM of the percentage of LT-HSCs in lineage-depleted BM of control and Meis1-deleted mice (n = 7). (B) Bar graph representing colony numbers in methylcellulose medium. Data are shown as means ± SEM of total colony numbers/10 000 cells (n = 4). (C) Line graphs of blood counts (vertical axis) over time (horizontal axis) since tamoxifen treatment. Identities of the various groups are indicated in the legend on the right, with 5 mice in each group. Also shown is a representative gel image of PCR on tail blood 12 weeks after vehicle or tamoxifen treatment. (D) Transplantation experiment demonstrating that the loss of LT-HSCs with Meis1 deletion was cell autonomous. Flow plots are as in panel A, but depict donor LT-HSCs (recipients were lethally irradiated). The bar graph shows peripheral blood chimerism in recipients.
Meis1 deletion leads to loss of LT-HSCs. Meis1-flox/CreER or control mice were treated with tamoxifen as described in the text. (A) Representative flow cytometry plots of CD48 and CD150 staining (gated on LSK fraction) showing significantly fewer LT-HSCs in the BMs of Meis1-deleted mice compared with controls. Bar graph depicts mean ± SEM of the percentage of LT-HSCs in lineage-depleted BM of control and Meis1-deleted mice (n = 7). (B) Bar graph representing colony numbers in methylcellulose medium. Data are shown as means ± SEM of total colony numbers/10 000 cells (n = 4). (C) Line graphs of blood counts (vertical axis) over time (horizontal axis) since tamoxifen treatment. Identities of the various groups are indicated in the legend on the right, with 5 mice in each group. Also shown is a representative gel image of PCR on tail blood 12 weeks after vehicle or tamoxifen treatment. (D) Transplantation experiment demonstrating that the loss of LT-HSCs with Meis1 deletion was cell autonomous. Flow plots are as in panel A, but depict donor LT-HSCs (recipients were lethally irradiated). The bar graph shows peripheral blood chimerism in recipients.
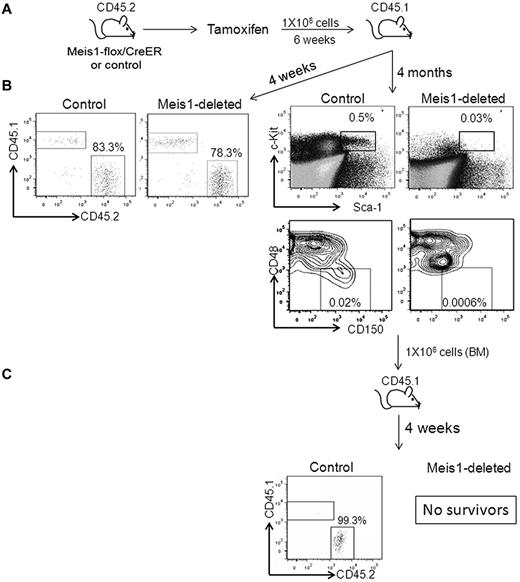
To validate that the effect on LT-HSCs was cell autonomous, we transplanted untreated Meis1-flox/CreER or control BM into lethally irradiated CD45.1 mice. Both the Meis1-flox and Rosa26CreER mice are of the CD45.2 background, allowing us to differentiate these donor cells by flow cytometry in the blood and BM of CD45.1 recipient mice. After engraftment (4 weeks), recipients were treated with tamoxifen and then analyzed 4 weeks later for HSC frequency and functional hematopoiesis. BM analysis showed a significant reduction in CD45.2 LT-HSCs in the Meis1-deleted mice compared with controls (Figure 1D). However, the production of differentiated elements in the peripheral blood was identical in the Meis1-deleted and control mice. These data strongly suggest that Meis1 functions to support LT-HSC maintenance but is dispensable for proliferation and differentiation of committed hematopoietic progenitors.
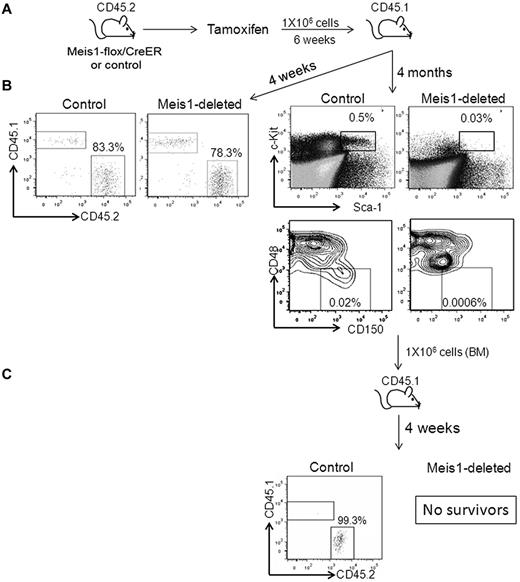
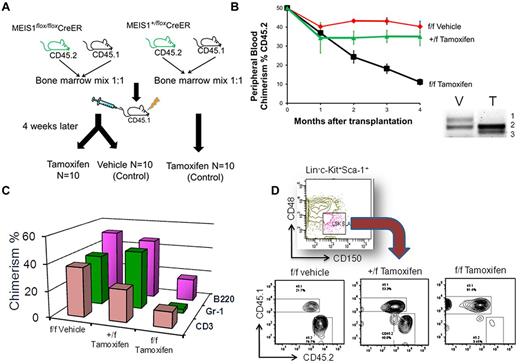
To determine the impact of Meis1-loss on cell-intrinsic HSC function, we treated Meis1-flox/CreER or control mice with tamoxifen and then transplanted whole BM cells into lethally irradiated CD45.1 mice (Figure 2). Four weeks after transplantation, peripheral blood chimerism was identical (approximately 80%) in the control and Meis1-deleted recipient mice, demonstrating that short-term engraftment and functional hematopoiesis were attained even in the absence of Meis1. We then treated these recipients with tamoxifen every 4 weeks (to prevent reemergence of the floxed-allele in the recipients of Meis1-flox/CreER). At 4 months after transplantation, Meis1-deleted BM showed near-complete loss of HSCs and multipotent progenitors compared with controls (Figure 2). Further, when transplanted into irradiated secondary recipients, the Meis1-depleted cells failed to reconstitute hematopoiesis, resulting in death of the mice from BM failure, whereas the control cells engrafted and rescued the recipients (Figure 2). We also performed a competitive transplantation assay in which we mixed Meis1flox/flox/CreER or Meis1flox/+/CreER BM in a 1:1 ratio with CD45.1 BM (Figure 3A). This mixture was then transplanted into lethally irradiated CD45.1 mice. After engraftment, recipients were treated with tamoxifen or vehicle. One week after tamoxifen treatment, peripheral blood PCR revealed complete recombination of the floxed allele and flow cytometry showed 35%-40% chimerism in both the Meis1-deleted and control conditions (Figure 3B). Serial peripheral blood analysis every 4 weeks showed a gradual loss of chimerism in the Meis1-deleted mice, whereas both the control groups (vehicle-treated Meis1flox/flox/CreER and tamoxifen-treated Meis1flox/+/CreER) showed maintenance of chimerism over the same time course. At 12 weeks after treatment (16 weeks after transplantation), chimerism in the Meis1-deleted group was reduced to < 10%. Analysis of peripheral blood lineages at this time showed loss of chimerism in myeloid, B-cell, and T-cell lineages (Figure 3C). BM analysis showed near-complete loss of chimerism in the LT-HSCs (Figure 3D). These results demonstrate that Meis1 functions in a cell-autonomous manner to promote the maintenance and function of LT-HSCs.
Meis1 deletion leads to loss of HSC function. (A) Meis1-flox/CreER or control mice were treated with tamoxifen, as described in the text, and transplanted into lethally irradiated CD45.1 mice. (B) Representative flow cytometry plots on the left depict peripheral blood chimerism at 4 weeks after transplantation. Representative flow cytometry plots on the right depict the percentages of LSKs and LT-HSCs in the BM of transplantation recipients at 4 months. Shown are donor cells. (C) Recipient mice were killed at 4 months and whole BM was transplanted into lethally irradiated secondary recipient mice. Shown are representative flow cytometry plots depicting chimerism in the BM of secondary recipients. The recipients of Meis1-deleted BM died of hematopoietic failure.
Meis1 deletion leads to loss of HSC function. (A) Meis1-flox/CreER or control mice were treated with tamoxifen, as described in the text, and transplanted into lethally irradiated CD45.1 mice. (B) Representative flow cytometry plots on the left depict peripheral blood chimerism at 4 weeks after transplantation. Representative flow cytometry plots on the right depict the percentages of LSKs and LT-HSCs in the BM of transplantation recipients at 4 months. Shown are donor cells. (C) Recipient mice were killed at 4 months and whole BM was transplanted into lethally irradiated secondary recipient mice. Shown are representative flow cytometry plots depicting chimerism in the BM of secondary recipients. The recipients of Meis1-deleted BM died of hematopoietic failure.
Meis1-deleted HSCs are deficient in maintenance of hematopoiesis. (A) Competitive repopulation assay was performed as described in the text. (B) Summary of peripheral blood CD45.2 chimerism (vertical axis) in recipient mice over time (horizontal axis) since transplantation. The groups are labeled to the right of the respective curve. Data are shown as means ± SEM (n = 5-8 per group). Also shown on the lower right is a representative PCR result on peripheral blood of recipient mice treated with vehicle (V) or tamoxifen (T). Bands labeled to the right represent, respectively, in numerical order floxed, wild-type, and deleted Meis1 alleles, showing that we achieved complete deletion of Meis1 by tamoxifen treatment. (C) Chimerism in the various lineages at 4 months. (D) Flow cytometry plots showing chimerism in the LT-HSC fraction of the BM of recipient mice at 4 months.
Meis1-deleted HSCs are deficient in maintenance of hematopoiesis. (A) Competitive repopulation assay was performed as described in the text. (B) Summary of peripheral blood CD45.2 chimerism (vertical axis) in recipient mice over time (horizontal axis) since transplantation. The groups are labeled to the right of the respective curve. Data are shown as means ± SEM (n = 5-8 per group). Also shown on the lower right is a representative PCR result on peripheral blood of recipient mice treated with vehicle (V) or tamoxifen (T). Bands labeled to the right represent, respectively, in numerical order floxed, wild-type, and deleted Meis1 alleles, showing that we achieved complete deletion of Meis1 by tamoxifen treatment. (C) Chimerism in the various lineages at 4 months. (D) Flow cytometry plots showing chimerism in the LT-HSC fraction of the BM of recipient mice at 4 months.
Meis1 deletion leads to loss of LT-HSC quiescence
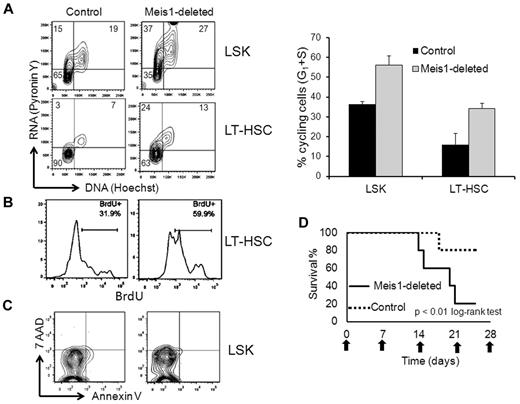
To determine the mechanism of HSC loss with Meis1 deletion, we studied the effect of Meis1 deletion on the survival and proliferation of LT-HSCs. Mice were treated with tamoxifen and 1 week later, BM was analyzed by flow cytometry. We used Hoechst/pyronin Y staining to ascertain the number of cycling LT-HSCS. As described previously, the majority of the control LT-HSCs (81.75% ± 3.25%, n = 2, Figure 4A) were in the G0 phase of the cell cycle.11 Conversely, with Meis1 deletion, there was a significant decrease in the proportion of quiescent (G0) LT-HSCs (56.10% ± 0.873%, n = 3) with a concomitant increase in the cycling (G1) cells (Figure 4A). The results were similar when the population analyzed was extended to LSK to include the short-term HSCs and multipotent progenitors. To corroborate that the increased cycling resulted in increased proliferation of stem cells, we injected tamoxifen-treated mice with BrdU and analyzed its incorporation by flow cytometry. As expected and as shown in Figure 4B, of the control LT-HSCs, 36.33% ± 3.02% (mean ± SEM, n = 3) incorporated BrdU, whereas this fraction of proliferating cells increased significantly with Meis1 deletion (53% ± 4.14%, n = 3). Conversely, annexin V staining did not show any differences between the control and Meis1-deleted cells (Figure 4C). These results demonstrate that loss of Meis1 induces cycling of LT-HSCs, leading to depletion of the stem-cell pool by proliferative exhaustion.
Meis1 deletion leads to loss of quiescence in LT-HSCs. (A) Meis1-flox/CreER and control mice were treated with tamoxifen and, 1 week later, BM was analyzed by flow cytometry for cell-cycle analysis. Flow cytometry plots depict results of pyronin Y (vertical axis, RNA stain) and Hoechst (horizontal axis, DNA stain) staining gated on the LSK and LT-HSC fractions. Left lower quadrant represents cells in G0, left upper quadrant G1, and upper right S phase. Data are summarized in the bar graph to the right (n = 2 control and 3 Meis1 deleted). (B) Mice were treated as in panel A and then injected with BrdU before being killed. Shown are representative histograms of anti-BrdU staining gated on the LT-HSCs, as defined previously. (C) Flow cytometry plots of annexin V and 7-amino-actinomycin D gated on the LSK fraction of control and Meis1-deleted mice. (D) Meis1-deleted and control mice were treated with weekly doses of 5-FU. Graph shows the percentage survival (vertical axis) over time (horizontal axis) with the horizontal-axis labels representing the dates of 5-FU injection.
Meis1 deletion leads to loss of quiescence in LT-HSCs. (A) Meis1-flox/CreER and control mice were treated with tamoxifen and, 1 week later, BM was analyzed by flow cytometry for cell-cycle analysis. Flow cytometry plots depict results of pyronin Y (vertical axis, RNA stain) and Hoechst (horizontal axis, DNA stain) staining gated on the LSK and LT-HSC fractions. Left lower quadrant represents cells in G0, left upper quadrant G1, and upper right S phase. Data are summarized in the bar graph to the right (n = 2 control and 3 Meis1 deleted). (B) Mice were treated as in panel A and then injected with BrdU before being killed. Shown are representative histograms of anti-BrdU staining gated on the LT-HSCs, as defined previously. (C) Flow cytometry plots of annexin V and 7-amino-actinomycin D gated on the LSK fraction of control and Meis1-deleted mice. (D) Meis1-deleted and control mice were treated with weekly doses of 5-FU. Graph shows the percentage survival (vertical axis) over time (horizontal axis) with the horizontal-axis labels representing the dates of 5-FU injection.
To further investigate the cycling-associated exhaustion of LT-HSCs, we compared the effect of 5-FU on control and Meis1-deleted mice. In this assay, animals were subjected to weekly sublethal doses of 5-FU, which kills proliferating cells, stimulating the cycling of quiescent HSCs to replenish hematopoiesis. One week after tamoxifen treatment, Meis1-flox/CreER and control mice were treated with 5-FU every 7 days. As shown by the survival curve in Figure 4D, the Meis1-deleted mice showed increased sensitivity to 5-FU, with the majority of the mice succumbing to 3 doses, whereas most of the control mice survived 5 doses. To determine whether these effects were cell autonomous, we repeated this experiment on mice that were transplanted with Meis1-flox/CreER or control BM. Meis1 deletion led to increased sensitivity to 5-FU treatment in transplantation recipients, with rapid development of pancytopenia (supplemental Figure 4).
Meis1 limits oxidative stress by regulating the hypoxia-response pathway
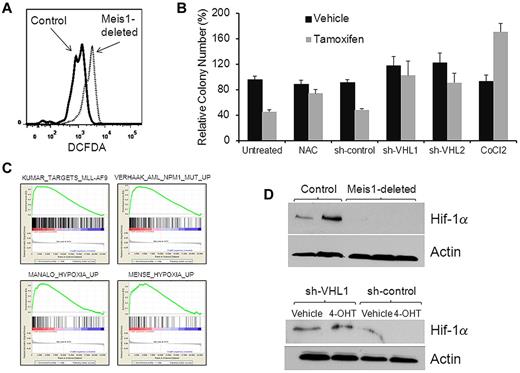
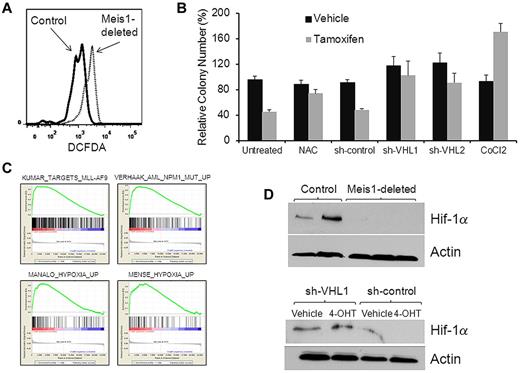
We next sought to determine the mechanisms by which Meis1 preserves quiescence in LT-HSCs. One of the critical determinants of HSC quiescence and function is the level of oxidative stress. High levels of ROS are known to induce loss of quiescence and exhaustion of LT-HSCs.19 To study the effect of Meis1 deletion on oxidative stress, we compared levels of ROS in control and Meis1-deplete LT-HSCs using the dye dichlorofluorescein diacetate. We found significantly higher levels of ROS in Meis1-deleted LT-HSCs compared with controls (Figure 5A). To corroborate that the accumulation of ROS was indeed causal in the hematopoietic defects associated with Meis1 deletion, we postulated that ROS scavenging would reverse these defects. Therefore, we deleted Meis1 in lineage-negative BM cells in the presence of the ROS scavenger N-acetylcysteine. In methylcellulose colony assays, N-acetylcysteine treatment prevented the inhibition of colony formation in Meis1-deleted cells, thus confirming that ROS accumulation played a contributory role in the loss of clonogenic potential (Figure 5B).
Meis1 regulates oxidative stress. (A) Representative flow cytometry plot showing results of dichlorofluorescein diacetate staining (a measurement of ROS levels) gated on LT-HSCs. (B) Bar graph depicting results of colony-formation assay performed with Meis1-flox/CreER lineage-depleted cells that were treated in vitro with vehicle (ethanol) or 4-OHT in the presence of the ROS scavenger N-acetyl cysteine (NAC), 1 of 2 shRNA constructs directed against VHL (shVHL1 and shVHL2), scrambled shRNA, or cobalt chloride (treatment conditions labeled on the bottom of the graph). Colony numbers (vertical axis) were normalized to those formed by vehicle-treated cells in each experiment. Data represent means ± SEM of triplicate colonies from 1 representative experiment. (C) Lineage-negative BM cells of Meis1-flox/CreER and control mice were treated with 4-OHT for 48 hours and then analyzed by whole genome microarrays. Depicted are gene-set enrichment analysis plots of the analysis showing enrichment in the control cells compared with Meis1-deleted of gene sets associated with leukemias that express high levels of Meis1 (top panel) and those associated with hypoxia-response (bottom panel). The gene sets are labeled on the top of each plot. (D) Western blot (top panel) showing levels of Hif-1α and actin (loading control) in whole BM of control (first 2 lanes) and Meis1-deleted mice. Bottom panel depicts Western blot results of Hif-1α and actin in vehicle- or 4-OHT–treated cells transduced with lentivirus expressing shRNA against VHL or a nontargeting construct.
Meis1 regulates oxidative stress. (A) Representative flow cytometry plot showing results of dichlorofluorescein diacetate staining (a measurement of ROS levels) gated on LT-HSCs. (B) Bar graph depicting results of colony-formation assay performed with Meis1-flox/CreER lineage-depleted cells that were treated in vitro with vehicle (ethanol) or 4-OHT in the presence of the ROS scavenger N-acetyl cysteine (NAC), 1 of 2 shRNA constructs directed against VHL (shVHL1 and shVHL2), scrambled shRNA, or cobalt chloride (treatment conditions labeled on the bottom of the graph). Colony numbers (vertical axis) were normalized to those formed by vehicle-treated cells in each experiment. Data represent means ± SEM of triplicate colonies from 1 representative experiment. (C) Lineage-negative BM cells of Meis1-flox/CreER and control mice were treated with 4-OHT for 48 hours and then analyzed by whole genome microarrays. Depicted are gene-set enrichment analysis plots of the analysis showing enrichment in the control cells compared with Meis1-deleted of gene sets associated with leukemias that express high levels of Meis1 (top panel) and those associated with hypoxia-response (bottom panel). The gene sets are labeled on the top of each plot. (D) Western blot (top panel) showing levels of Hif-1α and actin (loading control) in whole BM of control (first 2 lanes) and Meis1-deleted mice. Bottom panel depicts Western blot results of Hif-1α and actin in vehicle- or 4-OHT–treated cells transduced with lentivirus expressing shRNA against VHL or a nontargeting construct.
To determine the molecular pathways regulated by Meis1 in hematopoiesis, we compared the gene-expression profiles of Meis1-deleted and control cells. We treated lineage-negative BM cells of Meis1-flox/CreER and control mice with 4-OHT for 48 hours and analyzed RNA profiles by Affymetrix whole genome expression arrays. Gene-set enrichment analysis of the data showed that Meis1 deletion led to decreased expression of genes that are overexpressed in leukemias known to express high levels of Meis1—those associated with NPM1 mutations or with the MLL-AF9 fusion gene (Figure 5C).20-22 Interestingly, we also found several different hypoxia-related gene sets down-regulated in the Meis1-deleted cells; these included genes that were found to be up-regulated in response to hypoxia or those regulated by the hypoxia-response regulator Hif-1α.23,24 Recently, Meis1 was reported to bind the promoter region of Hif-1α, suggesting that Hif-1α might be a target of regulation by Meis1. Real-time quantitative RT-PCR on lineage-negative BM showed that Meis1 deletion led to decreased expression of Hif-1α mRNA (supplemental Figure 5), whereas Western blot on whole BM showed near-complete loss of the Hif-1α protein (Figure 5D). These findings are consistent with studies showing that LT-HSCs depend on precise control of Hif-1α levels, with decreased levels resulting in loss of HSC quiescence similar to the phenotype observed with Meis1 loss.11 Low Hif-1α levels lead to oxidative stress and to accumulation of ROS. These results led us to postulate that Meis1 deletion leads to hematopoietic defects via decreased Hif-1α levels and the resulting oxidative stress. Accordingly, these defects should be rescued by restoring Hif-1α levels in the Meis1-depleted cells. Therefore, we increased Hif-1α levels by shRNA knockdown of the VHL protein, the principal mediator of Hif-1α degradation.25 We transduced lineage-negative BM of Meis1-flox/CreER mice with 1 of 2 different VHL shRNA constructs or a scrambled shRNA and then incubated these cells with 4-OHT or vehicle (ethanol). As shown in Figure 5, VHL knockdown prevented the loss of the Hif-1α protein and the loss of colony formation caused by Meis1 deletion, whereas the scrambled shRNA was unable to do so. Real-time RT-PCR showed > 50% knockdown of VHL mRNA in the knockdown cells compared with scrambled controls (supplemental Figure 6). Further, chemical stabilization of Hif-1α by cobalt chloride (Figure 5C) or dimethyloxalylglycine (not shown) also prevented loss of colony formation.26 These results indicate that Meis1 regulates the hypoxia response pathway and limits ROS levels in LT-HSCs and that the hematopoietic defects of Meis1 deletion are at least partly mediated by oxidative stress and decreased Hif-1α levels.
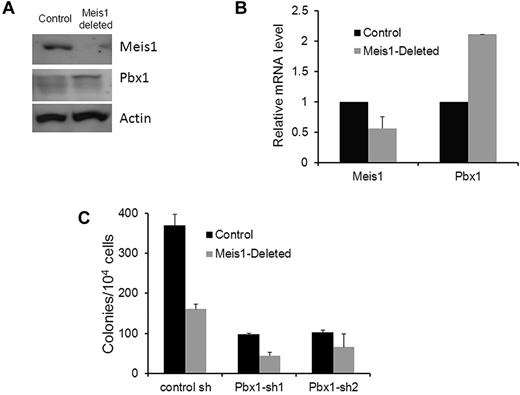
Feedback regulation of Pbx1 by Meis1
Because Meis1 and Pbx1 are found in the same nuclear complex and because Pbx1 was shown to regulate Meis1 in HSCs, we sought to determine the effect of Meis1 deletion on Pbx1. Interestingly, nuclear extracts of Meis1-deleted, lineage-depleted BM showed increased Pbx1 levels compared with controls (Figure 6A). Further, Pbx1 mRNA was also increased in response to Meis1 deletion (Figure 6B), suggesting the existence of compensatory feedback regulation of Pbx1 by Meis1. Therefore, overexpression of Pbx1 might dampen the effects of Meis1 loss. Consistent with this hypothesis, knockdown of Pbx1 in the context of Meis1 deletion led to a further reduction in colony formation in addition to that seen on Meis1 deletion alone (Figure 6C). As expected, Pbx1 knockdown by itself also led to significant reduction in colony formation. Our results demonstrate a feedback regulatory circuit within the Pbx1-Meis1 axis, with Pbx1 levels increasing in response to decreased levels of Meis1 or its effects.
Negative feedback of Pbx1 by Meis1. (A) Western blot showing Meis1, Pbx1, and actin (loading control) levels in control and Meis1-deleted lineage-negative BM. (B) Bar graph depicting Meis1 and Pbx1 mRNA levels in control and Meis1-deleted, lineage-negative BM. mRNA levels were measured by real-time quantitative RT-PCR using TaqMan primer/probes and normalized to β-actin (n = 2). (C) Lineage-negative BM cells were harvested from tamoxifen-treated Meis1-flox/CreER (Meis1-deleted) and Rosa26CreER (control) mice. Cells were transduced with lentivirus expressing 1 of 2 Pbx1-shRNAs or a control construct, and transduced cells were selected by culture in puromycin for 72 hours. Colony assay was then performed as described in the text. Bar graph depicts means ± SEM of triplicate colony counts from one representative experiment.
Negative feedback of Pbx1 by Meis1. (A) Western blot showing Meis1, Pbx1, and actin (loading control) levels in control and Meis1-deleted lineage-negative BM. (B) Bar graph depicting Meis1 and Pbx1 mRNA levels in control and Meis1-deleted, lineage-negative BM. mRNA levels were measured by real-time quantitative RT-PCR using TaqMan primer/probes and normalized to β-actin (n = 2). (C) Lineage-negative BM cells were harvested from tamoxifen-treated Meis1-flox/CreER (Meis1-deleted) and Rosa26CreER (control) mice. Cells were transduced with lentivirus expressing 1 of 2 Pbx1-shRNAs or a control construct, and transduced cells were selected by culture in puromycin for 72 hours. Colony assay was then performed as described in the text. Bar graph depicts means ± SEM of triplicate colony counts from one representative experiment.
Discussion
In the present study, we found that Meis1 is required for the normal function of HSCs in adult mice. In the absence of Meis1, the normally quiescent LT-HSCs enter the cell cycle and exhaust their capacity to maintain hematopoiesis, especially under stress. In addition, Meis1-depleted hematopoietic stem and progenitor cells show reduced clonogenic capacity. Levels of ROS are increased in Meis1-deficient HSCs, and gene-expression profiling studies showed that Meis1 regulates the hypoxia-response pathway. Finally, ROS scavenging or Hif stabilization led to reversal of the effects of Meis1 deletion, implicating oxidative stress as being at least partly responsible for the phenotype of Meis1-deficient hematopoiesis. Overall, our results identify a role for Meis1 in HSCs that is characterized by restricting the levels of ROS and limiting oxidative metabolism, thereby preserving quiescence and self-renewal capacity.
The reduced clonogenic capacity and competitive repopulation deficits we observed with Meis1 deletion are similar to those reported for fetal liver cells of Meis1−/− embryos. However, the overall impact of Meis1 deletion in adult mice was more subtle compared with that seen with constitutive deletion. We did not observe hemorrhaging in mice in which Meis1 was deleted in all adult tissues (ie, tamoxifen-treated Meis1-flox/RosaCreER). These data suggest that Meis1 plays a relatively more significant role in the development of the hematopoietic and vascular systems in the embryo than in the corresponding established systems of the adult. Similar results have been reported for other transcription factors (eg, RUNX1) that are critical during embryonic development of the hematopoietic system but less so in adulthood.27 Conversely, our results also indicate that the true impact of Meis1 deletion is more pronounced when the stem cells are under proliferative stress. Accordingly, steady-state hematopoiesis was not significantly affected by Meis1 loss, whereas stress hematopoiesis (ie, transplantation, 5-FU treatment, and in vitro colony formation) was significantly impaired. We conclude that Meis1 is likely critical in maintaining the repopulating capacity of stem cells.
Long-term self-renewal among the hematopoietic cells is largely restricted to quiescent stem cells, and several factors have been identified that regulate quiescence in LT-HSCs.28 One such factor is Pbx1, another TALE protein that forms hetero-oligomers with Meis1 and HOX proteins. Conditional deletion of Pbx1 in hematopoietic cells was shown previously to induce loss of quiescence and exhaustion of LT-HSCs.9 Studies in zebrafish also showed that knock down of Meis1 or Pbx1 results in identical phenotypes.29 Induced deletion of Pbx1 resulted in decreased expression of Meis1 in LT-HSCs, suggesting that the former regulates the latter. Our present data further support this notion, indicating that not only is Pbx1 required for normal Meis1 expression, but that this axis regulates HSC homeostasis via a negative feedback loop with a compensatory increase in Pbx1 levels in response to loss of Meis1. Previous gene-expression profiling of Pbx1-deficient cells identified several pathways dependent on Pbx1.9 Using gene-set enrichment analysis, we found a significant subset of Pbx1-dependent genes within our Meis1-dependent gene signature in lineage-depleted BM (supplemental Figure 7). Therefore, Meis1 and Pbx1 are complementary transcription factors in HSCs, with Pbx1 being an upstream regulator of this pathway. Conversely, it is possible that Pbx1 acts as a sensor of HSC homeostasis and is up-regulated in response to a decrease in HSC activity/number.30
Recent studies have shown that LT-HSCs reside in hypoxic niches, use anaerobic metabolism, and display higher expression of hypoxia-response genes than other hematopoietic cells.12 The hypoxic status of LT-HSCs is desirable because it prevents ROS production that could occur from oxidative phosphorylation.31 Loss of hypoxia response signaling (ie, deletion of Hif-1α) leads to loss of quiescence and exhaustion of LT-HSCs, which is associated with accumulation of ROS.11 Increased ROS levels are known to induce proliferation and exhaustion of HSCs.19 In the present study, we observed an increase in ROS levels in Meis1-deleted LT-HSCs; we also detected a decrease in Hif-1α mRNA and protein levels with Meis1 deletion and a decrease in expression of Hif-1α targets. Both of these mechanisms likely contributed to the loss of HSC maintenance because reversal of each separately rescued the loss of colony-forming ability by Meis1-deleted cells. Although it is likely that the accumulation of ROS in HSCs that we observed was secondary to decreased Hif-1α and the subsequent loss of hypoxia signaling, it is also possible that Meis1 deletion disrupts other mechanisms of limiting ROS independently of Hif-1α. Further, VHL knockdown stabilizes both Hif-1α and Hif-2α and, although the 2 share some downstream targets, there are distinctions in their functions.32 Mice lacking Hif-2α display pancytopenia and, although this effect is likely because of decreased production of exogenous erythropoietin, it is possible that Hif-2α can also preserve self-renewal, much like Hif-1α.33 Therefore, it is possible that the reversal of the effects of Meis1 deletion is mediated via Hif-2α alone or in combination with Hif-1α. Finally, the regulation of Hif-1α by Meis1 may be multifactorial. Although we observed a modest decrease in Hif-1α mRNA with Meis1 deletion, it is also possible that Meis1 exerts posttranscriptional and posttranslational regulatory controls that result in stabilization of Hif-1α. Overall, these results highlight some of the cellular and molecular functions of Meis1 in the maintenance of hematopoiesis, namely regulating hypoxia signaling and limiting oxidative stress, which in turn prevents proliferative exhaustion of LT-HSCs. These roles of Meis1 may also be operative in embryonic development. The defects in angiogenesis observed in Meis1−/− embryos mice are reminiscent of those seen in constitutive-Hif-1α−/− embryos.34,35 We conclude that it is possible that Meis1 is required for normal regulation of Hif-1α during development of the hematopoietic and vascular systems. This has implications not only in embryonic development, but also in the development of malignancies in which Meis1 is highly expressed, such as leukemias and certain tumors of the CNS.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr R. Keith Humphries (British Columbia Cancer Agency) for assistance with PCR detection of Meis1 deletion; Dr James P. Bridges (Cincinnati Children's Hospital Medical Center) for providing the anti–Hif-1α sera; and Drs James Mulloy and Jose Cancelas (Cincinnati Children's Hospital Medical Center) and Dr David Plas (University of Cincinnati) for guidance with the experiments and review of the manuscript.
This research was supported by grants from the National Institutes of Health (K08-CA122191), Cincinnati Children's Hospital Medical Center (trustee award), and Cancer Free Kids. The research flow cytometry core, comprehensive mouse and cancer core, and gene-expression microarray core facilities at Cincinnati Children's Hospital Medical Center provided valuable support.
National Institutes of Health
Authorship
Contribution: Z.U., J.P.C., J.R., and E.T. performed the experiments; L.T., N.G.C., and N.A.J. generated the Meis1-flox mice; H.L.G. helped with the experimental design; and A.R.K. supervised the study, analyzed the microarray data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ashish Kumar, Cincinnati Children's Hospital Medical Center, 3333 Burnet Ave, Cincinnati, OH 45236; e-mail: ashish.kumar@cchmc.org.