Abstract
IGHV3-21–using chronic lymphocytic leukemia (CLL) is a distinct entity with restricted immunoglobulin gene features and poor prognosis and is more frequently encountered in Northern than Southern Europe. To further investigate this subset and its geographic distribution in the context of a country (Italy) with both continental and Mediterranean areas, 37 IGHV3-21 CLLs were collected out of 1076 cases enrolled by different institutions from Northern or Central Southern Italy. Of the 37 cases, 18 were identified as homologous (hom)HCDR3–IGHV3-21 CLLs and were found almost exclusively (16 of 18) in Northern Italy; in contrast, 19 nonhomHCDR3–IGHV3-21 cases were evenly distributed throughout Italy. Clinically, poor survivals were documented for IGHV3-21 CLLs as well as for subgroups of mutated and homHCDR3–IGHV3-21 CLLs. Negative prognosticators CD38, ZAP-70, CD49d, and CD79b were expressed at higher levels in homHCDR3 than nonhomHCDR3–IGHV3-21 cases. Differential gene expression profiling (GEP) of 13 IGHV3-21 versus 52 non–IGHV3-21 CLLs identified, among 122 best-correlated genes, TGFB2 and VIPR1 as down- and up-regulated in IGHV3-21 CLL cases, respectively. Moreover, GEP of 7 homHCDR3 versus 6 nonhomHCDR3–IGHV3-21 CLLs yielded 20 differentially expressed genes, with WNT-16 being that expressed at the highest levels in homHCDR3–IGHV3-21 CLLs. Altogether, IGHV3-21 CLLs, including those with homHCDR3, had a peculiar global phenotype in part explaining their worse clinical outcome.
Introduction
The clinical course of B-cell chronic lymphocytic leukemia (CLL) can be in part foreseen by the presence of mutated (M) or unmutated (UM) immunoglobulin variable (IGV) genes.1 Analysis of IGV heavy chain (IGHV) gene usage has revealed expression of a biased IGHV repertoire in CLL compared with normal B cells.2,3 Moreover, a significant fraction of CLL expresses B-cell receptors (BCRs) characterized by nonrandom stereotyped combinations of IGHV-D-J rearrangements often associated with a restricted selection of IGV light chains.3-6 Such molecular peculiarities occur both in poor-prognosis UM CLL and in highly stable/indolent M CLLs.3-5,7,8 These observations, together with an IGHV mutation profile consistent with antigen-driven selection,9,10 led to the speculation that CLL pathogenesis may involve specific, though yet unidentified, (auto)antigens.11
Usage of the IGHV3-21 gene by CLL has been initially reported in about 10% of Scandinavian patients12,13 and subsequently confirmed in small CLL series from other Northern European countries.14,15 Interestingly, many IGHV3-21 rearrangements displayed unusually short and highly homologous IGHV complementarity determining region 3 (HCDR3), often associated with the IGV lambda (IGLV) chain IGLV3-21(Vλ2-14). Although IGHV3-21 CLL usually displays M IGHV genes, patients experience overall survivals similar to those of UM CLL.16
Recent studies reported a low incidence of IGHV3-21 CLL in Mediterranean countries, thus suggesting that the frequency of IGHV3-21 CLL may be related to geographic, ethnic, or environmental background.6,16 These reports also emphasized the grouping of IGHV3-21 CLL into 2 subsets according to the degree of homology in HCDR3 sequences: cases with a high degree of molecular homology seemed to have a more aggressive clinical behavior compared with nonhomologous cases.6,16 However, the prognostic value of HCDR3 homology among IGHV3-21 CLL remains still controversial.12,13,17
Italy may provide a valuable model to analyze IGHV3-21 distribution among CLL because the country includes both continental and Mediterranean areas. To conclusively dissect the issue of a biased geographic distribution and to provide a comprehensive biologic and clinical characterization of IGHV3-21 CLL, we investigated cases derived from 1076 unselected CLL patients enrolled by different reference centers located in Northern (N) or Central Southern (CS) Italy. Because IGHV3-21 CLL patients have a clinical behavior independent of the IGHV gene mutation status,3,12,13,16 we investigated the differential gene expression profile characterizing IGHV3-21 CLL versus non–IGHV3-21 cases. The resulting profile and the function of some differentially regulated genes integrated the information provided by the unique study published so far.18 Finally, we were able to identify a number of genes characterizing the differential expression signature of IGHV3-21 CLL with homologous versus nonhomologous HCDR3.
Patients, materials, and methods
Patients, healthy donors, and human cell lines
The study included peripheral blood (PB) samples from 1076 unselected CLL patients collected from the archives of the Clinical and Experimental Hematology Research Unit of the Aviano National Cancer Institute (n = 136); Division of Hematology of the Amedeo Avogadro University of Eastern Piedmont, Novara (n = 110); Department of Oncology and Hematology of the University of Modena-Reggio Emilia (n = 284); Hematology Unit of La Sapienza University, Rome (n = 228); S Eugenio Hospital, University of Tor Vergata, Rome (n = 127); Hematology Institute of the Catholic University, Rome (n = 95); and the Hematology Unit of the University of Siena (n = 96). According to the location of each center, 2 main geographic subgroups were identified: the N Italy subgroup, which included 530 patients recruited by the Aviano, Novara, and Modena-Reggio Emilia centers (N patients), and the CS Italy subgroup, with 546 patients recruited by the Siena center and the institutions located in Rome (CS patients) (Table 1). Median ages were 64.0 and 60.4 years, while male-female ratios were 1.6 and 1.4 for N and CS patients, respectively.
Normal B cells used in gene expression profiling (GEP) experiments were purified from PB samples from 13 healthy donors kindly provided by the Blood Bank of the Aviano National Cancer Institute.
For survival studies, we combined 3 single-institution series (Aviano, 69 patients; Modena-Reggio Emilia, 152; Roma Tor Vergata, 80; total, 301) in which the standard cutoff of 2% IGHV mutations proved to have prognostic value in the context of each single series (not shown); the overall mean follow-up of the 301 patients was 66 months (range, 3 to 289). Patient survival was analyzed using Kaplan-Meyer survival curves and log-rank test. Comparisons of frequencies between specific CLL subsets were made by applying the Pearson χ2 test with Yates continuity correction.
Analysis of IGHV, IGKV, and IGLV rearrangements and HCDR3 clustering
IGHV, IGV kappa (IGKV), and IGLV rearrangements were amplified from either reverse-transcribed total RNA or genomic DNA, as previously described.7,9 Purified amplicons were sequenced either directly or upon insertion into plasmid vectors and cloning.7,9 Sequences were aligned to the IgBLAST, VBASE, or ImMunoGeneTics directories and considered mutated if deviation from the corresponding germ line gene was at least 2%. For IGHD gene determination, a requirement of at least 7 matching nucleotides was used. HCDR3-driven clustering was performed by converting all in-frame IGHV rearrangements into amino acid (aa) sequences and by aligning HCDR3 sequences by means of the multiple sequence alignment software ClustalW.21 The length of HCDR3 was computed between codons 95 and 102, as described by Kabat.22 For consistency with previous studies,12,13,16,17 IGLV chains were also indicated according to the Kawasaki nomenclature.
Flow cytometry
All immunophenotypical determinations were performed at the Aviano National Cancer Institute. Sources, specificities, and fluorochrome combinations of monoclonal antibodies along with details on the working protocols were reported elsewhere.23 Detection of ZAP-70 was performed as reported using a cutoff point of 30% to define positivity.24,25
B-cell purification, RNA extraction, and gene expression profiling (GEP) analysis
Neoplastic and normal B cells (more than 95% pure), purified by Ficoll-Hypaque gradient (Pharmacia, Uppsala, Sweden) and positive immunoselection with anti-CD19–conjugated immunomagnetic beads (Miltenyi Biotec, Bologna, Italy), were employed as total RNA source. cDNA synthesis, cDNA purification, in vitro transcription of amino-allyl RNA (aRNA), aRNA dye coupling, and purification of dye-labeled aRNA were performed using the Amino Allyl MessageAmp II aRNA Amplification Kit (Ambion, Austin, TX) following manufacturer's guidelines. Cy3/Cy5 was from Amersham Biosciences (Amersham, United Kingdom). Cy3/Cy5 dye incorporation into aRNA yielded incorporation rates of 30 to 60 dye molecules per 1000 nucleotides by spectrophotometric analysis, as requested by the manufacturer.
GEP was performed by a dual labeling strategy using Cy3-labeled aRNA from pooled normal PB B cells of healthy donors as common reference and Cy5-labeled aRNA from purified CLL cells as tester. A mixture of 4 μg Cy3-labeled reference aRNA and 4 μg Cy5-labeled aRNA from each CLL was hybridized to Operon Human Genome Oligo Set Version 2.0 platforms (21 329 oligomers of 70 nucleotides; Operon Biotechnologies, Huntsville, AL) and spotted in duplicates onto MicroMax SuperChip I glass slides (Perkin Elmer Life Sciences, Boston, MA). Hybridization was performed for 18 hours at 48°C in a buffer containing 5× standard sodium cytrate (SSC), 0.1% sodium dodecyl sulfate (SDS), 25% formamide, and 100 μg/mL salmon sperm DNA using an automated HybArray 12 hybridization system (Perkin Elmer Life Sciences). Following washings, glass slides were analyzed by a Scan Array Lite scanner (Packard BioChip Technologies, Billerica, MA); images and data were analyzed using the GenePix Pro software (Axon Instruments, Foster City, CA). The final data set was analyzed by means of unsupervised and supervised clustering as detailed in Supplemental Materials and Methods, “Bioinformatics approach for analysis of gene expression profiling (GEP) of IGHV3-21 CLLs” (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Reverse transcriptase–polymerase chain reaction (RT-PCR) and quantitative-real-time RT-PCR (QRT-PCR)
Expression of TGFβ2, VIPR1, and WNT-16 was evaluated by both conventional and quantitative (Q) RT-PCR using, in the latter case, SYBR Green dye–containing reaction buffer (Applied Biosystems, Foster City, CA). Reactions were performed in the presence of 0.5 pmol/μL of primers specific for TGFβ2 (sense: 5′-AGAGGAGCGACGAAGAGTA-3′; antisense: 5′-AAGTCTGTAGGAGGGCAATAA-3′), VIPR1 (sense: 5′-GGCTCGGTGGGCTGTAAGG-3′; antisense: 5′-GACCAGGGAGACTTCGGCTTG-3′), and WNT-16 (sense: 5′-CGGGAGCCAGTTCAGA-CACGA-3′; antisense: 5′-CACTTGCTGAGCCGCCGTTCT-3′). Amplification protocol was 4 minutes (10 minutes for QRT-PCR) at 95°C for initial denaturation, 35 cycles (40 for QRT-PCR) of 15 seconds at 95°C, 30 seconds at 62°C (57°C for TGFβ2), and 90 seconds at 72°C, followed by final extension at 72°C for 5 minutes. Incorporation of SYBR Green dye into the PCR products was monitored in real time with an ABI PRISM 7700 sequence detection system (Applied Biosystems). Standard curves (10-fold dilution from 101 to 10−4 attomoles) were prepared for both target genes and the housekeeping gene β2-microgobulin (β2M). For each experimental sample, the amount of target and endogenous reference was determined from the appropriate standard curve, and the target amount was divided by the endogenous reference amount to obtain a normalized target value. Sequences of primers for β2M were previously reported.26
Results
Distribution of IGHV families in CLL from N and CS Italy
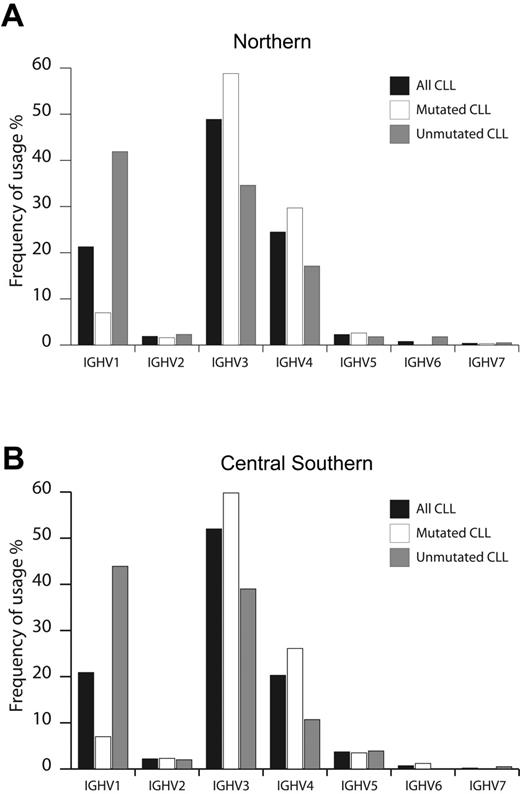
Of 1076 patients with CLL entering this study, 530 and 546 patients were enrolled by institutions located in N and CS Italy, respectively. As summarized in Figure 1, CLL used all 7 IGHV gene families, with overall similar frequencies in N and CS patients. In agreement with other smaller series,10,27,28 the IGHV3 family was most frequently used (N, 49%; CS, 52%), followed by IGHV1 (N, 21%; CS, 21%) and IGHV4 (N, 24%; CS, 20%) (Figure 1). Considering the 2% cutoff to discriminate between M and UM cases, 654 of the CLLs (60.8%) were classified as M, equally distributed among N (313 cases, 59.1%) and CS patients (341 cases, 62.5%). Similarly, 422 CLLs (39.2%) were classified as UM, equally distributed among N (217 cases, 40.9%) and CS patients (205 cases, 37.5%) (Table 1). The IGHV1 family was more frequently rearranged in UM CLL (N, 42%; CS, 44%), whereas IGHV3 was more represented among mCLL (59% both in N and CS patients; Figure 1). Finally, IGHV2, IGHV5, IGHV6, and IGHV7 were rarely expressed in both M and UM CLL from N and CS cohorts of patients (Figure 1).
Usage of IGHV gene families in CLL from Northern and Central Southern Italy. IGHV gene family usage was analyzed in CLL from Northern (530 cases) (A) and Central Southern Italy (546 cases) (B). Frequencies were calculated for the whole series (▪) and according to the IGHV mutational status (UM, ⊡; M, □).
Usage of IGHV gene families in CLL from Northern and Central Southern Italy. IGHV gene family usage was analyzed in CLL from Northern (530 cases) (A) and Central Southern Italy (546 cases) (B). Frequencies were calculated for the whole series (▪) and according to the IGHV mutational status (UM, ⊡; M, □).
Molecular features of IGHV3-21 CLLs: IGHV rearrangements
Among 1076 CLLs, IGHV3-21 was used in 37 cases (3.4%), all displaying an in-frame and potentially functional rearrangement. Table 2 summarizes the characteristics of IGHV3-21 rearrangements in our series. Twenty-five of 37 cases (67.6%) expressed an IGHV gene with more than 2% mutations, with a median mutation rate of 4.0% (range, 2.1% to 7.1%); 20 of 25 M IGHV3-21 CLLs had a “low” mutation burden of 2% to 5%.
A given IGHD segment was recognized in 28 of 37 IGHV3-21 rearrangements. IGHD5-24 and IGHD3-22 were the most common segments, detected in 4 rearrangements each. A specific IGHD segment was not assigned in 9 cases, 8 of them expressing the highly homologous short HCDR3 sequences described in detail below. Finally, the IGHJ6 gene was the most frequent IGHJ segment (20 of 37 cases), followed by IGHJ4 (9 of 37 cases).
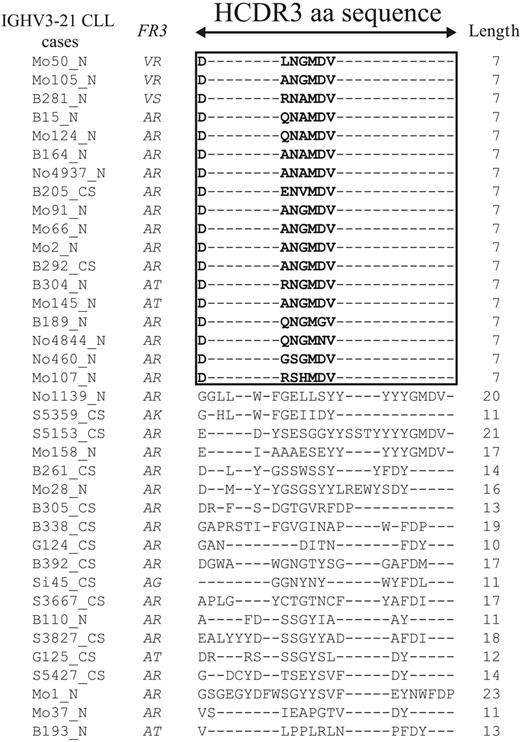
Cluster analysis of putative translated HCDR3 protein sequences allowed us to split IGHV3-21 CLL into 2 groups (Table 2; Figure 2). The first group (18 of 37 cases) displayed a highly homologous and short HCDR3 sequence (homHCDR3–IGHV3-21 CLL) characterized by the aa sequence DANGMDV in 6 of 18 cases (Figure 2). The remaining cases expressed a similar 7-aa stretch modified by 1 (6 cases) or 2 (5 cases) aa's; a single case (Mo107) had 3 aa changes in the conserved 7-aa sequence (Figure 2). Analyses of nucleotide sequences of homHCDR3–IGHV3-21 CLL often (8 of 18 cases) yielded an unidentified IGHD segment always associated with IGJH6 gene usage (Table 2). The second subset of IGHV3-21 CLLs (19 of 37 cases) expressed HCDR3 sequences usually longer (median length, 14 nucleotides; range, 11 to 23 nucleotides) and totally unrelated to each other (nonhomHCDR3–IGHV3-21 CLLs; Figure 2). Expression of longer HCDR3 sequences from this group may explain why identification of IGHD segments was possible in all but 1 case (Table 2).
Amino acid sequences of the HCDR3 in the IGHV3-21 cases. Alignment of the HCDR3 sequences belonging to 37 IGHV3-21 CLL cases was performed by converting all in-frame IGH rearrangements into aa sequences, selecting HCDR3 sequences only, and aligning them by means of the multiple sequence alignment software ClustalW.21 The length of HCDR3 was computed between codons 95 and 102, as described by Kabat. A black frame was drawn around the 18 sequences with the same length and similar aa composition. Missing aa's are indicated with dashes.
Amino acid sequences of the HCDR3 in the IGHV3-21 cases. Alignment of the HCDR3 sequences belonging to 37 IGHV3-21 CLL cases was performed by converting all in-frame IGH rearrangements into aa sequences, selecting HCDR3 sequences only, and aligning them by means of the multiple sequence alignment software ClustalW.21 The length of HCDR3 was computed between codons 95 and 102, as described by Kabat. A black frame was drawn around the 18 sequences with the same length and similar aa composition. Missing aa's are indicated with dashes.
We then investigated whether the 7-aa stretch expressed by homHCDR3–IGHV3-21 CLL was also employed by IGHV rearrangements other than IGHV3-21. HCDR3 sequences from IGHV3-21 CLLs were clustered along with HCDR3 sequences from 349 CLL cases representative of the whole series (Figure S1). Results of cluster analysis clearly identified a cluster that included all homHCDR3–IGHV3-21 CLLs along with 1 single non–IGHV3-21 case (case B383 rearranging IGHV3-66; Figure S1), suggesting that use of homologous HCDR3 characterized by the 7-aa stretch is very rare in non–IGHV3-21 CLL.
Molecular features of IGHV3-21 CLL: IGKV and IGLV rearrangements
Among 34 IGHV3-21 cases with available IGV light chain rearrangement data, a total of 44 rearrangements, including 32 IGL and 12 IGK, were found (Table 2). HomHCDR3–IGHV3-21 CLL showed the IGLV3-21(Vλ2-14)/IGLJ3 rearrangement either alone (9 cases) or in association with another potentially functional IGLV rearrangement (9 cases). These rearrangements included another IGLV3-21/Vλ2-14 (1 case), IGLV1-51/Vλ1-19 (3 cases), IGLV1-40/Vλ1-13 (2 cases) or, in the remaining cases, IGLV5-45/Vλ5-1, IGLV2-8/Vλ1-2, or IGLV6-57/Vλ1-22 (Table 2). Conversely, nonhomHCDR3–IGHV3-21 cases rearranged either IGLV (5 cases) or IGKV (11 cases) without any bias for specific genes. Two potentially functional IGV light chain rearrangements were amplified in a single case (Mo1).
Distribution of IGHV3-21 CLL in N and CS Italy
As summarized in Table 3, the percentage of IGHV3-21 CLLs was slightly higher in N (23 of 530, 4.3%) versus CS patients (14 of 546, 2.6%), although this difference did not reach statistical significance (P = .15). However, most homHCDR3–IGHV3-21 cases (16 of 18) were recruited by institutions located in N Italy. In particular, homHCDR3–IGHV3-21 CLLs represented 3.0% (16 of 530) of N cases and only 0.4% (2 of 546) of CS cases (P = .002; Table 3). A statistical significance (P = .003) was also reached when comparing the sole distribution of homHCDR3 and nonhomHCDR3 IGHV3-21 cases in the 2 main geographic areas (data not shown). Conversely, distribution of nonhomHCDR3–IGHV3-21 CLLs was not dissimilar in N and CS patient series (P = .39; Table 3).
Clinical features of IGHV3-21 CLL in Italy
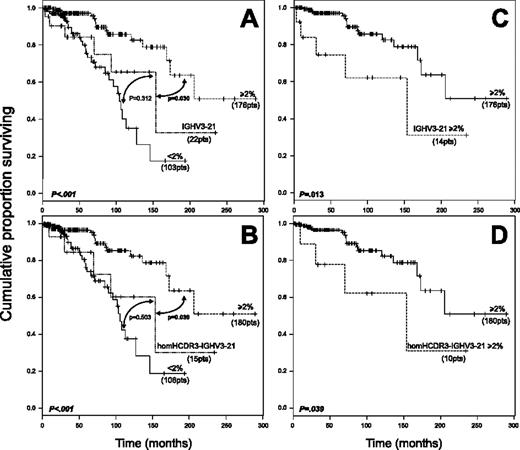
Survival data were analyzed in 301 patients, 279 with a non–IGHV3-21 CLL (176 with M and 103 with UM IGHV genes) and 22 with IGHV3-21 CLL (14 M and 8 UM). This latter group included 15 homHCDR3 and 7 nonhomHCDR3–IGHV3-21 CLLs. In the context of non–IGHV3-21 CLL (Figure 3A), a significantly longer survival was documented for M cases (median survival, not reached; 95% confidence interval [CI], 173 to not reached) compared with UM CLLs (median survival, 104 months; 95% CI, 90 to not reached). The survival curve of IGHV3-21 CLL (median survival, 154 months; 95% CI, 93 to not reached) was similar to that of UM non–IGHV3-21 cases (P = .312) and significantly different from M CLLs (P = .030; Figure 3A). An analogous trend was observed when only homHCDR3–IGHV3-21 CLLs were considered (Figure 3B). Moreover, both M IGHV3-21 CLL and M homHCDR3–IGHV3-21 CLLs displayed significantly shorter survivals when compared with M non–IGHV3-21 cases (Figure 3C-D). Finally, no differences were documented by comparing survival of UM (8 cases) versus M (14 cases) IGHV3-21 CLL as well as nonhomHCDR3– (7 cases) versus homHCDR3–IGHV3-21 CLL (15 cases) (data not shown).
Kaplan-Meier survival curve analysis in IGHV3-21 CLL patients. (A) Comparison of survival probabilities in IGHV3-21 patients (22 patients), patients with M IGHV gene configuration (at least 2%, 176 patients), and patients with UM IGHV gene configuration (less than 2%, 103 patients). (B) Comparison of survival probabilities in homHCDR3–IGHV3-21 CLLs (15 patients), M cases (180 patients), and UM cases (106 patients). (C) Kaplan-Meier curves comparing survivals in M IGHV3-21 cases (14 patients) versus M CLLs (176 patients). (D) Kaplan-Meier curves comparing survivals in M homHCDR3–IGHV3-21 CLLs (10 patients) versus all M CLLs (180 patients). Reported P values refer to the log-rank test.
Kaplan-Meier survival curve analysis in IGHV3-21 CLL patients. (A) Comparison of survival probabilities in IGHV3-21 patients (22 patients), patients with M IGHV gene configuration (at least 2%, 176 patients), and patients with UM IGHV gene configuration (less than 2%, 103 patients). (B) Comparison of survival probabilities in homHCDR3–IGHV3-21 CLLs (15 patients), M cases (180 patients), and UM cases (106 patients). (C) Kaplan-Meier curves comparing survivals in M IGHV3-21 cases (14 patients) versus M CLLs (176 patients). (D) Kaplan-Meier curves comparing survivals in M homHCDR3–IGHV3-21 CLLs (10 patients) versus all M CLLs (180 patients). Reported P values refer to the log-rank test.
Expression of immunophenotypic prognosticators by IGHV3-21 CLL
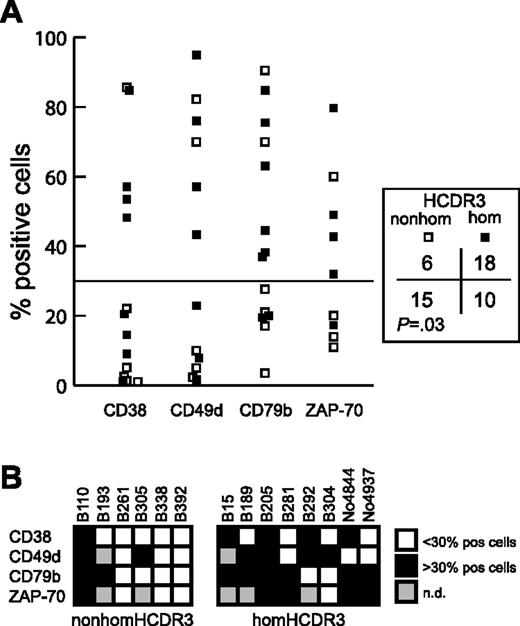
Expression data for the negative immunophenotypic prognosticators CD38, ZAP-70,25,29-32 CD49d, and CD79b 33,34 were available for 14 IGHV3-21 CLLs (8 homHCDR3 and 6 nonhomHCDR3–IGHV3-21 cases; 10 cases with M and 4 cases with UM IGHV genes). HomHCDR3–IGHV3-21 cases more frequently expressed these negative prognosticators above the cutoff value of 30% of positive cells23,24,34 as compared with nonhomHCDR3–IGHV3-21 CLL (P = .03) (Figure 4A). Moreover, while 3 of 6 nonhomHCDR3–IGHV3-21 CLLs expressed none of the negative prognosticators, all the homHCDR3–IGHV3-21 CLLs expressed at least 2 (7 of 8 cases) or 1 (all cases) negative prognosticators (Figure 4B). Of note, all 4 UM CLLs (cases B15, B205, B292 and No4937 in Figure 4B) had a homHCDR3–IGHV3-21 sequence and, with the only exclusion of CD79b for B292 and CD38/CD49d for No4937, expressed all the negative prognosticators above the established threshold (Figure 4B).
Expression of negative prognostic markers by homHCDR3–IGHV3-21 and nonhomHCDR3–IGHV3-21 CLLs. (A) The plot represents CD38, CD49d, CD79b, and ZAP-70 expression values in 8 homHCDR3–IGHV3-21 (▪) and 6 nonhomHCDR3–IGHV3-21 (□) CLLs. The horizontal line corresponds to the cutoff value of 30% of positive cells. The χ2 test reported on the right compares the number of cases (homHCDR3–IGHV3-21 or nonhomHCDR3–IGHV3-21 CLLs) in which the expression of each phenotypic marker was expressed above or below the 30% of positive cells threshold. (B) Grids indicating the expression of the 4 prognostic markers CD38, CD49d, CD79b, and ZAP-70 in the 6 nonhomHCDR3–IGHV3-21 (left grid) and in the 8 homHCDR3–IGHV3-21 (right grid) CLLs. Black or white boxes indicate expression of the marker above or below the 30% cutoff, respectively. Gray boxes indicate no data are available.
Expression of negative prognostic markers by homHCDR3–IGHV3-21 and nonhomHCDR3–IGHV3-21 CLLs. (A) The plot represents CD38, CD49d, CD79b, and ZAP-70 expression values in 8 homHCDR3–IGHV3-21 (▪) and 6 nonhomHCDR3–IGHV3-21 (□) CLLs. The horizontal line corresponds to the cutoff value of 30% of positive cells. The χ2 test reported on the right compares the number of cases (homHCDR3–IGHV3-21 or nonhomHCDR3–IGHV3-21 CLLs) in which the expression of each phenotypic marker was expressed above or below the 30% of positive cells threshold. (B) Grids indicating the expression of the 4 prognostic markers CD38, CD49d, CD79b, and ZAP-70 in the 6 nonhomHCDR3–IGHV3-21 (left grid) and in the 8 homHCDR3–IGHV3-21 (right grid) CLLs. Black or white boxes indicate expression of the marker above or below the 30% cutoff, respectively. Gray boxes indicate no data are available.
Differential gene expression profiling (GEP) of IGHV3-21 CLL
Thirteen IGHV3-21 (11 M and 2 UM) and 52 non–IGHV3-21 CLLs (35 M and 17 UM) were analyzed for GEP. An unsupervised approach was used in exploratory analyses to evaluate whether IGHV3-21 and non–IGHV3-21 CLLs could be separated by a hierarchic clustering (ie, had a distinguishable global GEP). This unsupervised analysis was performed on a total of 28 513 features, which passed the prefiltering steps aimed at eliminating all features that were either not expressed or overall not differentially modulated, as detailed in Supplemental Materials and Methods, “Bioinformatics approach for analysis of gene expression profiling (GEP) of IGHV3-21 CLLs.” The generated hierarchic clustering (Figure S2) clearly demonstrated that all 13 IGHV3-21 CLLs were intermingled among non–IGHV3-21 cases, thus indicating that the 2 subgroups shared the large part of the features investigated.
Therefore, to discover genes differentially expressed in IGHV3-21 versus non–IGHV3-21 CLL, we applied a supervised approach capable of detecting minimal differences in gene expression of otherwise related cell populations. In particular, to address this issue in the context of an unbalanced data set (13 IGHV3-21 versus 52 non–IGHV3-21 cases), we used an original modified version of significance analysis of microarrays (SAM)35,36 named multi-SAM (Supplemental Materials and Methods, “Bioinformatics approach for analysis of gene expression profiling (GEP) of IGHV3-21 CLLs.”). Using this approach, we were able to select 122 probes (115 genes) correlated with IGHV3-21 usage by CLL: 60 probes were up-regulated and 62 down-regulated in IGHV3-21 CLL. A hierarchic clustering generated by using these 122 best-correlated probes clearly split the 13 IGHV3-21 CLLs from all but 1 of the other non–IGHV3-21 cases (Figure 5A). A list of these 122 probes, including up-to-date gene names and position in cluster of Figure 5A, is reported in Table S1. Not surprisingly, IGLV3-21(Vλ2-14) was the gene up-regulated in IGHV3-21 CLLs with the highest score (Table S1). In this regard, by checking the portion of heat maps corresponding to both copies of IGLV3-21(Vλ2-14) genes (Figure 5A, bottom), an almost complete consistency between molecular and hybridization data could be detected.
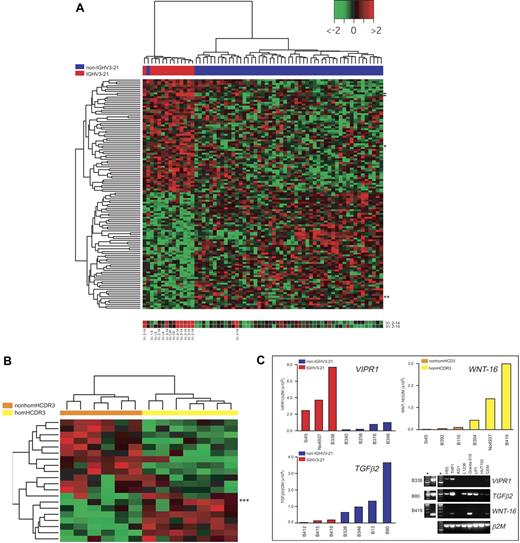
IGHV3-21–associated gene expression profiles in CLL. (A) Heat map generated with 122 probes found to be differentially expressed between IGHV3-21 patients (red bar under the tree) and non–IGHV3-21 patients (blue bar) by supervised analyses (Supplemental Materials). The color scale identifies relative gene expression changes normalized by the SD of 1 with 0 representing the mean expression level of a given gene. Expression for the 2 probes for the IGLV3-21(Vλ2-14) gene is reported below the heat map (location of these probes in the heat map is indicated by dashes); results of IGV light chain rearrangements, when available (ie, for all IGHV3-21 cases and 1 non–IGHV3-21 CLL), are reported below the corresponding hybridization spot. Location in the heat map of the VIPR1 and TGFβ2 genes are indicated by 1 and 2 asterisks, respectively. (B) Heat map generated with 20 genes found to be differentially expressed between homHCDR3–IGHV3-21 cases (yellow bar under the tree) and nonhomHCDR3–IGHV3-21 cases (orange bar) by supervised analyses (see “Materials and methods”). Location in the heat map of the WNT-16 gene is indicated by 3 asterisks. (C) Validation of microarray results by QRT-PCR. Three representative genes were selected to validate microarray results by QRT-PCR: VIPR1 and TGFβ2 were chosen to characterize non–IGHV3-21 versus IGHV3-21 CLLs (13 total cases), while WNT-16 was chosen among the genes differentially expressed by homHCDR3–IGHV3-21 and nonhomHCDR3–IGHV3-21 cases (6 cases). The incorporation of the SYBR Green dye into the PCR products was monitored in real time, and the resulting threshold cycles (Ct) (ie, the PCR cycle number corresponding to the beginning of the exponential growth of PCR products) were computed. Ct values were converted into attomoles by means of QRT-PCR experiments carried out with serial dilution of known concentrations of VIPR1-, TGFβ2-, WNT-16–, and β2M-specific amplicons. Results for each CLL case represent the values of the relative expression levels of VIPR1/β2M and TGFβ2/β2M in IGHV3-21 cases (red bars) and non–IGHV3-21 cases (blue bars) and WNT-16/β2M in homHCDR3–IGHV3-21 cases (yellow bars) and nonhomHCDR3–IGHV3-21 VH3-21 cases (orange bars). The panel on the bottom right represents ethidium bromide–stained agarose gels showing the expression of VIPR1, TGFβ2, and WNT-16 by conventional RT-PCR in 8 cell lines and in 3 representative CLL cases (B338, B80, and B419). Asterisks refer to molecular weight markers.
IGHV3-21–associated gene expression profiles in CLL. (A) Heat map generated with 122 probes found to be differentially expressed between IGHV3-21 patients (red bar under the tree) and non–IGHV3-21 patients (blue bar) by supervised analyses (Supplemental Materials). The color scale identifies relative gene expression changes normalized by the SD of 1 with 0 representing the mean expression level of a given gene. Expression for the 2 probes for the IGLV3-21(Vλ2-14) gene is reported below the heat map (location of these probes in the heat map is indicated by dashes); results of IGV light chain rearrangements, when available (ie, for all IGHV3-21 cases and 1 non–IGHV3-21 CLL), are reported below the corresponding hybridization spot. Location in the heat map of the VIPR1 and TGFβ2 genes are indicated by 1 and 2 asterisks, respectively. (B) Heat map generated with 20 genes found to be differentially expressed between homHCDR3–IGHV3-21 cases (yellow bar under the tree) and nonhomHCDR3–IGHV3-21 cases (orange bar) by supervised analyses (see “Materials and methods”). Location in the heat map of the WNT-16 gene is indicated by 3 asterisks. (C) Validation of microarray results by QRT-PCR. Three representative genes were selected to validate microarray results by QRT-PCR: VIPR1 and TGFβ2 were chosen to characterize non–IGHV3-21 versus IGHV3-21 CLLs (13 total cases), while WNT-16 was chosen among the genes differentially expressed by homHCDR3–IGHV3-21 and nonhomHCDR3–IGHV3-21 cases (6 cases). The incorporation of the SYBR Green dye into the PCR products was monitored in real time, and the resulting threshold cycles (Ct) (ie, the PCR cycle number corresponding to the beginning of the exponential growth of PCR products) were computed. Ct values were converted into attomoles by means of QRT-PCR experiments carried out with serial dilution of known concentrations of VIPR1-, TGFβ2-, WNT-16–, and β2M-specific amplicons. Results for each CLL case represent the values of the relative expression levels of VIPR1/β2M and TGFβ2/β2M in IGHV3-21 cases (red bars) and non–IGHV3-21 cases (blue bars) and WNT-16/β2M in homHCDR3–IGHV3-21 cases (yellow bars) and nonhomHCDR3–IGHV3-21 VH3-21 cases (orange bars). The panel on the bottom right represents ethidium bromide–stained agarose gels showing the expression of VIPR1, TGFβ2, and WNT-16 by conventional RT-PCR in 8 cell lines and in 3 representative CLL cases (B338, B80, and B419). Asterisks refer to molecular weight markers.
To elucidate the biologic functions of genes representing the differential expression signature of IGHV3-21 CLL, the gene identifiers for the 122 best-correlated probes were linked to 2 web-based bioinformatic tools for global analysis of gene function: “Onto-Express” and “Gene-Ontology Tree Machine.”37,38 Biologic process and molecular functions showing a significant enrichment for the genes found to be differentially expressed between IGHV3-21 and non–IGHV3-21 CLLs are summarized in Table S2. As an example, a group of 4 genes (RERG, ABI1, PMP22, TGFB2), all identified as negative regulators of cell proliferation, was selected among genes down-regulated in IGHV3-21 CLL cells along with a regulator of transcription (RUNX1). Conversely, genes up-regulated in IGHV3-21 CLLs were involved in positive regulation of cell proliferation (VIPR1), negative regulation of cell adhesion (RND1), as well as regulation of transcriptional (BRCA1) and oxidoreductase activities (HSD17B3, CYP3A5).
The differential expression of 2 of these genes, 1 up-regulated (VIPR1) and 1 down-regulated (TGFB2) in IGHV3-21 CLLs, was investigated by real-time quantitative PCR in representative CLL samples from both CLL subsets. As shown in Figure 5C, these experiments confirmed a higher expression of VIPR1, along with lower levels of TGFB2 transcripts, in IGHV3-21 CLL. Of note, both VIPR1- and TGFB2-specific mRNAs were detected in other human cell lines of different hematopoietic origin, although expression of VIPR1 seemed to be more restricted than that of TGFB2 (Figure 5C).
Differential GEP of homHCDR3–IGHV3-21 CLLs
A supervised analysis was performed to identify the differential GEP of homHCDR3–IGHV3-21 (7 cases) versus nonhomHCDR3–IGHV3-21 CLLs (6 cases).12,13,16 This comparison, made by applying a standard SAM35 statistical analysis with an adjusted P ≤ 10−2, yielded a list of 20 differentially expressed features (Table S3). Among these features, 12 were down-regulated and 8 up-regulated in homHCDR3–IGHV3-21 CLLs; in this latter group, the gene of known function with the highest score was WNT-16 (Table S3; Figure 5B). Real-time PCR experiments confirmed high WNT-16 expression in representative homHCDR3–IGHV3-21 CLL as well as in the Granta-519 B-cell line (Figure 5C).
Discussion
In the present report, we provide a comprehensive characterization of 37 IGHV3-21 CLLs collected out of a series of 1076 unselected cases enrolled by different reference centers located in N or CS Italy. The study addressed the geographic distribution of IGHV3-21 CLL in the context of different areas of the same country (ie, Italy) as well as the molecular, immunophenotypical, and clinical features of this CLL subset. The peculiar clinical behavior of IGHV3-21 CLL patients prompted us to investigate whether either IGHV3-21 or homHCDR3–IGHV3-21 CLLs were characterized by specific GEP explaining, at least in part, the disease's aggressiveness.12,13,16,17,39
Biased geographic distribution of IGHV3-21 CLL in Italy
An overrepresentation (about 10%) of IGHV3-21 CLL has been documented in several Northern European countries.12-15 Conversely, a multicenter study involving countries from Southern Europe, as well as studies on separate cohorts of patients, collectively indicated a significantly lower percentage of IGHV3-21 cases, ranging from 0% to about 3%.6,16,27,40
Our multicenter study reports a 3.4% incidence of IGHV3-21 CLL in Italy, a value similar to that observed in CLL collected from 4 Mediterranean countries (Greece, Italy, France, and Spain).6,16 Notably, our results point to heterogeneous distribution of homHCDR3–IGHV3-21 CLL between continental and Mediterranean areas of the same country (ie, Italy). Indeed, 16 of 18 (89%) homHCDR3–IGHV3-21 CLLs were enrolled by centers located in N Italy. Therefore, homHCDR3–IGHV3-21 CLL accounted for 3.0% (16 of 530) in N Italy but was virtually absent in CS Italy (2 of 546, 0.4%). A referral bias among N versus CS centers seems unlikely because (1) the frequency of IGHV usage by CLL cells overall was similar in N and CS patients and (2) the M/UM ratio (1.5 in the total series) was similar in N and CS cohorts of patients (1.44 and 1.66, respectively).
The incidence of homHCDR3–IGHV3-21 CLL in a smaller Italian series was 3 of 97 (3.1%)16 ; in this regard, it seems likely that the Italian series reported by the multicentric study of Southern European countries contains an unwanted bias, because such series was allegedly composed of patients recruited in N Italy. Curiously, the percentage of homHCDR3–IGHV3-21 CLLs detected by our study in CS Italy (0.4%) overlaps with that found in other Mediterranean areas, namely Greece and Spain.16 The reasons for the different incidence of homHCDR3–IGHV3-21 CLL in N versus CS Italy or, more in general, in typically Mediterranean areas versus continental regions are currently unclear.
Molecular characterization of IGHV3-21 CLL
As expected,6,12,13,16,17 almost 50% of IGHV3-21 cases displayed an IGHV gene rearrangement with a very short and homologous HCDR3 segment, often expressing the DANGMDV aa motif. HCDR3 cluster analysis documented that this 7-aa motif is very rarely used by IGHV rearrangements other than IGHV3-21. This observation lends further support to the putative involvement and to the high specificity of a common antigen responsible for the onset and behavior of IGHV3-21 CLL.3-5
All homHCDR3–IGHV3-21 CLLs expressed the combination IGLV3-21(Vλ2-14)/IGLJ3. The HCDR3 sequences of this light chain gene rearrangement always showed the conserved previously reported aa sequence.12,13,17 Of note, in 50% of cases (9 of 18 in our series), the functional rearrangement of IGLV3-21(Vλ2-14) coexisted with other functional IGLV rearrangement(s). This phenomenon seems to be circumscribed to homHCDR3–IGHV3-21 CLLs, because only 1 of 19 nonhomHCDR3–IGHV3-21 CLLs expressed 2 functional light chain rearrangements. Conceivably, a pairing between IGHV3-21/homHCDR3 and IGLV3-21(Vλ2-14) may be strongly favored, even if other IGLV rearrangements had primarily occurred in the B-cell clone.41,42
Clinical features and expression of immunophenotypic prognosticators in IGHV3-21 CLL
In agreement with previous studies evaluating the clinical impact of IGHV3-21 CLL versus non–IGHV3-21 cases,12,13 the survival of IGHV3-21 CLL in our series was comparable with that of UM patients. Differences in overall survival also held true when only the 15 homHCDR3–IGHV3-21 CLL cases were considered or when the 14 M IGHV3-21 cases and the 10 M homHCDR3–IGHV3-21 CLLs were compared with their M non–IGHV3-21 counterparts. Collectively, these findings suggest that we should withdraw IGHV3-21 cases when evaluating the prognostic impact of IGHV gene mutations in CLLs.
According to our data on overall survival, we failed to confirm the worse prognosis of homHCDR3–IGHV3-21 cases compared with nonhomHCDR3–IGHV3-21 CLLs suggested by one group6,16 but not by another.17 However, our results indicate that a set of well-established negative immunophenotypic prognosticators25,29-34 are more frequently expressed by homHCDR3–IGHV3-21 CLL than by nonhomHCDR3–IGHV3-21 cases. The low number of total cases, along with the presence of 4 of 8 UM CLLs in the homHCDR3–IGHV3-21 subset, prevents definitive conclusions on this matter. Notably, though, the finding of a differential gene expression profile distinguishing homHCDR3–IGHV3-21 from nonhom–IGHV3-21 CLL cells, characterized by the overexpression of genes allegedly enhancing the aggressiveness of CLL phenotype, could be in keeping with the association of homHCDR3–IGHV3-21 CLL with a poor-prognosis phenotype (see “GEP of IGHV3-21 CLL”).
GEP of IGHV3-21 CLL
Using an unsupervised approach, IGHV3-21 and non–IGHV3-21 CLL shared the large part of 28 513 features investigated. This finding is not surprising, because a similar approach was unable to split CLL into discrete categories.43,44 Nevertheless, subtle GEP differences of phenotypically related cell populations can be detected using supervised analyses.18,35,36,43 The multi-SAM approach used in this study had the additional advantage of overcoming the problem of an unbalanced representation of IGHV3-21 CLL (n = 13) versus non–IGHV3-21 CLL (n = 52) in our data set. The probes eventually selected for correlation with IGHV3-21 usage were 122, corresponding to 115 genes. As an internal control, IGLV3-21(Vλ2-14) fell among the best-ranked genes up-regulated in IGHV3-21 CLL, while probes related to IGHV3-21 were not available in the employed platform.
Genes down-regulated in IGHV3-21 CLL included 4 negative regulators of cell proliferation (RERG, ABI1, PMP22, TGFB2). In particular, TGFβ2, whose production by CLL has been previously demonstrated,45,46 had expression levels significantly lower in IGHV3-21 CLL cells by both GEP and RTQ-PCR validation experiments. Noteworthy, the down-regulated expression in IGHV3-21 CLL cells of 2 downstream molecules of the TGFβ signaling cascade (eg, MADH2/SMAD2 and AML1/RUNX1)45 has been demonstrated by Falt et al18 and in this study, respectively. In addition, the gene coding for the TGFβ receptor I was selected as down-regulated in IGHV3-21 CLL by multi-SAM, although not at a ranking as high as to be included in the 122 best-correlated probes. Furthermore, the regulatory subunit 15B (PP1R15B) of protein phosphatase 1 (PP1), a gene inhibiting the TGFβ signaling pathway, was significantly up-regulated in IGHV3-21 CLL and included in the 122 best-correlated probes. Taken together, present data and data reported by Falt et al18 collectively demonstrate that the TGFβ growth inhibition pathway is significantly hampered in IGHV3-21 CLL.
Genes with up-regulated expression in IGHV3-21 CLL cells according to GEP and RTQ-PCR validation experiments included VIPR1. Expression of VIPRs has been well documented in cells belonging to the immune system,47 where it promotes cell survival through a block of Fas/FasL-induced apoptosis.47,48 Of note, VIPR1 belongs to the family of the G protein–coupled receptors,49 and its triggering by the specific ligand activates proteins of the Rho guanosine triphosphatase (Rho GTPase) family to transmit downstream signals.49,50 In this regard, the gene encoding for Rho family GTPase1 protein (RND1) was up-regulated in IGHV3-21 CLL, as it was another gene from the same family, the guanidine nucleotide exchange factor 1 (GEF1), the latter according to GEP data reported by Falt et al.18
GEP of homHCDR3–IGHV3-21 CLL
A supervised analysis aimed at identifying the differential GEP of homHCDR3–IGHV3-21 versus nonhomHCDR3–IGHV3-21 CLLs was performed by applying a standard SAM statistical analysis. By setting the threshold of significance for the adjusted P value at 10−3 as made for multi-SAM, no differentially expressed genes were selected (D.M., personal communication, May 2006), thus suggesting that homHCDR3–IGHV3-21 and nonhomHCDR3–IGHV3-21 CLLs are more closely related than IGHV3-21 and non–IGHV3-21 cases. However, by lowering the stringency of the analysis and setting the statistical significance at an adjusted P value of less than 10−2, we identified as few as 20 genes that differentiated homHCDR3–IGHV3-21 from nonhomHCDR3–IGHV3-21 CLL by cluster analysis. Among the genes of known function differentially expressed with the highest score, WNT-16 was found to be up-regulated in homHCDR3–IGHV3-21 CLL, as also validated by RTQ-PCR.
Members of the WNT gene family encode for secreted glycoproteins known to regulate normal B-cell differentiation and survival51 and have been found to be active and overall up-regulated in CLL.44,52,53 In particular, WNT-3, WNT-6, WNT-10, and WNT-16 were comparably expressed in UM and M CLLs, while WNT-5b and WNT-14 levels were higher in UM CLL cells.44,52 The up-regulated expression of WNT-16 in homHCDR3–IGHV3-21 was paralleled by that of WNT-3, a gene selected as up-regulated in IGHV3-21 CLL cells by multi-SAM, although not at a ranking as high as to be included in the 122 best-correlated probes. Because an uncontrolled WNT signaling contributes to apoptosis impairment both in CLL and acute leukemias,52,53 the up-regulation of selected WNT members in IGHV3-21 and homHCDR3–IGHV3-21 CLL provides novel molecular evidence for the worse prognosis of this disease subset.
Authorship
Contribution: R.B., M.D.B., D.B., and M.D. performed part of the IGHV gene mutation and gene expression profiling studies and contributed to data analyses and writing of the manuscript; D.C., F.F., D.G.E., R. Maffei, E.M.G., and M.T. performed part of the IGHV gene mutation studies and contributed to writing of the manuscript; A.Z. performed immunophenotypical studies and contributed to writing of the manuscript; R.R., A.G., R.F., R. Marasca, and G.G. provided part of the IGHV gene sequences and contributed to writing of the manuscript; G.D.P., M.I.D.P., P.B., and L.L. provided clinical data of patients and some of the CLL cases; D.M., G.P., and R.C. performed all the bioinformatic analyses of gene expression profiling; and V.G. coordinated the study and data analyses and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Valter Gattei, Clinical and Experimental Hematology Research Unit, Centro di Riferimento Oncologico, I.R.C.C.S., Via Pedemontana Occidentale, 12, Aviano (PN), Italy; e-mail: vgattei@cro.it.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Supported in part by Ministero della Salute (Ricerca Finalizzata IRCCS and Alleanza Contro il Cancro), Rome; Programmi di Ricerca di Interesse Nazionale (PRIN) and Fondo per gli Investimenti per la Ricerca di Base (FIRB), Ministero dell'Università e della Ricerca (MUR), Rome; Associazione Italiana contro le Leucemie linfomi e mieloma (AIL)-Venezia, Pramaggiore group (M.D.B., fellowship); Novara-AIL Onlus, Novara; Ricerca Scientifica Applicata Regione Piemonte, Torino; Associazione Italiana per la Ricerca sul Cancro (AIRC), Milan; Piano di Ateneo per la Ricerca (PAR) 2005, University of Siena, Italy; The Leukemia & Lymphoma Society (White Plains, NY); Hairy Cell Leukemia Research Foundation (Shaumburg, IL); European Hematology Association 2004-Clinical Research Grant; and The Swedish Cancer Society.
References
Supplemental data
HCDR3 sequences from a total of 349 cases of known IGHV rearrangement enrolled by different institutions were converted into aa sequences and clustered by means of the multiple sequence alignment software ClustalW (available at the EMBL-EBI, European Bioinformatics Institute website, www.ebi.ac.uk/clustalw/; accessed June 2006), which was used also to draw the dendrogram. The length of HCDR3 was computed between codons 95 and 102, as described by Kabat.21 Only IGHV rearrangements for IGHV3-21 CLL cases are indicated (arrows next to their corresponding branch). The enlargement in the upper part of the figure corresponds to the area of the tree containing homHCDR3–IGHV3-21 cases and the only case with non–IGHV3-21 rearrangement. The dotted line indicates the minimum distance that characterized the cluster containing the homHCDR3–IGHV3-21 cases. The inset represents IGHV gene family usage of the 349 reported cases. The histogram indicates the frequencies for each IGHV family, computed for the whole series (black columns) and according to the IGHV mutational status (UM, gray columns; M, white columns).