Abstract
Gene expression profiles were examined in 33 adult patients with T-cell acute lymphocytic leukemia (T-ALL). Nonspecific filtering criteria identified 313 genes differentially expressed in the leukemic cells. Hierarchical clustering of samples identified 2 groups that reflected the degree of T-cell differentiation but was not associated with clinical outcome. Comparison between refractory patients and those who responded to induction chemotherapy identified a single gene, interleukin 8 (IL-8), that was highly expressed in refractory T-ALL cells and a set of 30 genes that was highly expressed in leukemic cells from patients who achieved complete remission. We next identified 19 genes that were differentially expressed in T-ALL cells from patients who either had a relapse or remained in continuous complete remission. A model based on the expression of 3 of these genes was predictive of duration of remission. The 3-gene model was validated on a further set of T-ALL samples from 18 additional patients treated on the same clinical protocol. This study demonstrates that gene expression profiling can identify a limited number of genes that are predictive of response to induction therapy and remission duration in adult patients with T-ALL. (Blood. 2004;103:2771-2778)
Introduction
Acute lymphocytic leukemia (ALL) has been studied extensively for more than 4 decades. As a result, much is known about the cellular origin of this leukemia and the genetic mechanisms that lead to malignant transformation.1-3 Based on the expression of lineage-specific antigens and the presence of lineage-specific gene rearrangements, ALL cells are known to be derived from either B- or T-cell precursors.4 In B-lineage ALL, malignant cells often have additional specific genetic abnormalities, which have a significant impact on the clinical course of the disease. In contrast, few molecular abnormalities have been detected in T-cell ALL (T-ALL) and no cytogenetically defined prognostic subgroups have been identified.
The introduction of microarray technology5-7 has made it possible to simultaneously quantify expression of thousands of genes within well-defined cellular populations. This method has been used to examine gene expression profiles of malignant cells and recent studies have identified signatures characteristic of various hematologic and nonhematologic tumors.8-15 Gene expression profiles have also identified sets of genes that classify subsets of patients with distinct outcomes.13,15-17 Further applications of this technology will determine whether gene expression profiles can be used to identify distinct genetic pathways of malignant transformation in different cell types and whether new targets for therapy can be identified.18
In the current study, we used oligonucleotide microarrays to determine gene expression profiles in leukemia cells from 33 adult patients with newly diagnosed T-ALL. Analysis of ALL cells included the uniform characterization of immunologic markers, as well as cytogenetic and molecular abnormalities. These experiments identified distinct sets of genes associated with response to therapy and long-term maintenance of disease remission. The prognostic significance of 3 of these genes was confirmed by subsequent analysis of an independent set of 18 adult patients with T-ALL treated on the same clinical protocol.
Patients, materials, and methods
Patient characteristics
Patients were enrolled in the Italian Gruppo Italiano Malattie Ematologiche dell'Adulto (GIMEMA) multicenter clinical trial 0496 for adult patients with newly diagnosed ALL.19 Written informed consent was obtained from all patients prior to therapy. The present study was approved by the institutional review board of the university “La Sapienza” of Rome. Pretreatment leukemia samples from bone marrow or peripheral blood or both were centrally processed at the University “La Sapienza” of Rome, where mononuclear cells were isolated by density gradient centrifugation, cryopreserved with 10% dimethyl sulfoxide, and stored in vapor-phase liquid nitrogen. Thirty-three samples containing more than 90% blasts were used for gene expression analysis. Leukemia samples from 18 additional patients were used to test the 3-gene model developed from gene expression profiling. Conventional immunophenotypic, cytogenetic, and molecular diagnostic studies were uniformly carried out on all samples.20 Immunophenotypic analysis was performed using monoclonal antibodies specific for the following antigens: TdT, HLA-DR, CD7, CD19, CD22, CD10, CD14, CD33, CD13, CD61, CD34, CD2, myeloperoxidase, and surface (s) CD3. For further characterization, expression of the following panel of T-cell-associated antigens was used to establish the degree of differentiation of leukemic blasts: cytoplasmic (cy) CD3, CD5, CD1a, CD4, and CD8, and samples were classified according to the criteria of the European Group of Immunological Characterization of Leukemia (EGIL).21 Immunophenotypic and cytogenetic characteristics of all 51 ALL samples are summarized in Table 1.
Of the 33 patients evaluated by gene expression profiling, 6 were refractory to induction chemotherapy, 2 died during induction chemotherapy, and 25 achieved complete remission (CR). Of these, 16 had a relapse within a median period of 12.5 months (range, 3-43 months); 8 are in continuous complete remission (CCR) with a median follow-up of 41 months (range, 12-62 months); and 1 underwent stem cell transplantation and was not considered for outcome analysis. All 18 patients analyzed exclusively by quantitative polymerase chain reaction (PCR) achieved CR after induction therapy; 13 experienced a relapse within a median period of 10 months (range, 6-19 months), and 5 are in CCR (median follow-up, 30 months; range, 21-58 months).
RNA extraction and oligonucleotide microarray methods
Cryopreserved leukemia cells were rapidly thawed; total RNA was extracted using TRizol reagent (Gibco, Grand Island, NY) and purified using SV total RNA isolation system (Promega, Madison, WI), with minor modifications. HGU95aV2 gene chips (Affymetrix, Santa Clara, CA) were used to determine gene expression profiles. The detailed protocol for sample preparation and microarray processing is available on the manufacturer's website (http://www.affymetrix.com/support/technical/manual/expression_manual.affx). CEL files (Affymetrix cell intensity file) are available at http://bioconductor.org/Docs/Papers/2002/Chiaretti/.
Real-time quantitative PCR analysis
Real-time quantitative reverse transcription-PCR (RT-PCR) analysis was performed using an ABI PRISM 7700 sequence detection system and the SYBR green I dye (PE Biosystems, Foster City, CA) method as previously described.22 Real-time PCR conditions were as follows: 1 cycle at 50°C for 2 minutes, 1 cycle at 95°C for 10 minutes, 1 cycle at 95°C for 15 seconds, 1 cycle at 60°C for 1 minute, for a total of 40 cycles. For each sample, CT values for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin were determined for normalization purposes. For each run of samples, a correction factor was calculated by dividing by the minimum GAPDH and β-actin values and then averaging the adjusted control values for the samples. This value was used to correct each sample for difference in RNA content. ΔCT was calculated by subtracting the mean of one group of samples from the mean of the other group of samples. Primers were designed using Primer Express 1.0 software (PE Biosystems). The following primers were used: 5′AHNAK: 5′-ATGCTCCAGGGCTCAACCT-3′; 3′AHNAK: 5′-CGTGCCCCAACGTTAAGCTT-3′; 5′TTK: 5′-TGCCACCACAAGATGAGAA-3′; 3′TTK: 5′-GACTCTTCCAAATGGGCATGA-3′; 5′IL-8: CAATGCGCCAACACAGAAAT-3′; 3′ IL-8: 5′-TCTCCACAACCCTCTGCACC-3′; 5′CD2: 5′-CCAGCCTGAGTGCAAAATTCA-3′; 3′CD2: 5′-GACAGGCTCGACACTGGATTC-3′; 5′β-actin: 5′-AGTACTCCGTGTGGATCGGC-3′; 3′β-actin: 5′-CTTGCTGATCCACATCTGCTG-3′; 5′ GAPDH: 5′-CCACCCATGGCAAATTCC-3′; 3′GAPDH: 5′-GATGGGATTTCCATTGATGACA-3′.
Statistical methods
Affymetrix gene expression data were processed and analyzed with dChip (www.dchip.org), which uses an invariant set normalization method. Model-based expressions were computed for each array and probe set.23 Experimental data quality assessment is provided by dChip software. Hierarchical clustering was used as described by Eisen et al.24 All other analyses were carried out using the R language (http://cran.r-project.org/).25
Filtering criteria were defined to select genes for further analysis. Gene selection criteria required the expression level to be higher than 100 in more than 20% of the samples and the ratio of the SD to the mean expression across samples to be between 0.7 and 10 (dChip default values). To select genes that were differentially expressed in subgroups of interest, different statistical tests (eg, the t test) were applied and genes selected based on the nominal P values attained. Genes associated with response to therapy and outcome were selected using either a t test (refractory versus CR) or a proportional hazards model (duration of CR).
Several methods of analysis used cross-validation (CV), a statistical procedure that can be used for estimating the error rate of a prediction method, model selection, or variable selection.26 The basis of CV is to repeatedly partition the data into 2 groups. One group (the training set) is used to estimate the parameters of a statistical model and the second group (the test set) is used to assess the predictive capability of the model. One form of CV is leave-one-out CV, that is, the size of training set versus the test set are n-1:1. In this method, each observation is used as the test set, in turn, and the remaining n-1 observations are the training set. Leave-one-out CV was used for analysis of all gene expression data.
The CV estimation of prediction error rate is obtained by averaging the observed error rate for each of the test sets. To select a model by CV we first determined the CV error rate for all models of interest and then selected the model with the smallest CV error rate. To assess the error rate it is important that variable selection is performed within each CV step. Finally, CV can be used for variable selection by selecting the best performing (according to some criterion) variables for each training set and then selecting those appearing more often. We deemed a gene a reliable predictor if it was selected in more than 75% of the training sets. We found this process a valuable mechanism for excluding genes whose apparent effect was only due to a limited number of samples.
Linear discriminant analysis26 was used to classify patients into specific groups and classification rates were estimated by CV. Proportional hazard models26 were used to explore the relationship between the duration of CCR and the genes of interest. Kaplan-Meier and log-rank tests were used to estimate the probability of CCR in patients with available follow-up information.
Results
Hierarchical clustering reflects the degree of differentiation of leukemic cells
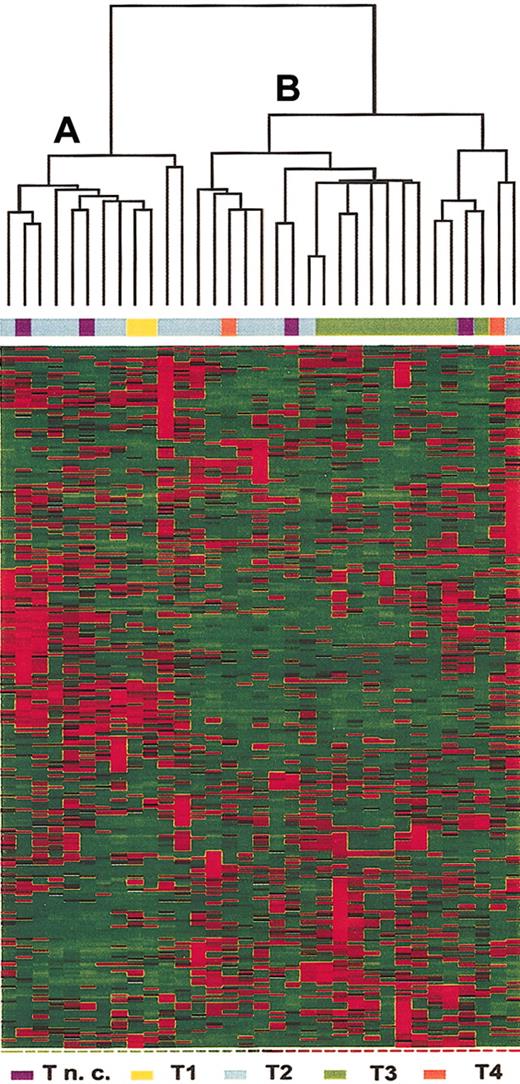
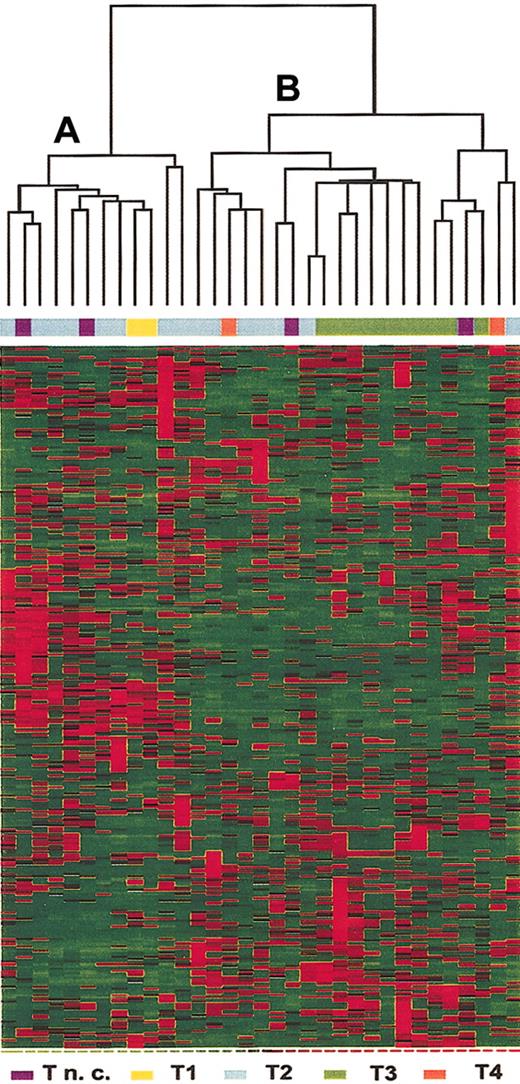
Gene selection criteria were applied to identify genes that were differentially expressed in T-ALL cells from adult patients. As shown in Figure 1, 313 genes (see Supplemental Table S1 on the Blood website; click on the Data Set link at the top of the online article) were selected and hierarchical clustering of the 33 samples identified 2 major groups. Group A included most of the samples classified as relatively undifferentiated T-ALL (2 of 2 T1, 8 of 15 T2, and 2 samples not further subclassified). Group B included most of the leukemias with phenotypic evidence of further T-cell differentiation (7 of 15 T2, 10 of 10 T3, and 2 of 2 T4, and 2 samples not further subclassified). The samples without further phenotypic subclassification were excluded. A Fisher exact test for association between clusters (group A and B) and levels of T-cell differentiation was performed and proved highly significant (P < .01).
Hierarchical clustering of the 33 T-ALL samples based on expression of 313 selected genes. Each column represents a sample and each row represent a gene. Relative levels of gene expression are depicted with a color scale where red represents the highest level of expression and green represents the lowest level. Unsupervised clustering identified 2 subsets of samples, A and B.
Hierarchical clustering of the 33 T-ALL samples based on expression of 313 selected genes. Each column represents a sample and each row represent a gene. Relative levels of gene expression are depicted with a color scale where red represents the highest level of expression and green represents the lowest level. Unsupervised clustering identified 2 subsets of samples, A and B.
Gene expression profile associated with response to therapy
To identify genes associated with response to treatment, we compared expression profiles of T-ALL samples from 25 patients who achieved CR and 6 patients who were refractory to induction therapy. CV for feature selection (a t test with P cut-off of .05 was the criterion used to select a gene as best performing) was used to identify 34 genes deemed reliable predictors of response to therapy (Figure 2). Prediction of response to treatment was assessed using linear discriminant analysis (LDA).
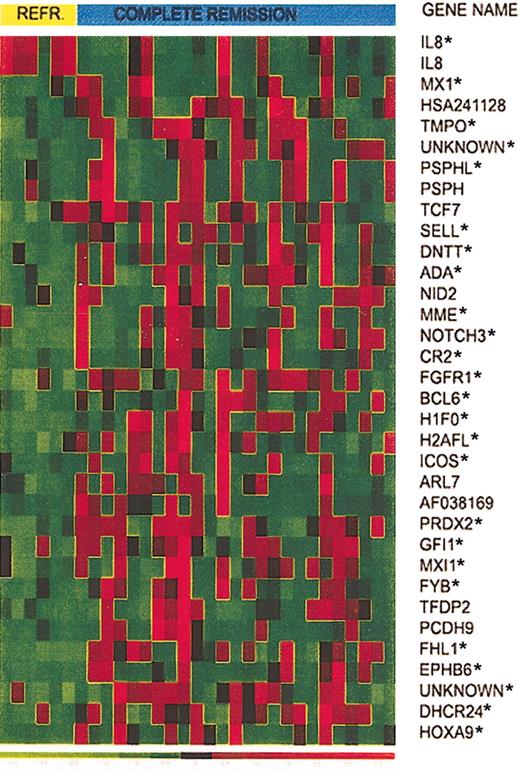
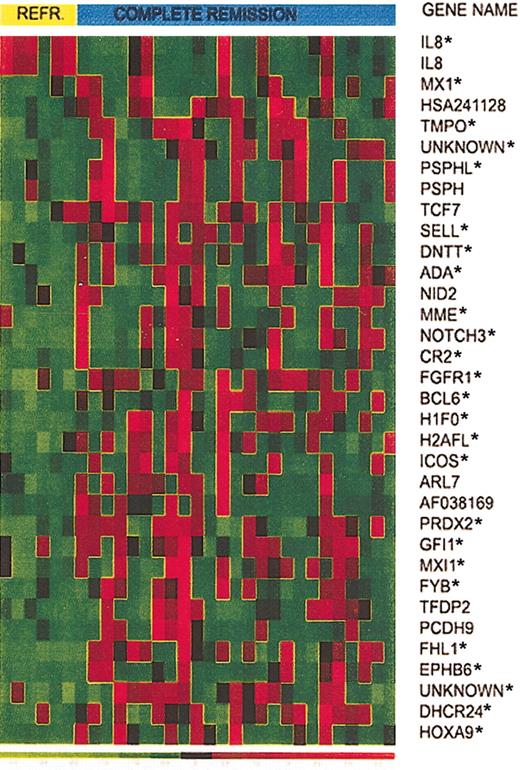
Expression of 34 selected genes in responding versus refractory T-ALL patients. Expression of 34 selected genes in T-ALL from 6 patients who did not respond to induction therapy and 25 patients who achieved CR. Asterisks identify the top 25 genes that provide the smallest prediction error.
Expression of 34 selected genes in responding versus refractory T-ALL patients. Expression of 34 selected genes in T-ALL from 6 patients who did not respond to induction therapy and 25 patients who achieved CR. Asterisks identify the top 25 genes that provide the smallest prediction error.
CV for model selection was used to determine the number of genes to use in the LDA. To do this, we left each observation out, in turn. On the test set (n - 1), we ranked the 31 reliable predictors by t test P value. We then created models with k predictors (from k = 1 to k = 28, ordered by P value; beyond 28 numerical problems were encountered) and for each model predicted the outcome of the held-out sample. Then for each k, over all CV samples, the prediction error rate was estimated. The minimum error rate occurred at k = 25 genes (identified by the asterisk in Figure 2), where 19 of the 25 responders and 4 of the 6 refractory patients were correctly predicted.
Whereas the profile of patients who achieved CR was heterogeneous, the refractory group showed a homogeneous pattern, characterized by the high expression of interleukin 8 (IL8) and reduced expression of the remaining genes. Genes expressed at lower levels in the refractory group include a set of genes belonging to the histone family (H1F0 and H2AFL) and several genes that have a role in cell adhesion (CR2, SELL) and cell cycle progression (GFI1, BCL6). This pattern suggests impaired cell proliferation in T-ALL cases that did not respond to therapy. Another low-expressed gene in this group is Max-interacting protein 1 (MXI1), a transcriptional repressor of myc, which is thought to be a tumor suppressor gene. CD10, often considered a marker of better survival in children with pre-B and T-ALL, and recently shown to be characteristic of cycling, apoptosis prone cells,27-29 was expressed at lower levels in the refractory group.
Gene expression profile associated with long-term outcome
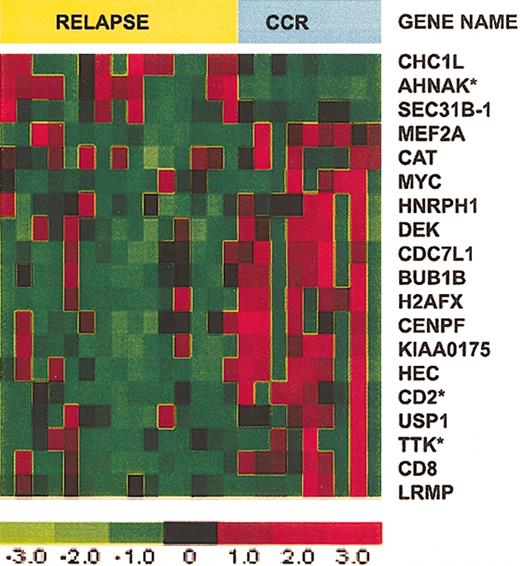
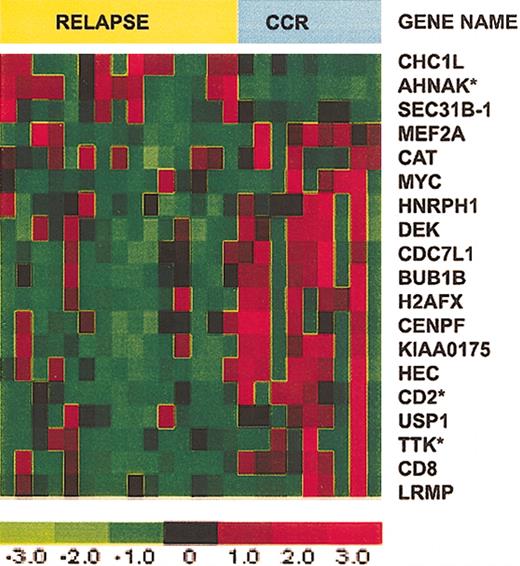
Our next analysis compared patients who had a relapse within 2 years with patients who were in CCR for at least 2 years. Patients in CCR whose follow-up was less than 2 years and the single patient who underwent stem cell transplantation were excluded from this analysis. Eight patients were in CCR, 16 experienced a relapse, and 2 patients had a relapse more than 2 years after achieving CR; these latter samples were included in the CCR group. CV for feature selection, with selection based on a univariate Cox model P cut-off of .05, identified 19 reliable predictors (Figure 3). BUB1B, TTK, and CENPF, which play a role in mitotic assembly and mitotic checkpoint, were selectively expressed in T-ALL cells from patients with a long duration of CCR. CD2 was also more highly expressed in this group. Among the few genes with increased expression in the relapse group, we identified AHNAK, which encodes an unusually large 700-kDa protein with poorly defined function.30
Expression of 19 selected genes in T-ALL patients who relapsed versus those who remained in CCR. Expression of 19 selected genes in T-ALL from 8 patients who remained in CCR for more than 2 years and 16 patients who experienced a relapse less than 2 years after achieving CR. Asterisks identify the top 3 genes that provide the smallest prediction error.
Expression of 19 selected genes in T-ALL patients who relapsed versus those who remained in CCR. Expression of 19 selected genes in T-ALL from 8 patients who remained in CCR for more than 2 years and 16 patients who experienced a relapse less than 2 years after achieving CR. Asterisks identify the top 3 genes that provide the smallest prediction error.
CV for model selection was used to determine the number of genes to use in the multivariate Cox model. To do this, we left out each observation in turn. On the test set (n - 1), we used the 19 reliable predictors and a forward selection algorithm to fit models with between 1 and 5 genes. We used each model (k from 1 to 5) to predict the status of the held out sample. Then for each k, over all CV samples, we estimated the prediction error rate. Models with k = 1 and k = 3 tied for the minimum error rate. In both cases we identified genes that were selected more than 75% of the time: for k = 1 this was AHNAK and for k = 3 AHNAK, CD2, and TTK satisfied this criterion. The mean expression values determined by microarray for AHNAK, CD2, and TTK in the CCR and relapsed groups are shown in Table 2.
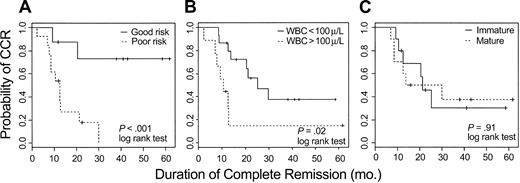
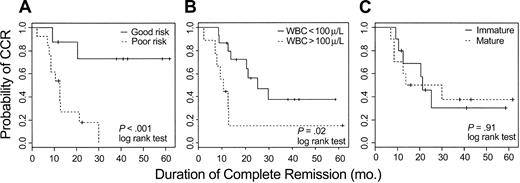
The clinical impact of this analysis was examined by categorizing the 24 evaluable patients into good- or poor-risk cohorts based on expression values for AHNAK, TTK, and CD2. For each patient, in turn, the remaining 23 patients were used to construct a Cox model on the basis of their outcome and observed values of AHNAK, CD2, and TTK. The probability of being in CCR at 2 years was predicted for the held-out patient, based exclusively on the observed values of these 3 genes; if that probability exceeded .5 a patient was defined as good risk; otherwise the patient was defined as poor risk. Using the 3-gene model, 11 patients were classified as good risk; 6 remain in CCR and 5 had a relapse after 9, 12, 14, 30, and 43 months. Thirteen patients were classified as poor risk; 11 have experienced a relapse and 2 remain in CCR. Kaplan-Meier curves comparing duration of remission for the good and poor risk groups determined by this 3-gene model are shown in Figure 4A (log-rank test < .01).
Kaplan-Meier plots estimating probability of maintaining CR for adult T-ALL. (A) Twenty-four evaluable patients were assigned to either good-risk or poor-risk T-ALL based on expression of AHNAK, CD2, and TTK as measured by oligonucleotide microarrays. (B) Kaplan-Meier plots based on the WBC count at diagnosis. (C) Kaplan-Meier plots based on the degree of T-lineage differentiation of the leukemic cell (immature = T1-T2; mature = T3-T4 of EGIL classification).
Kaplan-Meier plots estimating probability of maintaining CR for adult T-ALL. (A) Twenty-four evaluable patients were assigned to either good-risk or poor-risk T-ALL based on expression of AHNAK, CD2, and TTK as measured by oligonucleotide microarrays. (B) Kaplan-Meier plots based on the WBC count at diagnosis. (C) Kaplan-Meier plots based on the degree of T-lineage differentiation of the leukemic cell (immature = T1-T2; mature = T3-T4 of EGIL classification).
In contrast to pre-B ALL, in which certain molecular aberrations are associated with different outcome, no such well-defined molecular rearrangements are clearly established for T-ALL. Clinical markers such as high white blood cell (WBC) count (> 100 000/μL) at presentation and the degree of differentiation, determined by flow cytometry, of the leukemic blasts have been reported to be associated with outcome.31 We therefore also evaluated these conventional approaches for predicting outcome. As shown in Figure 4B-C, both conventional parameters appeared to be less predictive than the 3-gene expression model.
Real-time quantitative PCR analysis
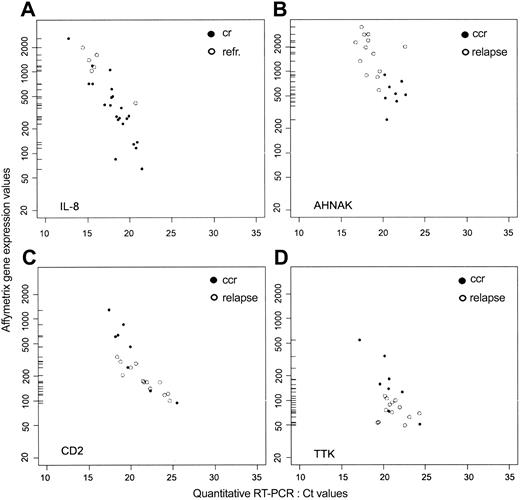
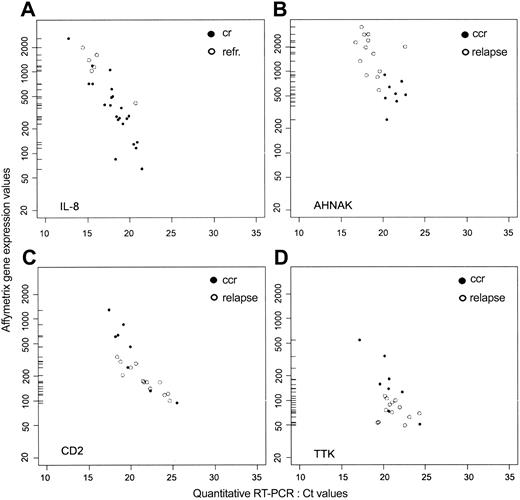
To confirm the results of microarray analysis, quantitative RT-PCR was performed on 4 genes, IL-8, AHNAK, TTK, and CD2. IL-8, the only gene that was more highly expressed in refractory leukemic cells, was quantified in 24 samples (6 refractory and 18 CR); AHNAK, TTK, and CD2 were quantified in 21 samples (7 CCR and 14 relapse). As shown in Figure 5, there was a significant correlation (Pearson correlation coefficient) between the RT-PCR and microarray data for each gene. Because high CT values from quantitative RT-PCR correspond to low values of gene expression from microarray analysis, we found a negative correlation for each gene: IL-8 = -0.85; AHNAK = -0.62; CD2 = -0.70; and TTK = -0.58 (Figure 5A-D). CD2 expression was also evaluated by flow cytometry. Results in Table 3 show a high degree of correlation between all 3 methods used to assess CD2 expression in these samples. Taken together, these results support the validity of the expression data obtained by oligonucleotide microarrays for this set of 4 genes.
Gene expression values in adult T-ALL measured by oligonucleotide microarray and quantitative RT-PCR. For IL8, ○ represents refractory patients, and • represents patients who achieved CR. For AHNAK, CD2, and TTK, ○ represents patients who experienced a relapse, and • represents patients who remain in CCR.
Gene expression values in adult T-ALL measured by oligonucleotide microarray and quantitative RT-PCR. For IL8, ○ represents refractory patients, and • represents patients who achieved CR. For AHNAK, CD2, and TTK, ○ represents patients who experienced a relapse, and • represents patients who remain in CCR.
Validation of the 3-gene model for predicting long-term outcome
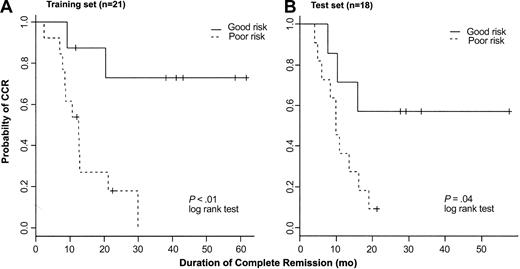
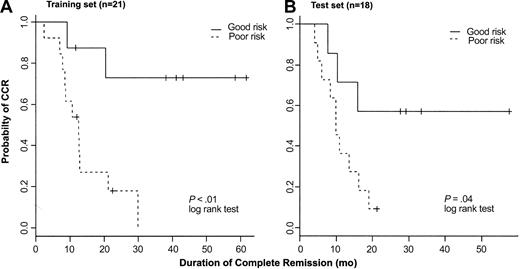
To further test the predictive model for outcome based on expression of AHNAK, CD2, and TTK, we used the quantitative RT-PCR data obtained from the 21 samples (training set) to fit a Cox proportional hazard model. To assess the consistency of the 3-gene model based either on array data or quantitative RT-PCR, we conducted CV to estimate the classification errors on both datasets of the 21 T-ALL samples. For the array data, 9 patients were classified as good risk and 6 remain in CCR; 12 patients were classified as poor risk and 11 have experienced a relapse. The overall error rate based on oligonucleotide array data is 19%. Using quantitative RT-PCR, 8 patients were classified as good risk and 5 remain in CCR; 13 patients were classified as poor risk and 11 have had a relapse. The overall error rate using RT-PCR data was 24%. Receiver operator characteristics (ROCs) were also examined, confirming the reliability of this model (data not shown). Figure 6A shows the Kaplan-Meier curves for the 2 prognostic groups based on RT-PCR data (P < .01). These results are similar to results shown in Figure 4 based on oligonucleotide array data. Given the limited sample size, results based on both of these methods are very consistent with each other.
Kaplan-Meier plots. Kaplan-Meier plots represent probability of maintaining CR in a training set of 21 patients (A) and a test set of 18 patients (B) with T-ALL treated on the same clinical protocol. Patients were assigned to either good-risk or poor-risk T-ALL based on expression of AHNAK, CD2, and TTK as measured by RT-PCR.
Kaplan-Meier plots. Kaplan-Meier plots represent probability of maintaining CR in a training set of 21 patients (A) and a test set of 18 patients (B) with T-ALL treated on the same clinical protocol. Patients were assigned to either good-risk or poor-risk T-ALL based on expression of AHNAK, CD2, and TTK as measured by RT-PCR.
Subsequently, we analyzed T-ALL samples from 18 additional patients treated on the same clinical protocol (test set). Because of the limited amount of RNA available from these samples, only quantitative RT-PCR was performed to measure the gene expression of AHNAK, CD2, and TTK. In the 18-sample test set, 7 patients were classified as good risk and 11 as poor risk. Four of 7 good-risk patients remain in CCR and 10 of 11 poor-risk patients have had a relapse. The overall error rate in the test set was 22%. Kaplan-Meier plots showing probability of maintaining CR for each prognostic group are shown in Figure 6B (P = .04).
Lack of predictive value of gene expression associated with long-term outcome in pediatric T-ALL
Two recent reports have identified distinct sets of genes associated with long-term outcome in T-ALL in pediatric patients. Yeoh et al15 identified a set of 7 genes (CD44, KIAA0056, FLJ38984, TBXAS1, GRAP2, PRSAP2, HNRPH2) that were differentially expressed in T-ALL in a group of 34 pediatric patients who were either in CCR or had a relapse after achieving CR. In a second study, Ferrando et al32 found that high-level HOX11 expression in pediatric T-ALL was significantly associated with maintenance of long-term remission. Thus, we specifically examined the levels of expression of these selected genes measured by oligonucleotide arrays in adult T-ALL samples. Using expression values for the 7 genes reported by Yeoh et al, each of our 24 evaluable adult T-ALL patients was categorized into either good or poor prognostic groups using a model based on the remaining 23 samples. Twelve samples were categorized as good risk and 12 were poor risk; the overall error rate of the model based on the 7 pediatric T-ALL genes was 41%.
We also examined whether expressions of AHNAK, CD2, and TTK were associated with outcome in the pediatric T-ALL patients described by Yeoh et al.15 Clinical outcome was available for 36 pediatric patients (26 CCR and 8 relapses). Expression of CD2 in T-ALL cells was significantly associated with outcome (P = .03, t test), but expression of the other 2 genes was not significantly different in leukemia cells from pediatric patients who remained in CCR compared with those who had a relapse after therapy.
Assessment of HOX11 demonstrated that this gene was highly expressed in 6 of 33 adult patients in our test set; one died in CR, one was refractory to induction chemotherapy, and 3 had relapses. In contrast to pediatric patients where HOX11 expression was associated with a favorable outcome, only one of 6 adult patients with high-level HOX11 expression remains in CCR.
Discussion
The goal of this study was to identify a gene expression signature in adult T-ALL that would have prognostic value in terms of response to induction therapy and long-term outcome. We took advantage of the ongoing policy of characterizing and storing pretreatment leukemia cells from all patients enrolled in the Italian adult ALL multicenter GIMEMA protocols. Our first approach was to use nonspecific filtering criteria to identify genes that were differentially expressed in adult T-ALL samples; a large set of 313 genes was identified and clustering of the 33 patients using these genes identified 2 groups of patients. Further inspection of these groups showed that this clustering mainly reflected the stage of differentiation of the T-ALL blasts.33 One group primarily included T-ALL with a more differentiated phenotype, whereas the other group included most of the samples with a more immature T-cell phenotype. Although 2 different groups of patients were identified in this analysis, this classification had no correlation with response to induction chemotherapy or with long-term outcome.
To identify genes associated with response to induction therapy, additional filtering criteria were established and 31 genes were selected. Classification errors were assessed by leave-one-out CV, that is, leaving one sample out, selecting the top k genes, and using the top k genes to predict the class (CR versus refractory) for the left-out sample. The misclassification rate was estimated to be 26%, or the correct prediction rate 74%. The best prediction occurred at k = 25, but only one of these genes, IL-8, was more highly expressed in refractory T-ALL. All of the remaining genes were more highly expressed in the leukemia cells that responded to induction therapy. This result was independently confirmed by quantitative RT-PCR.
IL-8 is a chemoattractant cytokine reported to play a role in several hematologic malignancies. In chronic lymphocytic leukemia (CLL), increased IL-8 mRNA expression has been associated with prolonged survival of CLL cells,34,35 and increased serum levels of IL-8 were found in patients with CLL.34,36 More recently, Liu et al37 demonstrated increased plasma IL-8 levels in patients with acute myeloid and acute lymphocytic leukemia (AML and ALL, respectively), showing an inverse correlation between CR rate and IL-8 levels. Our results are consistent with these earlier observations and show a significant difference in IL-8 mRNA expression by microarray and quantitative PCR in refractory T-ALL.
Induction of CR was associated with increased expression of a diverse set of genes. Many of these genes play a role in the transition from G1 through the S phase of the cell cycle. Many of the agents used for leukemia induction chemotherapy act in a cell cycle-dependent fashion and the increased expression of genes associated with cell cycle progression in these blasts is therefore consistent with the increased susceptibility of highly proliferating cells to treatment. The expression of these genes in leukemia cells that were sensitive to chemotherapy was very heterogeneous. In contrast, low expression of these genes was uniformly found in T-ALL cells from patients who were refractory to treatment.
Comparison of profiles in leukemia cells from patients who remained in CCR for more than 2 years or relapsed within this period identified 19 genes that were differentially expressed in these 2 groups. Three genes had increased expression in leukemia cells from patients who experienced a relapse and 16 genes were more highly expressed in blasts from patients who remained in remission. Leave-one-out CV was then used to identify those genes that were most consistently associated with duration of remission. This analysis identified 3 genes, AHNAK, CD2, and TTK, that were highly predictive and together correctly classified 71% of outcomes. Individually, AHNAK expression was a better predictor of long remission duration, whereas CD2 and TTK were better predictors of short remission duration (data not shown). Of these genes, AHNAK had increased expression in blasts from patients who subsequently had a relapse, whereas CD2 and TTK had increased expression in patients who remained in remission. Using microarray expression values for these 3 genes in leukemia cells, we categorized patients into either poor- or good-risk groups and found that maintenance of long-term CR was significantly different in the 2 groups. Similar results were obtained using data derived from quantitative RT-PCR. Furthermore, these 3 genes were also evaluated by quantitative RT-PCR in an additional set of 18 patients. Analysis of this independent test set of patients who were treated on the same protocol confirmed the significant association of the expression of these 3 genes with long-term clinical outcome.
AHNAK is an unusually large 700-kDa protein encoded by an intronless gene located on chromosome 11q12-13.38 Although the cellular function of AHNAK is unknown, it can become highly phosphorylated and appears to be involved in several signal transduction pathways.30 The expression of AHNAK is regulated in a cell cycle-dependent manner, with highest levels of RNA and protein occurring during the G1 or G0 phase, and subsequent decrease in proliferating cells. AHNAK phosphorylation also appears to be cell cycle dependent with highest levels of unphosphorylated protein occurring in the G1 phase. AHNAK is localized in the cytoplasm at low Ca2+ concentrations, but translocates to the plasma membrane in response to increased extracellular calcium.39,40 AHNAK was originally identified by differential display as a gene with high-level expression in pheochromocytoma, melanoma, and some leukemia cell lines but low-level expression in neuroblastoma and Burkitt lymphoma. Although recent studies have begun to characterize the involvement of AHNAK in several functional pathways, the function of this protein in leukemia cells has not previously been investigated.
TTK, initially isolated from a T-cell cDNA library,41 is a dual-function kinase that possesses serine/threonine and tyrosine kinase activity. TTK mRNA levels are elevated in proliferating cells compared to resting cells, with high expression in the S phase and a peak in G2/M.42 TTK has been implicated in the regulation of multiple cell cycle-related processes, including the duplication of the spindle pole body and the correct segregation of chromosomes during cell division.43 This gene was generally expressed at very low levels in most of the T-ALL samples and higher levels of expression were primarily found in the subset of T-ALL that remained in prolonged remission. These observations suggest that impaired expression or function of TTK may contribute to a more aggressive phenotype in T-ALL. When combined with up-regulation of AHNAK gene expression, the down-regulation of TTK in T-ALL cells may contribute to the maintenance of a relatively quiescent state and subsequent resistance to intensive and prolonged combination chemotherapy.
CD2 is a type I transmembrane glycoprotein that is expressed by natural killer (NK) cells, as well as T cells.44 This antigen was originally identified as the surface receptor for sheep red blood cells (E-rosette receptor) and the expression of this antigen by ALL cells provided the first evidence that these leukemias were derived from the T-cell lineage. This surface antigen is expressed relatively early in T-cell differentiation45,46 and is involved in cell adhesion and formation of the T-cell synapse.47,48 Cross-linking of CD2 receptors on T and NK cells also induces activation of these cells, resulting in either cell proliferation or apoptosis.49-51 Increased levels of CD2 gene expression measured by microarray analysis correlated well with CD2 mRNA levels measured by quantitative RT-PCR and surface expression of this antigen determined by flow cytometry. Previous studies by Uckun et al examined the clinical significance of CD2 expression in a large cohort of pediatric patients with T-ALL,52 reporting that high levels of CD2 antigen determined by flow cytometry were associated with a better outcome. Our analysis of CD2 also found that high-level gene expression was associated with improved outcome in adult patients, confirming these earlier observations, and extending them to adult patients with T-ALL.
Because the number of samples analyzed is relatively small, we have been cautious in our statistical approach and have relied on CV as a basis for determining many of the components of our final model and conclusions. In all cases of model selection, the process of feature selection was carried out separately for each CV iteration. When using CV for gene selection we used a preselected value for calling a feature a reliable predictor. The strong concordance between estimates of mRNA abundance between the RT-PCR data and the microarray data were reassuring. Moreover, the strong relationship between duration of remission and the mRNA levels of AHNAK, TTK, and CD2 held regardless of the method used to assay mRNA abundance. Finally, the association of expression of these 3 genes with long-term clinical outcome was confirmed by analysis of an independent set of 18 adult patients with T-ALL who were treated on the same clinical protocol.
The present study investigates, for the first time, the identification of gene expression profiles associated with both short-term and long-term outcome in adult patients with T-ALL. Whereas approximately 70% of pediatric patients with both B- and T-lineage ALL have excellent long-term response to intensive combination chemotherapy, adult patients have a much less favorable outcome. In B-lineage ALL, the poor prognosis of adult patients is partly due to the presence of specific genetic translocations, such as the BCR/ABL or ALL1/AF4 gene rearrangements.1 In contrast, the poor prognosis of patients with T-ALL has thus far not been attributed to specific molecular rearrangements. Recently, expression of HOX1132 has been associated with good prognosis in pediatric patients with T-ALL. A related gene, HOX11L253 was associated with poor prognosis in these patients. HOX11L2 is not represented in the HGU95aV2 platform, but probes for HOX11 are present and this allowed us to evaluate the association of this gene with outcome in adult patients with T-ALL. In contrast to pediatric patients, expression of HOX11 determined by microarray did not correlate with a more favorable outcome in adult patients.
In an extensive analysis of 360 pediatric patients with ALL, Yeoh et al specifically examined gene expression patterns in 34 patients with T-ALL.15 HOX11 was not identified as a prognostic marker in these patients, but a distinct set of 7 genes was found to be selective expressed in T-ALL cells at high risk for relapse. Thus, we specifically examined the association of these 7 genes with long-term outcome in our cohort of adult patients. Expression of these genes was not significantly associated with remission duration in adult T-ALL. Conversely, of the 3 genes that were most predictive of remission duration in our adult patients, only CD2 was significantly associated with outcome in the pediatric T-ALL patients. As noted previously, this is in agreement with prior studies.52 Given the marked differences in the biology and response to treatment between pediatric and adult ALL, it is not surprising that different genes are associated with outcome in these patients. Taken together, these observations emphasize the important differences between adult and childhood T-ALL and suggest that the genetic mechanisms of resistance to chemotherapy in these diseases are likely to be very different.
The findings that a single gene was closely associated with resistance to first-line treatment and that 3 genes were highly predictive of outcome in uniformly treated adults with T-ALL need to be verified in larger cohorts of adult patients enrolled in different treatment protocols. However, this limited set of genes can be evaluated with simpler methods such as quantitative RT-PCR, histochemistry, or flow cytometry. Further characterization of genes associated with poor prognosis identified by gene expression profiling in adult T-ALL may also lead to a better understanding of the differences between adult and pediatric ALL and to the identification of genetic abnormalities that contribute to the malignant phenotype of these cells. A further definition of the functional role of these genes may additionally lead to the identification of new therapeutic targets for these patients.
Prepublished online as Blood First Edition Paper, December 18, 2003; DOI 10.1182/blood-2003-09-3243.
Supported by National Institutes of Health grant CA66996, Ted and Eileen Pasquarello Research Fund, Associazione Italiana per la Ricerca sul Cancro (AIRC), Istituto Superiore di Sanita', Ministero dell'Istruzione, Università e della Ricerca Scientifica, Progetto FIRB (Fondo per gli Investimenti della Ricerca di Base), and Associazione per le Leucemie Acute dell'adulto “Cristina Bassi” e Fondazione Cassa di Risparmio di Genova e Imperia.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.