Abstract
The role of Meis1 in leukemia is well established, but its role in hematopoietic stem cells (HSCs) remains poorly understood. Previously, we showed that HSCs use glycolytic metabolism to meet their energy demands. However, the mechanism of regulation of HSC metabolism, and the importance of maintaining this distinct metabolic phenotype on HSC function has not been determined. More importantly, the primary function of Meis1 in HSCs remains unknown. Here, we examined the effect of loss of Meis1 on HSC function and metabolism. Inducible Meis1 deletion in adult mouse HSCs resulted in loss of HSC quiescence, and failure of bone marrow repopulation after transplantation. While we previously showed that Meis1 regulates Hif-1α transcription in vitro, we demonstrate here that loss of Meis1 results in down-regulation of both Hif-1α and Hif-2α in HSCs. This resulted in a shift to mitochondrial metabolism, increased reactive oxygen species production, and apoptosis of HSCs. Finally, we demonstrate that the effect of Meis1 knockout on HSCs is entirely mediated through reactive oxygen species where treatment of the Meis1 knockout mice with the scavenger N-acetylcystein restored HSC quiescence and rescued HSC function. These results uncover an important transcriptional network that regulates metabolism, oxidant defense, and maintenance of HSCs.
Introduction
Hematopoietic stem cells (HSCs) are defined by their abilities to self-renew and to differentiate into all blood cell types.1,2 Much of the advancement in HSC therapy is credited to decades of pioneering work that led to the development of HSC enrichment techniques based on staining of cell-surface antigens or vital dyes followed by fluorescence-activated cell sorting (FACS).3-5 However, little is known about metabolic characteristics of HSCs, its regulation, or how the metabolic phenotype may influence HSC function.
In 1978, the concept of the special microenvironment, or niche, of HSCs was introduced.6 Since then, it has become clear that the niche plays a crucial role in self-renewal and differentiation of HSCs.7,8 One of the hallmarks of the HSC niche is its low oxygen tension, hence the term “hypoxic niche.”9 Numerous studies indicate that this low oxygen environment is not only tolerated by HSCs, but is also essential for their function.10 We recently demonstrated that HSCs rely on glycolysis and have lower rates of oxygen consumption,11 which may be crucial for survival of HSCs within hypoxic bone marrow niches.
In the mitochondria, oxygen is used as the terminal electron acceptor for the respiratory chain, and in the absence of oxygen the proton gradient generated by the respiratory chain collapses and mitochondrial ATP production declines. Under these hypoxic or anoxic conditions, energy production is derived from cytoplasmic glycolysis through the fermentation of glucose, and in the final step of anaerobic glycolysis, pyruvate is converted to lactate to replenish NAD+. Anaerobic glycolysis produces 18 times less ATP than mitochondrial oxidative phosphorylation,12 which may be well suited for quiescent cells, but certainly cannot sustain cells with high-energy demands.
The energy advantage of mitochondrial oxidative phosphorylation over glycolysis is, unfortunately, not without deleterious consequences, as the mitochondrion is considered a major source of reactive oxygen species (ROS) production.13,14 ROS are believed to be important mediators of aging, and of numerous degenerative diseases, including HSC dysfunction and senescence.15 In fact, within the HSC compartment, the repopulation capacity is localized to only those HSCs with low levels of free radicals.16 Therefore, the glycolytic metabolic phenotype of HSCs may not only protect them against hypoxic insults, but may also serve to minimize oxidant damage that result from mitochondrial oxidative phosphorylation.
Hypoxia-inducible factor-1α (Hif-1α) is a major transcriptional regulator of hypoxic response. Hif-1α mediates the metabolic switch from aerobic mitochondrial metabolism, to anaerobic cytoplasmic glycolysis17-19 by increasing both the expression,20 and kinetic rate21 of key glycolysis enzymes. Moreover, Hif-1α inhibits the use of pyruvate by the mitochondria,22,23 and inhibits mitochondrial biogenesis.24 Takubo and colleagues recently demonstrated that Hif-1α is enriched in HSCs, and that loss of Hif-1α results in HSC dysfunction,25 while our group recently showed that Meis1 is required for optimum transcriptional activation of Hif-1α in HSCs ex vivo.11
Meis1, which is a 3-amino-acid loop extension homeodomain protein, plays an important role in leukemogenesis as well as normal hematopoiesis. Meis1 was first identified as a common viral integration site in myeloid leukemic cells of BXH-2 mice,26 and it is also frequently up-regulated in human primary acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL) samples.27 Moreover, overexpression of Meis1 accelerates the initiation of AML in murine models.28,29 In normal hematopoiesis, Meis1 is expressed in the most primitive hematopoietic populations and is down-regulated on differentiation.30-32 Targeted Meis1 knockout causes lethality by embryonic day 14.5 with multiple hematopoietic and vascular defects.33,34 Moreover, Pbx-1, a cofactor of Meis1, has been shown to regulate self-renewal of HSCs by maintaining their quiescence.35 However, the role of Meis1 regulating the function and metabolism if HSCs remain poorly understood.
In the current report, we show that Meis1 regulates both HSC metabolism and oxidant stress response, through transcriptional regulation of Hif-1α and Hif-2α, respectively.
Methods
Mouse breeding and genotyping
Meis1 knockout (KO) mice were genotyped with Meis1 For1: 5′-CCAAAGTAGCCACCAATATCATGA-3′ and Meis1 Rev: 5′-AGCGTCACTTGGAAAAGCAATGAT-3′ primers. Wild-type (WT) allele is determined by a 332-bp-long PCR product and mutant allele determined by a 440-bp long PCR product on 1.2% agarose gel. HSC-specific deletion of Meis1 was achieved by following crosses of Meis1f/f with Scl-Cre-ERT mice.4 Scl-Cre mice were genotyped using Scl-Cre-ER primer 1: 5′-GAACCTGAAGATGTTCGCGAT-3 and Scl-Cre-ER primer 2: 5′-ACCGTCAGTACGTGAGATATC-3. To generate Meis1−/− mice, Meis1f/f;Scl-Cre-ERT+ mice were injected intraperitoneally with tamoxifen (40 mg/kg, T5648-1G; Sigma-Aldrich) daily for 14 days. We used age-matched tamoxifen-injected Meis1+/+;Scl-Cre-ERT(+) or Meis1f/f;Scl-Cre-ERT(−) mice as controls (Meis1+/+ mice). Genotyping of Cre-deleted Meis1 locus (Meis1 exon8 deleted) was performed using Meis1 For2: 5′-CATTGACTTAGGTGTATGGGTGTC-3′ and Meis1 Rev: 5′-AGCGTCACTTGGAAAAGCAATGAT-3′ primers. Cre-deleted Meis1 locus gives rise to a 261-bp product while wt and mutant (nondeleted) shows no amplicon.
Dr Joseph A. Garcia (UT Southwestern) kindly provided the Hif-1α floxed mice. HSC-specific deletion of Hif-1α was achieved by following crosses of Hif-1αf/f with Scl-Cre-ERT mice and tamoxifen injections (40 mg/kg for 14 days). We used age-matched tamoxifen-injected Hif-1α+/+;Scl-Cre-ERT(+) or Hif-1αf/f;Scl-Cre-ERT(−) mice as controls (Hif-1α+/+ mice).
Flow cytometry
Donor bone marrow (BM) cells were isolated from 8- to 12-week-old Meis1+/+ or Meis1−/− mice (or Hif-1α+/+ and Hif-1α−/− mice). Lin−Sca1+Kit+Flk2−CD34− cells (long-term HSCs [LT-HSCs]) were isolated by staining with a biotinylated lineage cocktail (anti-CD3, anti-CD5, anti-B220, anti–Mac-1, anti-Gr-1, anti-Ter119; StemCell Technologies) followed by streptavidin-PE/Cy5.5, anti–Sca-1–FITC, anti–Kit-allophycocyanin (APC), anti–Flk-2–PE, and anti-CD34–PE. For analyzing repopulation of mouse HSCs, peripheral blood cells of recipient CD45.1 mice were collected by retro-orbital bleeding, followed by lysis of red blood cells and staining with anti-CD45.2–FITC, anti-CD45.1–PE, anti-Thy1.2–PE (for T-lymphoid lineage), anti-B220–PE (for B-lymphoid lineage), anti–Mac-1–PE, or anti–Gr-1–PE (cells costaining with anti-Mac–1 and anti–Gr-1 were deemed to be of the myeloid lineage) monoclonal antibodies (BD Pharmingen). The “percent repopulation” shown in all figures was based on the staining results of anti-CD45.2–FITC and anti-CD45.1–PE. In all cases, FACS analysis of the above-listed lineages was also performed to confirm multilineage reconstitution as previously described.11
For analyzing LT-HSCs in the peripheral blood by flow cytometry, peripheral blood of Meis1−/− or Meis1+/+ mice was collected by retro-orbital bleeding and stained with LT-HSCs markers as described in the previous paragraph.
The cell-cycle analysis with Hoechst 33342 and pyronin Y staining was performed as we described.36 Samples were immediately analyzed by flow cytometry (FACSAria; BD Biosciences). To examine the apoptosis, Lin−Kit+Sca-1+ cells were stained with PE-conjugated anti–annexin V and 7AAD according to the manufacturer's manual (BD Pharmingen). To study the apoptosis in LT-HSCs, bone marrow cells were stained for HSC markers Sca-1-PE/Cy5.5, C-Kit-APC, CD34-PE, and Flk2-PE after lineage depleting and stained with FITC-conjugated anti–annexin V according to the manufacturer's manual (eBioscience).
Competitive reconstitution analysis
The indicated numbers of CD45.2 donor cells from Meis1+/+ or Meis1−/− mice were mixed with 1 × 105 freshly isolated CD45.1 competitor BM cells and the mixture was injected intravenously via the retro-orbital route into each of a group of 6- to 8-week-old CD45.1 mice previously irradiated with a total dose of 10 Gy. To measure reconstitution of transplanted mice, peripheral blood was collected at the indicated time points posttransplantation, and the presence of CD45.1+ and CD45.2+ cells in lymphoid and myeloid compartments was measured as described.37
Homing
BM Lin− cells were labeled with 5- (and -6) carboxyfluorescein succinimidyl ester (CFSE), and 3 × 106 cells were transplanted into indicated strains of lethally irradiated mice. After 16 hours, the total number of CFSE+ cells in the BM, spleen, or liver was determined by flow cytometry. When CFSE+ LSK (CFSE+Lin−Sca1+Kit+) cells (HSCs) were analyzed, the BM cells were stained with a biotinylated lineage cocktail followed by streptavidin-PE/Cy5.5, anti–Sca-1–PE, and anti-Kit–APC before analysis, as described.36
Colony-forming assays
Normal BM cells were diluted to the indicated concentration in IMDM with 2% FBS, and were then seeded into methylcellulose medium M3434 (StemCell Technologies), for CFU-GM and BFU-E colony formation according to the manufacturer's instructions.
Real-time PCR
Total RNA was isolated from Meis1−/− and Meis1+/+ HSCs using the RNeasy Mini Kit (QIAGEN) according to the manufacturer's instructions. cDNA was synthesized using SuperScript II RT (Invitrogen). Predesigned primers (Table 1) from the National Institutes of Health (NIH) mouse primer depot (http://mouseprimerdepot.nci.nih.gov/) were ordered from Integrated DNA Technologies. Real-time PCR was performed with SyberGreen (Applied Biosystems) on an ABI Prism 7700 Sequence Detector (Applied Biosystems). β-actin was used as control to normalize results.
List of primers used for real-time PCR
| Gene . | Forward primer . | Reverse primer . |
|---|---|---|
| β-actin | GAACCCTAAGGCCAACCGTGAAAGAT | ACCGCTCGTTGCCAATAGTGATG |
| p1638 | GGGTTTCGCCCAACGCCCCGA | TGCAGCACCACCAGCGTGTCC |
| p1938 | GTTTTCTTGGTGAAGTTCGTGC | TCATCACCTGGTCCAGGATTC |
| Meis1 | GTTGTCCAAGCCATCACCTT | ATCCACTCGTTCAGGAGGAA |
| Hif-1α | CGGCGAGAACGAGAAGAA | AAACTTCAGACTCTTTGCTTCG |
| Hif-2α | ATCACGGGATTTCTCCTTCC | GGTTAAGGAACCCAGGTGCT |
| Hif-3α | TGTGAACTTCATGTCCAGGC | GCAATGCCTGGTGCTTATCT |
| Gene . | Forward primer . | Reverse primer . |
|---|---|---|
| β-actin | GAACCCTAAGGCCAACCGTGAAAGAT | ACCGCTCGTTGCCAATAGTGATG |
| p1638 | GGGTTTCGCCCAACGCCCCGA | TGCAGCACCACCAGCGTGTCC |
| p1938 | GTTTTCTTGGTGAAGTTCGTGC | TCATCACCTGGTCCAGGATTC |
| Meis1 | GTTGTCCAAGCCATCACCTT | ATCCACTCGTTCAGGAGGAA |
| Hif-1α | CGGCGAGAACGAGAAGAA | AAACTTCAGACTCTTTGCTTCG |
| Hif-2α | ATCACGGGATTTCTCCTTCC | GGTTAAGGAACCCAGGTGCT |
| Hif-3α | TGTGAACTTCATGTCCAGGC | GCAATGCCTGGTGCTTATCT |
PCR array
The Mouse Hypoxia Signaling Pathway RT2 Profiler PCR Array was performed as described previously.11 RNA was extracted from Meis1−/− and Meis1+/+ LT-HSCs (Lin−Sca1+Kit+Flk2−CD34−cells) using the RNeasy Mini Kit (QIAGEN) according to manufacturer's instructions. cDNA was retrotranscribed by using SuperScript II RT (Invitrogen). Mouse Real-Time Syber Green PCR Master Mix (SuperArray) and Cell Cycle primer sets (SABiosciences) used for real-time PCR on an ABI Prism 7700 Sequence Detector (Applied Biosystems). The data were analyzed using the ΔΔCt method. Fold change was calculated as difference in gene expression between Meis1−/− and Meis1+/+ LT-HSC samples.
Generation of luciferase reporter vectors
Conserved Meis1 motifs in Hif-2α gene were determined using a genome browser (http://genome.ucsc.edu/). A 779-bp-long DNA fragment containing conserved Meis1 sites (located next to start codon sequence, human chr2:46 525 052-46 525 064) from the Hif-2α promoter was amplified by PCR from mouse genomic DNA with the following primers: conserved Meis1 site: pHif-2α-F, 5′-GGGCTAAACGGAACTCCAGG-3′ and pHif-2α-R, 5′-CATAGGAACGCTCTCGGAAAGAC-3′. PCR fragments were subcloned into the pCR2.1-TOPO vector (Invitrogen) according to the manufacturer's instructions. pHif2a-TOPO and E1b-pGL2 vectors were digested with XhoI and KpnI. Then, the PCR fragments containing conserved Meis1 sites were cloned into E1b-pGL2 to generate the pHif2a-pGL2 luciferase reporter vector. To test Meis1 site specificity, Meis1-binding sites (TGAC) at the TGACAGCTGACAA (Meis1 binding site is indicated in bold) sequence were mutated to TGGCGGCCGCCAA (NotI site insertion) using iProof High-Fidelity DNA Polymerase (Bio-Rad) from the Hif-2α-pGL2 vector with the following primers: Mut-Hif2a-F, 5′-AAgcggccgcCAAGGAGAAAAAAAGGTAAGCGGG-3′ and Mut-Hif2a-R, 5′-AAgcggccgcCATTGTCGCCGTGGCCCTC-3′. The Hif-2α-Mut-pGL2 reporter was generated after NotI digestion and ligation of the PCR product. Transcriptional activation of Hif-2α by Meis1 was evaluated using a luciferase reporter system (Promega) as described previously.11
Metabolic assays
Metabolic assays are carried out as described previously11 with some modifications.
Oxygen consumption assays
Meis1+/+ and Meis1−/− HSCs were separated flowcytometrically as described in “Flow cytometry.” Equal numbers of cells (5-10 × 104 cells/well) were incubated for 6 hours in the provided 384-well plate (BD Oxygen Biosensor System) and sealed to prevent air exchange before measurement. Culture media lacking cells was used as a negative control and sodium sulfite (100mM) was used as a positive control. Oxygen consumption is presented as relative units.
ATP assays
Meis1+/+ and Meis1−/− HSCs were sorted as described and centrifuged at 1200g for 10 minutes. At least 50 000 cells were used for each single ATP measurement. Fifty microliters of ATP standards (10−6-10−12M) and 50 μL of cell lysates were quantified using the ATP Bioluminescence Assay Kit CLS II (Roche) using Fluostar Optima plate reader (BMG Labtech). Finally, data were normalized to cell count and protein content.
Glycolytic flux assay
13C-lactate production, end product of glycolysis, was measured as described previously11 using glycolytic flux medium supplemented with 10mM D-[1-6-13C]-glucose (Cambridge Isotope Labs) to allow up to all of the glucose-derived lactate pool to be labeled on C-3. A minimum of 50 000 cells were cultured in 40 μL of flux medium overnight. Then, the cells were pelleted and supernatant collected and prepared for gas chromatography–mass spectrometry. Lactate abundance was determined by monitoring m/z at 117 (unenriched), 118 (lactate containing 13C from glucose), and 119 (internal standard) as described previously.11
Measurement of ROS
Bone marrow cells from Meis1+/+ and Meis1−/− mice were isolated as described in “Flow cytometry.” After lineage depletion (lineage depletion kit; BD Biosciences), cells were incubated with 1μM 5-(and-6)-carboxy-2′,7′-dichlorofluorescein diacetate (carboxy-DCFDA; Invitrogen) for 30 minutes in a 37°C water bath in the dark. Then, cells were stained for HSC markers Sca-1-PE/Cy5.5, C-Kit-APC, CD34-PE, and Flk2-PE and assayed with a flow cytometer.
NAC administration in vivo
Six-week-old Meis1f/f;Scl-Cre-ERT+ mice were injected intraperitoneally with tamoxifen (40 mg/kg) daily for 14 days. After tamoxifen injections, animals were treated daily for 2 weeks with N-acetyl-L-cysteine (NAC; 100 mg/kg; Sigma-Aldrich) by subcutaneous administration or provided in drinking water (500 mg × kg−1 × d−1 in drinking water as described previously39,40 ) and were subsequently analyzed by flow cytometry. We used age-matched tamoxifen-injected Meis1+/+;Scl-Cre-ERT(+) or Meis1f/f;Scl-Cre-ERT(−) mice as controls (Meis1+/+ mice).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assay (ChIP) assays were performed to evaluate the in vivo binding of Meis1 to its consensus sequence in the Hif-2α gene. The assays were done in Kasumi-1 cells, a hematopoietic progenitor cell line, using the ChIP kit (Upstate) as described previously.11 Meis1 antibody (Santa Cruz Biotechnology) and normal goat IgG (Santa Cruz Biotechnology) were used. The DNA isolated from input chromatin fragments and from the precipitated chromatin fragments by anti-Meis1 antibody or control IgG was subjected to real-time PCR using primers flanking the consensus Meis1-binding sites on Hif-2α promoter: 5′-GTTCCTCCCAGTCACCTTTCTCC-3′ and 5′-TCTCCCAACCACTCTCGGTC-3′.
Measurement of blood cell counts (CBC/differential)
Approximately 50 μL of peripheral blood collected by retro-orbital bleeding from Meis1+/+ and Meis1−/− mice post 1 month and 3 months of tamoxifen injections in K3-EDTA tubes. Samples are submitted to University of Texas Southwestern Medical Center Animal Resources Center (UTSW ARC) Diagnostic Laboratory and analyzed with the HemaVet 950FS analyzer. The following parameters are reported for the sample submitted: white blood count (WBC), neutrophil count, lymphocyte count, monocyte count, eosinophil count, basophil count, red blood cell count, hemoglobin, HCT, MCV, MCH, MCHC, platelet count, and MPV.
Generation of the Meisf/f;Scl-Cre+; rtTA; TRE-Hif-1α mice and rescue analysis by viral Hif-1α and Hif-2α overexpression
To determine the role of Hif-1α in Meis1 KO HSCs, inducible HSC-specific, Hif-1α transgenic mice were generated. The transgenic construct was made by cloning Hif-1α cDNA (Open Biosystems) into the pTRE-Tight vector (Clontech), which makes it responsive to rtTA regulatory proteins in the Tet-On system. We crossed TRE-Hif-1α mice with rtTA mice which has a stop-codon flanked by loxP sequence. Cre-mediated removal of the stop region by deletion of the loxP flanked sequence allows expression of rtTA as described.41 Therefore, sequential administration of tamoxifen will result in Meis1 deletion, and concomitant deletion of the stop-codon, which allows for Hif-1α overexpression on administration of doxycycline. We generated Meisf/f;Scl-Cre+; rtTA; TRE-Hif-1α mice by a series of crosses to overexpress in Hif-1α in HSC-specific manner after 14 days of tamoxifen (1 mg/d/mice) and providing doxycycline in drinking water (500 mg/L). HSCs from these mice were either used for in vivo repopulation studies, or for in vitro colony forming assay.
We also used a viral strategy to overexpress Hif-1α or Hif-2α in HSCs (generated by cloning into XZ201 vector) and lentivirus-expressing Hif-2α (kindly provided by Dr Joseph Garcia, UT Southwestern). Fetal liver cells were harvested from Meis1f/f;Scl-Cre+ pups at P1 and infected with either lentivirus-expressing Hif-2α or retrovirus-expressing Hif-1α with IRES-GFP. Then, infected cells were transplanted into CD45.1+ recipient host mice. The repopulation was measured 2 months posttransplantation before induction of deletion of Meis1 with tamoxifen injections. The repopulation analysis performed at indicated time points post-tamoxifen treatments either measuring GFP+ cells or CD45.2+ donor cells (n = 3-5).
Finally, in a separate set of experiments, we used cobalt chloride (CoCl2 100μM), which is a known stabilizer of both Hif-1α and Hif-2α, in methocult culture after deletion of Meis1 to examine the effects of stabilization of Hif-1α and Hif-2α on colony formation assay after Meis1 deletion. After 14 days of tamoxifen injection into aged matched Meis1+/+;Scl-Cre+ (Control) and Meis1f/f;Scl-Cre+ (Meis1 KO) mice, we performed colony-forming assays using Methocult medium supplemented with 100μM CoCl2. Same number of bone marrow cells (20 × 103 cells/plate) was plated as recommended by manufacturer and colonies were quantified after 12 days of culture.
Statistical analysis
Results are expressed as mean ± SEM, and a 2-tailed Student t test was used to determine the level of significance. P < .05 was considered statistically different.
Results
Deletion of Meis1 in LT-HSCs
Because the global loss of Meis1 is embryonic lethal,33,34 we sought to pursue an inducible deletion of Meis1 to study the role of Meis1 in adult HSCs. Meis1fl/fl mice with loxp flanking exon 8 were crossed with transgenic mice expressing the tamoxifen-inducible Cre recombinase under the control of stem cell leukemia (Scl) HSC enhancer, which drives deletion in HSCs.4 Upon tamoxifen treatment, exon 8 is deleted which results in the loss of Meis1 expression in HSCs (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). To verify deletion of Meis1, we performed genotyping analysis and quantitative RT-PCR in peripheral blood cells and phenotypic LT-HSCs (Lin−Sca1+Kit+Flk2−CD34−) in the bone marrow after tamoxifen treatment. Meis1 in peripheral blood cells was deleted 14 days after tamoxifen treatment, and Meis1 mRNA level in LT-HSCs was markedly decreased to approximately 10% of control values (supplemental Figure 1B).
Meis1 is required for the maintenance of LT-HSCs
To explore the role of Meis1 in LT-HSCs, we first examined the frequencies of phenotypic LT-HSCs in control (Meis1+/+) and Meis1 conditional KO (Meis1−/−) mice. As shown in Figure 1A, 7 days after tamoxifen treatment, there was a 4.5-fold increase of HSC frequency in Meis1−/− mice compared with Meis1+/+ controls (0.0092% vs 0.002%). FACS analysis revealed robust reduction of granulocyte-monocyte progenitors (GMPs) and common myeloid progenitors (CMPs) and mild reduction in the total number of megakaryocyte/erythroid progenitors (MEPs; supplemental Figure 1C). Meanwhile, by using a colony-forming assay, we demonstrated that Meis1−/− mice had much lower primitive myeloid progenitor cells (CFU-GEMM), but no change in percentage of differentiated myeloid progenitor cells (CFU-GM), or erythroid progenitor cells (BFU-E; Figure 1B, supplemental Figure 1D). No difference was detected in the distribution of T, B, myeloid, and erythroid lineages either in BM or peripheral blood of Meis1−/− (supplemental Figure 1E-F).
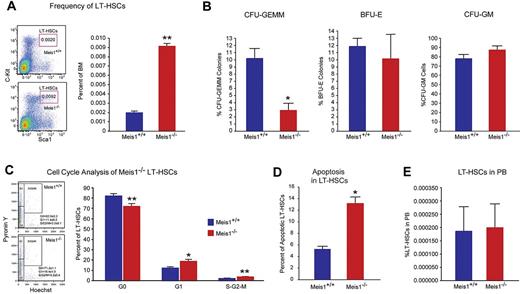
Meis1 Deletion in LT-HSCs results in apoptosis and loss of quiescence. (A) Left panel: Representative flow cytometry profile of LT-HSCs (Lin−Sca-1+Kit+Flk2−CD34−) of bone marrow (BM) cells are shown for control Meis1+/+ and mutant Meis1−/− mice. Numbers in the FACS plots indicate percentages among total BM cells. Right panel: Quantification of LT-HSCs demonstrates significantly higher number of HSCs in Meis1−/− BM (n = 6). (B) The in vitro methylcellulose colony formation assay was performed at the time of sacrifice after tamoxifen injections with BM cells of control and Meis1−/− mice. CFU-GEMM colonies representing most undifferentiated progenitors type of colonies derived from Meis1+/+ and Meis1−/− BM cells demonstrates decreased percentage of CFU-GEMM. Quantification of BFU-E and CFU-GM colonies derived from Meis1−/− cells shows no differences (n = 3). (C) Left panel: Representative FACS analysis of Pyronin Y/Hoecst staining on LT-HSCs (Lin−Sca-1+Kit+CD150+CD48−) of Meis1+/+ and Mesi1−/− mice. Numbers in the FACS plots indicate percentages among LT-HSCs. Right panel, The quantification of G0, G1, or S/G2/M phase in Meis1+/+ and Meis1−/− LT-HSCs (n = 6). (D) Quantification of apoptosis in Meis1+/+ and Meis1−/− LT-HSCs (n = 3). (E) Quantification of LT-HSCs in peripheral blood (PB) of Meis1+/+ and Meis1−/− mice (n = 6); *P < .05, **P < .01.
Meis1 Deletion in LT-HSCs results in apoptosis and loss of quiescence. (A) Left panel: Representative flow cytometry profile of LT-HSCs (Lin−Sca-1+Kit+Flk2−CD34−) of bone marrow (BM) cells are shown for control Meis1+/+ and mutant Meis1−/− mice. Numbers in the FACS plots indicate percentages among total BM cells. Right panel: Quantification of LT-HSCs demonstrates significantly higher number of HSCs in Meis1−/− BM (n = 6). (B) The in vitro methylcellulose colony formation assay was performed at the time of sacrifice after tamoxifen injections with BM cells of control and Meis1−/− mice. CFU-GEMM colonies representing most undifferentiated progenitors type of colonies derived from Meis1+/+ and Meis1−/− BM cells demonstrates decreased percentage of CFU-GEMM. Quantification of BFU-E and CFU-GM colonies derived from Meis1−/− cells shows no differences (n = 3). (C) Left panel: Representative FACS analysis of Pyronin Y/Hoecst staining on LT-HSCs (Lin−Sca-1+Kit+CD150+CD48−) of Meis1+/+ and Mesi1−/− mice. Numbers in the FACS plots indicate percentages among LT-HSCs. Right panel, The quantification of G0, G1, or S/G2/M phase in Meis1+/+ and Meis1−/− LT-HSCs (n = 6). (D) Quantification of apoptosis in Meis1+/+ and Meis1−/− LT-HSCs (n = 3). (E) Quantification of LT-HSCs in peripheral blood (PB) of Meis1+/+ and Meis1−/− mice (n = 6); *P < .05, **P < .01.
The increase of HSC frequency may result from cell-autonomously accelerated proliferation or the compensatory effect of increased apoptosis or mobilization in Meis1−/− HSCs. To address this concern, we first examined the cell cycle of HSCs by using Hoechest 33342 and pyronin Y staining, and we found that only approximately 71% of Meis1−/− LT-HSCs were in the G0 compartment, which is much lower than that of Meis1+/+LT-HSCs (∼ 83%; Figure 1C). This indicated that Meis1−/− HSCs were much less quiescent and prone to proliferation. Next, we examined apoptosis status in LT-HSCs (Lin−Sca1+Kit+Flk2−CD34−) by using annexin V. We detected more apoptotic cells in Meis1−/− LT-HSCs compared with Meis1+/+ counterpart (Figure 1D, supplemental Figure 1G for LSK cells). Finally, to examine HSC mobilization, we performed LT-HSC staining in peripheral blood in Meis1+/+ and Meis1−/− mice, which did not show any difference in HSC frequency (Figure 1E). These data suggest that the increased apoptosis in Meis1−/− LT-HSCs may result in increase of HSC frequency and decrease of quiescence in BM.
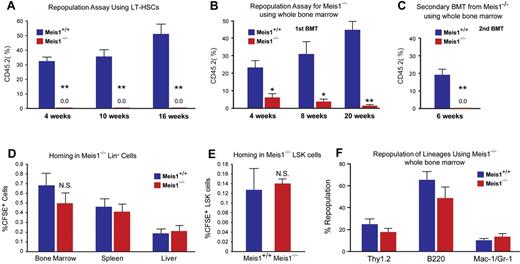
To further evaluate the function of Meis1 in HSCs in vivo, we performed competitive bone marrow transplantation with Meis1+/+ and Meis1−/− HSCs. Two weeks after tamoxifen treatment, 150 Lin−Sca1+Kit+Flk2−CD34− Meis1+/+ or Meis1−/− CD45.2 HSCs, together with 1 × 105 CD45.1 competitors, were administered into CD45.1 recipients through retro-orbital injection. Repopulation was examined at 4, 10, 16 weeks after transplantation. Strikingly, as shown in Figure 2A, we could not detect any engraftment with Meis1−/− HSC donors, indicating a severe impairment of HSC repopulation ability after loss of Meis1. A repeat transplantation experiment with total BM cells showed similar results (Figure 2B). The impaired repopulation of Meis1-deficient LT-HSCs (Figure 2A) and progressive decline in repopulation of Meis1-deficient cells in primary transplant recipients (Figure 2B) suggested a defect in self-renewal. Therefore, we performed secondary transplantation experiments to confirm the reduction of function Meis1-deficient LT-HSCs in the bone marrow of primary transplant recipients. Eight weeks after primary transplantation, total bone marrow from primary recipients from whole transplanted mice were harvested and transplanted into secondary recipients. Meis1-deficient cells from primary recipients (0.5-1 × 106 cells) were unable to repopulate secondary recipients (Figure 2C). The decreased repopulation may result from defects in HSC maintenance, or homing, or increased apoptosis. To exclude the possibility that the decreased engraftment is caused by defect of homing in Meis1−/− HSCs, we labeled Meis1+/+ or Meis1−/− Lin− cells with CFSE and injected into lethally irradiated recipients. Sixteen hours later, we examined CFSE+ or LSK CFSE+ cells in spleen, liver, and BM. No significant difference in homed total CFSE+ or LSK CFSE+ cells was detected between Meis1+/+and Meis1−/− donors (Figure 2D-E). In addition, quantification of lineage repopulation of Meis1+/+ and Meis1−/− donors demonstrates no lineage-specific defects (Figure 2F). Taken together, these data provide strong evidence that Meis1 plays a crucial role in the maintenance of HSCs and prevention of apoptosis in HSCs.
Impaired repopulation in Meis1−/− LT-HSCs. Repopulation assays: (A) LT-HSCs (Lin−Sca1+Kit+Flk2−CD34−; 150 cells) from either control (Meis1+/+) or mutant Meis1−/− CD45.2 mice were transplanted into irradiated CD45.1 hosts in competition with BM from CD45.1 mice (1 × 105 cells). Quantification of flow cytometry profile of peripheral blood of bone marrow recipient mice up to 16 weeks for percentage of CD45.2+ cells demonstrates total loss of bone marrow reconstitution of Meis1−/− LT-HSCs (n = 5). (B) Repopulation assay with whole bone marrow from either control Meis1+/+ or mutant Meis1−/− CD45.2 mice were transplanted into irradiated CD45.1 mice demonstrates significantly impaired repopulation in mice transplanted with Meis1−/− cells (n = 5). (C) Analysis of repopulation after second bone marrow transplantation (BMT) from first BMT mice demonstrates complete loss of repopulation (n = 5). Homing assays: (D) BM Lin− cells (3 × 106 cells) were transplanted into irradiated mice and quantified for CFSE+ cells in different tissues. Quantification of percentage of CFSE+ cells in BM, spleen, and liver show no difference between Meis1+/+ and Meis1−/− mice (n = 5). (E) Quantification of percentage of CFSE+ LSK cells in BM of Meis1+/+ and Meis1−/− mice (n = 5). (F) Quantification of repopulation of lineages for Meis1−/− using whole bone marrow from first BMT mice demonstrates no defects in lineage repopulation (n = 5); *P < .05, **P < .01.
Impaired repopulation in Meis1−/− LT-HSCs. Repopulation assays: (A) LT-HSCs (Lin−Sca1+Kit+Flk2−CD34−; 150 cells) from either control (Meis1+/+) or mutant Meis1−/− CD45.2 mice were transplanted into irradiated CD45.1 hosts in competition with BM from CD45.1 mice (1 × 105 cells). Quantification of flow cytometry profile of peripheral blood of bone marrow recipient mice up to 16 weeks for percentage of CD45.2+ cells demonstrates total loss of bone marrow reconstitution of Meis1−/− LT-HSCs (n = 5). (B) Repopulation assay with whole bone marrow from either control Meis1+/+ or mutant Meis1−/− CD45.2 mice were transplanted into irradiated CD45.1 mice demonstrates significantly impaired repopulation in mice transplanted with Meis1−/− cells (n = 5). (C) Analysis of repopulation after second bone marrow transplantation (BMT) from first BMT mice demonstrates complete loss of repopulation (n = 5). Homing assays: (D) BM Lin− cells (3 × 106 cells) were transplanted into irradiated mice and quantified for CFSE+ cells in different tissues. Quantification of percentage of CFSE+ cells in BM, spleen, and liver show no difference between Meis1+/+ and Meis1−/− mice (n = 5). (E) Quantification of percentage of CFSE+ LSK cells in BM of Meis1+/+ and Meis1−/− mice (n = 5). (F) Quantification of repopulation of lineages for Meis1−/− using whole bone marrow from first BMT mice demonstrates no defects in lineage repopulation (n = 5); *P < .05, **P < .01.
Role of Meis1 in regulating Hif-1α for glycolytic metabolism of LT-HSCs
We have shown previously that the Meis1 gene acts upstream of Hif-1α by regulating its transcriptional activity by an enhancer located in the first intron of Hif-1α.11 We further examined whether hypoxia-inducible factors (Hif-1α, Hif-2α, and Hif-3α) are regulated by Meis1 in LT-HSCs. Quantitative real-time PCR analysis demonstrated that mRNA levels of both Hif-1α and Hif-2α were down-regulated but no change in Hif-3α levels was observed in Meis1−/− LT-HSCs (Figure 3A). Moreover, we found that several downstream targets of Hif-1α and/or Hif-2α are down-regulated in Meis1−/− LT-HSCs as determined by quantitative PCR (qPCR) array (supplemental Figure 2). To explore the role of Hif-1α in the regulation of metabolism of LT-HSCs, we used the same inducible Cre system to delete Hif-1α specifically in HSCs (supplemental Figure 3A). Hif-1αfl/fl mice with loxp flanking exon 2 were crossed with transgenic mice expressing the tamoxifen-inducible Cre recombinase under the control of stem cell leukemia (Scl) HSC enhancer. Upon tamoxifen treatment, exon 2 is deleted resulting in the loss of Hif-1α expression in HSCs (supplemental Figure 3A). To verify deletion of Hif-1α, we performed quantitative RT-PCR in LT-HSCs (Lin−Sca1+Kit+ Flk2−CD34−) in bone marrow after tamoxifen treatment. Hif-1α mRNA level in LT-HSCs was dramatically decreased, approximately 400-fold lower than that of control (Hif-1+/+) after 14 days of tamoxifen treatment (supplemental Figure 3B). We showed previously that LT-HSCs primarily rely on cytoplasmic glycolysis rather than mitochondrial oxidative phosphorylation.11 However, the role of Hif-1α in HSC metabolism was not examined before. Here, we demonstrate that HSC-specific deletion of Hif-1α results in increased mitochondrial respiration as shown by increased oxygen consumption (supplemental Figure 3C), and decreased glycolytic flux measured by the rate of glucose-derived 13C lactate production (supplemental Figure 3D).
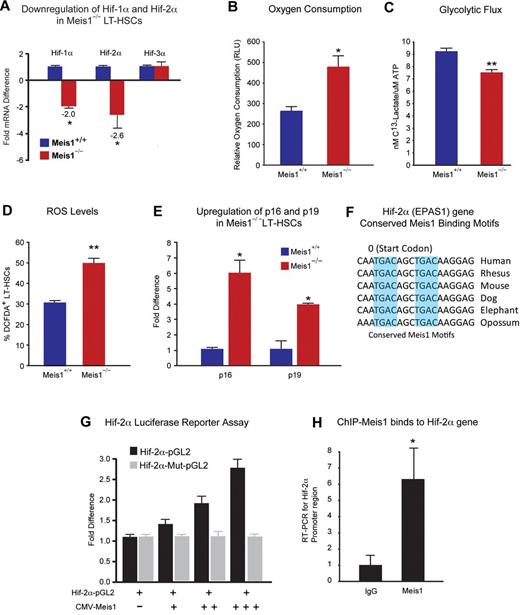
Metabolic regulation of LT-HSCs by Meis1. (A) RT-PCR histogram demonstrates the significant down-regulation of Hif-1α and Hif-2α (EPAS1), but not Hif-3α after Meis1 deletion in LT-HSCs (n = 3). (B) Measurement of oxygen consumption rate for 6 hours demonstrates significantly higher aerobic phosphorylation in Meis1−/− LT-HSCs compared with Meis1+/+ LT-HSCs (n = 3). (C) Quantification of labeled lactated in glycolytic flux assay demonstrates that Meis1−/− LT-HSCs are less glycolytic (n = 3). (D) Measurement of reactive oxygen species (ROS) in LT-HSCs as determined by quantification of the percentage of DCFDA+ LT-HSCs in Meis1+/+ and Meis1−/− mice (n = 3). (E) RT-PCR of p16 and p19 demonstrates association of higher level of ROS with up-regulation of p16 and p19 in Meis1−/− LT-HSCs (n = 3). (F) Figure shows conserved consensus Meis1 motifs found on Hif-2α (EPAS1) gene. Note the duplex Meis1-binding motifs found next to each other and conserved till Opossum. (G) Luciferase reporter assays demonstrate dose-dependent transcriptional activation of Hif-2α by Meis1 (n = 3). (H) Real-time PCR with primers flanking the consensus Meis1-binding sequence after ChIP assay demonstrating in vivo binding of Meis1 to Hif-2α promoter (n = 3); *P < .05, **P < .01.
Metabolic regulation of LT-HSCs by Meis1. (A) RT-PCR histogram demonstrates the significant down-regulation of Hif-1α and Hif-2α (EPAS1), but not Hif-3α after Meis1 deletion in LT-HSCs (n = 3). (B) Measurement of oxygen consumption rate for 6 hours demonstrates significantly higher aerobic phosphorylation in Meis1−/− LT-HSCs compared with Meis1+/+ LT-HSCs (n = 3). (C) Quantification of labeled lactated in glycolytic flux assay demonstrates that Meis1−/− LT-HSCs are less glycolytic (n = 3). (D) Measurement of reactive oxygen species (ROS) in LT-HSCs as determined by quantification of the percentage of DCFDA+ LT-HSCs in Meis1+/+ and Meis1−/− mice (n = 3). (E) RT-PCR of p16 and p19 demonstrates association of higher level of ROS with up-regulation of p16 and p19 in Meis1−/− LT-HSCs (n = 3). (F) Figure shows conserved consensus Meis1 motifs found on Hif-2α (EPAS1) gene. Note the duplex Meis1-binding motifs found next to each other and conserved till Opossum. (G) Luciferase reporter assays demonstrate dose-dependent transcriptional activation of Hif-2α by Meis1 (n = 3). (H) Real-time PCR with primers flanking the consensus Meis1-binding sequence after ChIP assay demonstrating in vivo binding of Meis1 to Hif-2α promoter (n = 3); *P < .05, **P < .01.
To further examine the role of Hif-1α in HSCs, we examined expression of Hif-2α and Hif-3α in Hif-1α−/− LT-HSCs. We found that deletion of Hif-1α in LT-HSCs results in a profound increase in Hif-2α mRNA levels (> 120-fold), with no significant change in the levels of Hif-3α mRNA (supplemental Figure 3E). This marked compensatory up-regulation of Hif-2α in Hif-1α−/− LT-HSCs is in stark contrast to the down-regulation of Hif-2α in the Meis1−/− LT-HSCs.
HSC-specific deletion of Hif-1α also demonstrated hematopoietic defects similar (supplemental Figure 4) to global deletion (Mx-1-Cre) in the bone marrow shown by Takubo and colleagues,25 including the increased frequency of HSCs (supplemental Figure 4A), decreased myeloid progenitors (supplemental Figure 4B), loss of quiescence (supplemental Figure 4C), and similar multilineage defects (supplemental Figure 4F-G).
Meis1 is a transcriptional activator of Hif-2α
Down-regulation of Hif-1α and Hif-2α in Meis1−/− LT-HSCs (Figure 3A) led us to examine their metabolic profile and ROS levels. We found that Meis1−/− LT-HSCs have increased rates of oxygen consumption (Figure 3B) and decreased glycolytic flux (Figure 3C). In addition, Meis1−/− LT-HSCs showed significantly higher ROS levels compared with control Meis1+/+ LT-HSCs (Figure 3D). This increased ROS was associated with increased expression of p16Ink4a and p19Arf in Meis1−/− LT-HSCs, which are known to induce HSC senescence and apoptosis, respectively (Figure 3E). Therefore, the increased ROS production in Meis1−/− LT-HSCs is compounded by the down-regulation of the master antioxidant gene Hif-2α. Moreover, measurement of peripheral blood counts of Meis1−/− mice post 1 month and 3 months of tamoxifen treatments demonstrates decreased red blood cells, white blood cells, as well as platelets, similar to the phenotype observed in Hif-2α−/− mice42 (supplemental Figure 5). To determine the mechanism of down-regulation of Hif-2α in Meis1−/− LT-HSCs, we identified 2 Meis1 consensus-binding motifs in the promoter of the Hif-2α gene (Figure 3F). Using a Hif-2α–pGL2 reporter which includes the conserved Meis1-binding sites, we demonstrate a dose-dependent activation of Hif-2α by Meis1 expression vector (CMV-Meis1; Figure 3G). In addition, this activation demonstrates dependence on binding of Meis1 to its consensus-binding sequences in the Hif-2α promoter because mutation of the seed sequences (Hif-2α–Mut-pGL2) completely abolished the activation of Hif-2α by Meis1. Given down-regulation of Hif-2α in Meis1−/− LT-HSCs (Figure 3A), these results indicate that Meis1 is required for optimal transcriptional activation of Hif-2α in LT-HSCs. We also confirmed the in vivo binding of Meis1 to its conserved sequences in the Hif-2α gene by ChIP assays in Kasumi1 cells as determined by real-time PCR after immunoprecipitation with Meis1 antibody (Figure 3H).
Effect of ROS scavenging on the Meis1−/− phenotype
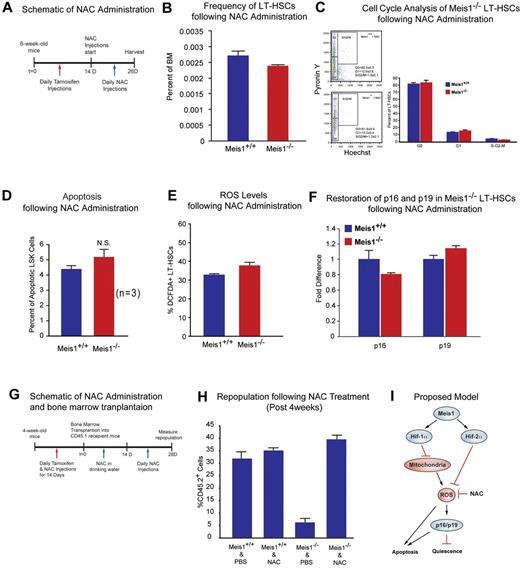
To study effects of ROS observed in Meis1−/− mice, we used NAC, an antioxidant, in an attempt to rescue the Meis1 phenotype by scavenging of ROS in Meis1 HSCs. After 14 days of tamoxifen injection, we administered NAC intraperitoneally for 12 days (Figure 4A). Mice where then harvested and HSCs were isolated for analysis of HSC frequency, HSC cell-cycle status, apoptosis rate, ROS levels, and expression of redox-sensitive cell-cycle regulators p16Ink4a and p19Arf. Here, we show that NAC administration rescues the Meis1−/− phenotype. We found that frequency of HSCs in Meis1−/− mice become similar to Meis1+/+ mice (Figure 4B). Flow cytometric analysis of cell cycle of Meis1+/+ and Mesi1−/− LT-HSCs, which were both injected with NAC, shows similar numbers of G0 cells in Meis1−/− LT-HSCs (Figure 4C). Quantification of apoptosis in Meis1+/+ and Meis1−/− mice shows an increased apoptosis in Meis1−/− mice; however, this increase in the number of apoptotic cells was not statistically significant (P = .051; Figure 4D). In addition, quantification of ROS after NAC treatments restored ROS to Meis1+/+ control levels in Meis1−/− LT-HSCs (Figure 4E). Finally, scavenging ROS restored the transcript levels of p16Ink4a and p19Arf in HSC to Meis1+/+ control levels (Figure 4F). While these effects are most likely attributable to the antioxidant effect of NAC, other unknown mechanisms of NAC may also contribute to this rescue.
Effect of ROS scavenging on the Meis1−/− phenotype. (A) Schematic of NAC administration. We performed daily IP injections for tamoxifen for 14 days followed by daily NAC injections up to 12 days. (B) Flow cytometry profile of LT-HSCs (Lin−Sca-1+Kit+Flk2−CD34−) of Meis1+/+ and Meis1−/− mice after 12 days NAC administration. Note the number of HSCs in Meis1−/− mice is now similar to Meis1+/+ values (n = 3). (C) Left panel, FACS plot of Pyronin Y/Hoechst staining of LT-HSCs. Right panel: Quantification of flow cytometric analysis of cell cycle of Meis1+/+ and Mesi1−/− LT-HSCs demonstrates restored numbers of G0 cells in Meis1−/− cells which indicates restored quiescence of LT-HSCs (n = 3). (D) Quantification of apoptosis in Meis1+/+ and Meis1−/− LSK cells (Lin−Sca-1+Kit+) showing persistent trend toward an increase in the number of apoptotic cells, which was not statistically significant (P = .059; n = 3). (E) Quantification of ROS in Meis1+/+ and Meis1−/− LT-HSCs (Lin−Sca-1+Kit+Flk2−CD34−) after NAC treatment showing only a modest increase in ROS in Meis1−/− HSCs (n = 3). (F) Real-time PCR of HSCs isolated from Meis1+/+ and Meis1−/− HSCs after NAC treatment demonstrating no change in p16 and p19 transcripts (n = 3). (G) Schematic of NAC administration and bone marrow transplantations. We performed daily IP injections for tamoxifen and NAC for 14 days followed by bone marrow transplantation. Then, NAC is provided in drinking water for 2 weeks and administrated another 2 weeks. Repopulation was examined at 4 weeks after transplantation. (H) Analysis of repopulation after NAC treatments of BMTs from Meis1+/+ and Meis1−/− mice demonstrates restoration of repopulation defect after Meis1 deletion (n = 5). (I) Schematic of proposed model is demonstrating how Meis1 regulates metabolism and maintenance of HSCs through its role on Hif-1α and Hif-2α.
Effect of ROS scavenging on the Meis1−/− phenotype. (A) Schematic of NAC administration. We performed daily IP injections for tamoxifen for 14 days followed by daily NAC injections up to 12 days. (B) Flow cytometry profile of LT-HSCs (Lin−Sca-1+Kit+Flk2−CD34−) of Meis1+/+ and Meis1−/− mice after 12 days NAC administration. Note the number of HSCs in Meis1−/− mice is now similar to Meis1+/+ values (n = 3). (C) Left panel, FACS plot of Pyronin Y/Hoechst staining of LT-HSCs. Right panel: Quantification of flow cytometric analysis of cell cycle of Meis1+/+ and Mesi1−/− LT-HSCs demonstrates restored numbers of G0 cells in Meis1−/− cells which indicates restored quiescence of LT-HSCs (n = 3). (D) Quantification of apoptosis in Meis1+/+ and Meis1−/− LSK cells (Lin−Sca-1+Kit+) showing persistent trend toward an increase in the number of apoptotic cells, which was not statistically significant (P = .059; n = 3). (E) Quantification of ROS in Meis1+/+ and Meis1−/− LT-HSCs (Lin−Sca-1+Kit+Flk2−CD34−) after NAC treatment showing only a modest increase in ROS in Meis1−/− HSCs (n = 3). (F) Real-time PCR of HSCs isolated from Meis1+/+ and Meis1−/− HSCs after NAC treatment demonstrating no change in p16 and p19 transcripts (n = 3). (G) Schematic of NAC administration and bone marrow transplantations. We performed daily IP injections for tamoxifen and NAC for 14 days followed by bone marrow transplantation. Then, NAC is provided in drinking water for 2 weeks and administrated another 2 weeks. Repopulation was examined at 4 weeks after transplantation. (H) Analysis of repopulation after NAC treatments of BMTs from Meis1+/+ and Meis1−/− mice demonstrates restoration of repopulation defect after Meis1 deletion (n = 5). (I) Schematic of proposed model is demonstrating how Meis1 regulates metabolism and maintenance of HSCs through its role on Hif-1α and Hif-2α.
To further evaluate the effect of ROS in engraftment defect observed after Meis1 deletion, we performed another bone marrow transplantation from Meis1+/+ and Meis1−/− mice treated with and without NAC. Two weeks after tamoxifen treatment and NAC administration, 1 × 105 BM cells from Meis1+/+ or Meis1−/− CD45.2, together with 1 × 105 CD45.1 competitors, were injected into CD45.1 recipients through the retro-orbital complex (Figure 4G). After bone marrow transplantation, NAC is provided in drinking water for 2 weeks and injected daily for 2 more weeks. Repopulation was examined at 4 weeks posttransplantation. As shown in Figure 4H, scavenging of ROS by NAC administration restored engraftment defects in Meis1−/− donors. In conclusion, scavenging ROS, through administration of NAC, rescues the Meis1 phenotype in HSC by decreasing levels of ROS, thereby normalizing p16Ink4a and p19Arf expression and restoring HSC quiescence and repopulation defects.
Effect of HIFs on stem cell function in Meis1−/− phenotype
Meis1 KO BM cells treated with 100μM CoCl2 resulted in restoration of total number of colonies to the WT levels (supplemental Figure 6A). Moreover, we found that Hif-1α overexpression using the Meis1f/f;Scl-Cre+; rtTA; TRE-Hif-1α mice strategy, which resulted in marked up-regulation of Hif-1α in HSCs, resulted in restoration of the BFU-E and total number of colonies to the WT levels, as well as partial restoration of number of mixed colonies (CFU-GEMM) which is a measure of most undifferentiated hematopoietic progenitors (supplemental Figure 6C). In addition, we attempted to rescue repopulation defect in Meis1−/− HSCs using viruses that express Hif-1α and Hif-2α (supplemental Figure 6D) or using Meisf/f;Scl-Cre+; rtTA; TRE-Hif-1α (supplemental Figure 6E); however, these strategies failed to rescue the in vivo reconstitution capacity up to 4 months after Meis1 deletion. In retrospect, these results are not entirely surprising in light of recent results demonstrating a narrow beneficial dose range of Hif-1α on HSC function, where overstabilization of Hif-1α results in worsening HSC function43 which might explain worsening of the HSC repopulation defect in Meis1 KO HSCs in vivo (supplemental Figure 6D-E).
In summary, we demonstrate that Meis1 deletion results in a shift in HSC metabolism toward oxygen consumption, with the resultant increase in ROS production. This phenotype is compounded by down-regulation of the oxidant stress response gene Hif-2α, and as a result Meis1 deletion results in HSC dysfunction and apoptosis.
Discussion
In the current report, we demonstrate that Meis1 functions upstream of a transcriptional network that regulates HSC metabolism and oxidant defense. The Meis1 deletion-induced metabolic shift and oxidant injury is compounded by the down-regulation of Hif-2α, which is in stark contrast to the marked up-regulation of Hif-2α in HSC after Hif-1α deletion. This phenotype is associated with up-regulation of p16Ink4a and p19Arf, loss of quiescence, increased apoptosis, and marked HSC dysfunction.
Endothelial PAS domain protein 1 (EPAS1), also known as Hif-2α,44 is closely related to Hif-1α in structure and is likewise activated during hypoxia.20 While Hif-1α is a master regulator of metabolism, Hif-2α is a master regulator of oxidant stress response45 and is induced by ROS.46 It is involved in regulation of numerous antioxidant genes that minimize the oxidant damage that results from mitochondrial respiration.45 Hif-2α−/− mice are pancytopenic,42,47 and have high levels of oxidative stress,45 which suggests that Hif-2α is required for normal hematopoiesis. Our results indicate that Hif-2α is markedly up-regulated after Hif-1α deletion, which is an indication of a robust antioxidant response to Hif-1α deletion, likely secondary to a shift toward oxidative metabolism, with the subsequent increase in ROS.46 In stark contrast, we show that Hif-2α is down-regulated after Meis1 KO in HSCs, which may partially explain the severity of the Meis1−/− phenotype compared with the Hif-1α−/− phenotype.
Delicate control of ROS levels in HSCs is crucial for HSC maintenance, where elevated ROS levels in HSCs is associated with defects in HSC self-renewal and increased apoptosis. Regulation of ROS in HSCs is a highly complex process that involves regulation of the metabolic phenotype of HSCs, as well as regulation of antioxidant defense mechanisms. The contribution of oxidative metabolism to ROS production has been extensively studied.48 It is estimated that 2% of all electrons flowing through the mitochondrial respiratory chain result in the formation of oxygen free radicals. Electrons leaking from the respiratory chain interact with oxygen, partially reducing it to superoxide anion (O2−·).48 Even though O2−· itself is not a strong oxidant, it is the precursor of most other ROS.14 ROS overwhelm the natural antioxidant defense mechanisms over time, and result in widespread cellular damage.48 ROS can also induce the cell-cycle regulators p16Ink4a and p19Arf, which cause loss of quiescence and apoptosis of HSCs.49 Our results indicate that Meis1 regulates ROS production in HSC through regulation of both the metabolic phenotype (through Hif-1α) and oxidant defense mechanisms (through Hif-2α). Finally, although our ROS scavenging studies indicate that the effect of loss of Meis1 on HSC cell cycle and engraftment defect are entirely mediated through an increase in ROS production, rescue studies using Hif-1α and/or Hif-2α overexpression or stabilization failed to fully rescue the phenotype. These results support previous reports which indicate that the precise dose of Hif-1α is necessary for optimal HSC function.25 Although our results implicate Hif-1α and Hif-2α in the HSC phenotype after Meis1 deletion, there certainly may be additional ROS modulators that are targeted by Meis1.
In the current report, we highlight the role of Meis1 in a transcriptional network that regulates HSC metabolism and antioxidant defense. These results implicate Meis1 as an important regulator of the redox state of HSCs, which may be echoed by its role in leukemogenesis. Therefore, it would be important for future studies to determine the role of Meis1 in regulation of leukemia stem cell metabolism, survival, and self-renewal. Our findings suggest that HSCs are endowed with redundant mechanisms for regulation of their metabolic phenotype, rather than being solely dependent on environmental signals, such as the hypoxic microenvironment. Deciphering the role of these transcriptional networks in regulating HSC fate and function may provide valuable clues for understanding HSC disease and malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Keith Humphries, Michelle Miller, and Patty Rosen for sharing some initial data on characterization of the KO line genotyping, Dr Joachim R. Goethert at Universitaetsklinikum Essen for providing the Scl-Cre-ERT mice, and Dr Joseph A. Garcia for Hif-1α floxed mice.
This work is supported by grants from the American Heart Association (AHA; GIA12060240 [H.A.S.]), the Gilead Research Scholars Program in Cardiovascular Disease (H.A.S.), National Institutes of Health (NIH) grant R01HL115275 (H.A.S.), NIH grant K01 CA 120099 (C.Z.), and Cancer Prevention and Research Institute of Texas (CPRIT) RP100402 (C.Z.).
National Institutes of Health
Authorship
Contribution: F.K. designed and performed the research, analyzed data, and wrote the manuscript; J.Z., S.T., N.G.C., N.A.J., and R.J.D. performed research and analyzed data; and C.Z. and H.A.S. designed and supervised research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for F.K. is Texas Institute of Biotechnology, Edu&Res, North American College, Houston, TX.
Correspondence: Hesham A. Sadek, MD, PhD, Department of Internal Medicine, Division of Cardiology, UT Southwestern Medical Center, NB10.222, 6000 Harry Hines Blvd, Dallas, TX 75390; e-mail: hesham.sadek@utsouthwestern.edu; or Chengcheng Zhang, PhD, Departments of Physiology and Developmental Biology, UT Southwestern Medical Center, NB10.222, 6000 Harry Hines Blvd, Dallas, TX 75390; e-mail: alec.zhang@utsouthwestern.edu.
References
Author notes
F.K. and J.Z. contributed equally to this project.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal