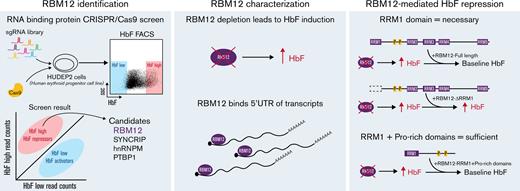
Key Points
A CRISPR/Cas9 screen targeting RNA binding proteins identifies RBM12 as a novel HbF regulator.
RBM12-mediated HbF repression is BCL11A-independent and reliant on its RRM1 domain.
Abstract
The fetal-to-adult hemoglobin transition is clinically relevant because reactivation of fetal hemoglobin (HbF) significantly reduces morbidity and mortality associated with sickle cell disease (SCD) and β-thalassemia. Most studies on the developmental regulation of the globin genes, including genome-wide genetics screens, have focused on DNA binding proteins, including BCL11A and ZBTB7A/LRF and their cofactors. Our understanding of RNA binding proteins (RBPs) in this process is much more limited. Two RBPs, LIN28B and IGF2BP1, are known posttranscriptional regulators of HbF production, but a global view of RBPs is still lacking. Here, we carried out a CRISPR/Cas9-based screen targeting RBPs harboring RNA methyltransferase and/or RNA recognition motif (RRM) domains and identified RNA binding motif 12 (RBM12) as a novel HbF suppressor. Depletion of RBM12 induced HbF expression and attenuated cell sickling in erythroid cells derived from patients with SCD with minimal detrimental effects on cell maturation. Transcriptome and proteome profiling revealed that RBM12 functions independently of major known HbF regulators. Enhanced cross-linking and immunoprecipitation followed by high-throughput sequencing revealed strong preferential binding of RBM12 to 5′ untranslated regions of transcripts, narrowing down the mechanism of RBM12 action. Notably, we pinpointed the first of 5 RRM domains as essential, and, in conjunction with a linker domain, sufficient for RBM12-mediated HbF regulation. Our characterization of RBM12 as a negative regulator of HbF points to an additional regulatory layer of the fetal-to-adult hemoglobin switch and broadens the pool of potential therapeutic targets for SCD and β-thalassemia.
Introduction
The hemoglobin molecule is a tetramer composed of 2 α- and 2 β-type globin chains. Different isoforms of α- and β-type globin genes are expressed during development, thus changing the composition of hemoglobin. The β-type globin genes comprise 1 embryonic (HBE, encoding ε-globin), 2 fetal (HBG1/2, encoding γ-globin), and 2 adult (HBD and HBB, encoding δ- and β-globin, respectively) genes.1,2 During late gestation until birth, HBG1/2 are the predominant forms expressed, giving rise to fetal hemoglobin (HbF,α2γ2). After birth, HBG1/2 expression is silenced, whereas the levels of HBB and HBD increase and then remain the primarily expressed β-type globin genes throughout life. The mechanisms controlling this fetal-to-adult switch have been under investigation for decades, with the interest in this process fueled by the recognition that reversal of the switch benefits patients with β-hemoglobinopathies such as sickle cell disease (SCD) and β-thalassemia.3 Two transcription factors, BCL11A and ZBTB7A/LRF, are the major direct repressors of the HBG1/2 genes in adulthood.4-6 These factors, along with a host of cofactors, are themselves subject to regulation,7-9 in some cases via posttranscriptional mechanisms.10
Posttranscriptional gene regulation has been implicated in various developmental contexts, including hematopoiesis.11-16 Posttranscriptional mechanisms diversify the cellular proteome through control of messenger RNA (mRNA) splicing, stabilization, localization, and translation via RNA binding proteins (RBPs).17 RBPs have been studied in the context of the fetal-to-adult switch. For example, the expression of RBPs LIN28B and IGF2BP1 is developmentally controlled, with predominant expression in fetal erythroid cells followed by silencing in adult cells. Forced expression of either RBP in adult cells markedly induces HBG1/2 production by posttranscriptionally lowering BCL11A levels.10,18,19 Aside from these examples, our knowledge of RBPs in HbF regulation is sparse. To begin to fill this void, we carried out a CRISPR/Cas9-based genetic screen targeting a collection of RBPs for their ability to alter HBG1/2 levels. We discovered RNA binding motif 12 (RBM12) as a novel repressor of HbF expression. RBM12 is a nuclear RBP that functions in pathways not involving the major known HbF regulators. It binds mRNAs in a selective pattern with a strong bias toward 5′ untranslated regions (UTRs). Remarkably, only 1 of RBM12’s 5 RNA recognition motifs (RRMs) is essential for HbF repression, and the addition of a proline-rich linker region to that RRM restores its HbF repressive activity. Thus, RBM12 emerges as a novel HbF repressor, providing an underexplored view into the regulation of the fetal-to-adult hemoglobin switch.
Materials and methods
CRISPR/Cas9 screen
The single guide RNA (sgRNA) library, targeting 341 RBPs, contains a total of 3095 sgRNAs with 3 to 6 sgRNAs per RRM and RNA methyltransferase domain. The library was cloned into the LRG2.1T lentiviral vector, which was used to transduce the human erythroid progenitor cell line HUDEP2 stably expressing spCas9.20,21 After expansion and differentiation, the top and bottom 10% of HbF-expressing cells were collected via fluorescence-activated cell sorting, and the representation of the sgRNAs enriched in those populations was assessed via sequencing on a MiSeq instrument. Candidates were identified if the ratio of the normalized read counts of sgRNAs in the HbF high vs low populations was ≥1.5 in a majority of the sgRNAs.
Mouse fetal liver cultures
Experiments were approved by the institutional animal care and use committee of the Children’s Hospital of Philadelphia (protocol IAC 21-000660). Mouse fetal livers at embryonic day 14.5 were dissected from pregnant wild-type (WT) C57BL/6J females that were crossed with Cas9/Cas9 males. Hematopoietic progenitors were isolated using the EasySep mouse Hematopoietic Progenitor Cell Enrichment Kit (no. 19856; StemCell Technologies). Cells were transduced with retrovirus harboring sgRNAs targeting Rbm12 along with Rosa26 control and selected with 1 μg/mL puromycin for 48 hours in StemPro34 with supplement (no. 10639011; ThermoFisher Scientific). Cells were differentiated for 48 hours and harvested for quantitative reverse transcription polymerase chain reaction (qRT-PCR) and Western blotting.
Sickling assay
Peripheral blood mononuclear cells were isolated using Ficoll-Paque Premium (GE17-5442-02; Sigma) density gradient centrifugation on apheresis waste product collected from deidentified patients with SCD. CD34+ cells were purified via magnetic-assisted cell sorting using ultrapure CD34 Microbeads (MACS CD34+ purification kit; no. 130-100-453; Miltenyi) and magnetic columns (no. 130-042-201; Miltenyi). Cells were differentiated for a total of 21 days, sorted for enucleated RBCs via Hoechst 33342 exclusion, and exposed to 2.5% O2 for 2 hours. Following fixation with 2% glutaraldehyde, sickled cells were quantified manually via bright-field microscopy in a blinded fashion.
eCLIP-seq
Enhanced cross-linking and immunoprecipitation followed by high-throughput sequencing (eCLIP-seq) was performed following the standard operating procedure from ENCODE as described by Van Nostrand et al22 (details provided in supplemental Materials and Methods). Briefly, RBP-RNA complexes were cross-linked via UV exposure (254 nm, 400 mJ/cm3), fragmented via RNaseI digestion, and immunoprecipitated with anti-RBM12 antibody (sc-514259; Santa Cruz Biotechnology) coupled to magnetic beads (10003D; ThermoFisher Scientific). Samples were washed, and RNA barcode adapters were ligated, resolved on standard protein gels, and transferred to nitrocellulose membranes. RNA was isolated from a region encompassing 75 kDa above the expected size of the RBP, purified, and reverse-transcribed. Complementary DNA libraries were prepared with Illumina index primers and sequenced on a NextSeq2000 instrument.
RBM12 overexpression
V5 C-terminally tagged RBM12 constructs were rendered conditional via fusion to the estrogen receptor ligand-binding domain (RBM12-ER-V5) and inserted into a lentiviral vector.4,23 Lentivirus was transduced into HUDEP2 cells, which underwent selection in 10 μg/mL of blasticidin for 4 days. RBM12-ER-V5 was activated by addition of 10 nM 4-hydroxytamoxifen.
Results
A CRISPR/Cas9 screen targeting RPBs identifies novel candidate HbF repressors
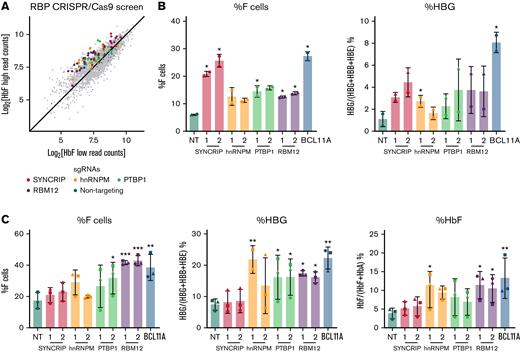
To identify RBPs that influence HbF levels, we carried out a CRISPR/Cas9 screen in the adult human erythroblast line HUDEP2,20,21 targeting 341 RBPs via their RRM and RNA methyltransferase domains (supplemental Figure 1A). The RRM is one category of RNA binding domain that confers binding to target transcripts.24 Potential HbF repressors were called if a majority of the sgRNAs targeting the RBP were enriched in the HbF high population with HbF activators enriched in the HbF low population. As expected, the nontargeting (NT) control sgRNAs were evenly distributed among the 2 populations. The sgRNAs against IGF2BP1 were enriched in the HbF low population, consistent with IGF2BP1’s functions as an HbF activator (supplemental Figure 1B).19 Four RBPs (SYNCRIP, hnRNPM, PTBP1, and RBM12) emerged from this screen as potential HbF repressors (Figure 1A). Results were validated by depleting each of these RBPs with 2 independent sgRNAs in HUDEP2 cells. Depletion of each candidate RBP led to a 2- to 4-fold increase in the proportion of erythroid cells expressing HbF, termed F-cells (as measured by flow cytometry), along with a 2- to 4-fold increase in HBG1/2 mRNA (Figure 1B). Among these, SYNCRIP depletion led to the strongest HbF induction.
CRISPR/Cas9-based screen identified SYNCRIP, hnRNPM, PTBP1, and RBM12 as candidate HbF regulators. (A) Scatterplot displaying RBP-focused CRISPR/Cas9 screening results. Each dot represents an sgRNA. Highlighted are NT control sgRNAs and sgRNAs targeting candidate HbF regulators. (B) HUDEP2 validation results. Controls are an NT sgRNA and a BCL11A sgRNA targeting exon 2 of BCL11A. Each candidate was targeted with 2 independent sgRNAs (n = 2 biological replicates). Plotted are means ± standard deviation. Left: Percentage of F cells quantified via HbF flow cytometry. Right: qRT-PCR of HBG1/2 mRNA as percentage of total β-like globin transcripts. Results were normalized to HPRT1. ∗P < .05 by Student’s t test. (C) CD34+ HSPC validation result. Controls are NT and a BCL11A sgRNA targeting the +58 erythroid enhancer of BCL11A. Each candidate was targeted with 2 independent sgRNAs (n = 3 biological replicates). Plotted are means ± standard deviation. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 by Student’s t test. Left: Percentage of HbF+ cells quantified via HbF flow cytometry. Center: qRT-PCR of HBG1/2 mRNA as percentage of total β-like globin transcripts. Results were normalized to HPRT1. Right: Percentage of HbF relative to NT quantified via high-performance liquid chromatography.
CRISPR/Cas9-based screen identified SYNCRIP, hnRNPM, PTBP1, and RBM12 as candidate HbF regulators. (A) Scatterplot displaying RBP-focused CRISPR/Cas9 screening results. Each dot represents an sgRNA. Highlighted are NT control sgRNAs and sgRNAs targeting candidate HbF regulators. (B) HUDEP2 validation results. Controls are an NT sgRNA and a BCL11A sgRNA targeting exon 2 of BCL11A. Each candidate was targeted with 2 independent sgRNAs (n = 2 biological replicates). Plotted are means ± standard deviation. Left: Percentage of F cells quantified via HbF flow cytometry. Right: qRT-PCR of HBG1/2 mRNA as percentage of total β-like globin transcripts. Results were normalized to HPRT1. ∗P < .05 by Student’s t test. (C) CD34+ HSPC validation result. Controls are NT and a BCL11A sgRNA targeting the +58 erythroid enhancer of BCL11A. Each candidate was targeted with 2 independent sgRNAs (n = 3 biological replicates). Plotted are means ± standard deviation. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 by Student’s t test. Left: Percentage of HbF+ cells quantified via HbF flow cytometry. Center: qRT-PCR of HBG1/2 mRNA as percentage of total β-like globin transcripts. Results were normalized to HPRT1. Right: Percentage of HbF relative to NT quantified via high-performance liquid chromatography.
RBM12 participates in HbF silencing in primary human erythroid cells
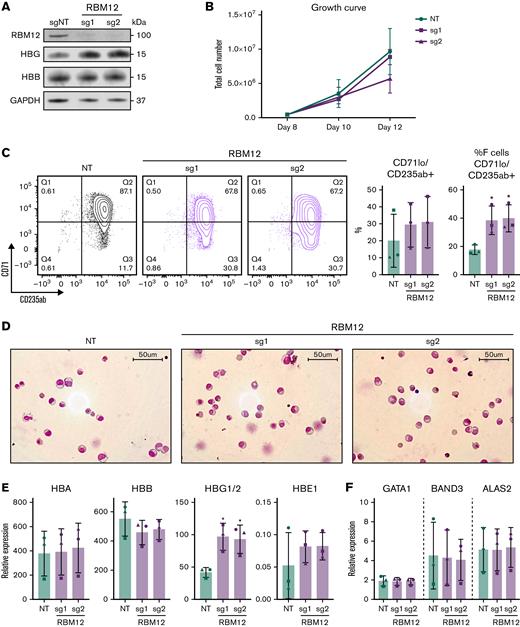
Although the HUDEP2 cell line provides a powerful screening platform, any hits require validation in primary human erythroblasts. We thus depleted each of the 4 candidate RBPs via CRISPR/Cas9 ribonucleoprotein electroporation of human CD34+ hematopoietic stem and progenitor cells (HSPCs) isolated from 3 healthy donors. As part of a 3-phase erythroid culture system,25 cells underwent erythroid differentiation for 12 to 15 days prior to analysis. In HUDEP2 cells and CD34+ HSPCs, ≥40% depletion of SYNCRIP, PTBP1, and RBM12 at the protein level was achieved (supplemental Figure 1C). Although hnRNPM protein persisted, the sgRNA cutting efficiency (assayed via Tracking of Indels by DEcomposition, TIDE)26 was >50% in CD34+ cells, suggesting that the edits resulted in a dysfunctional protein. In contrast to the HUDEP2 validation results, depletion of SYNCRIP increased HbF production only minimally as assayed by flow cytometry, qRT-PCR, and high-performance liquid chromatography (Figure 1C). Loss of PTBP1 or hnRNPM increased HbF levels, but at the cost of cellular maturation and fitness (supplemental Figure 2A-C). Depleting RBM12 induced HbF expression >2-fold at the mRNA and protein levels, along with an increase in the proportion of F-cells. Importantly, RBM12 depletion did not significantly compromise cellular maturation (Figure 2). Because of the consistent HbF induction along with maintenance of cellular maturation upon depletion in primary human culture, we focused our efforts on characterizing RBM12 in the context of HbF regulation.
RBM12 depletion in primary human CD34+HSPCs does not impair erythroid maturation. CRISPR/Cas9 depletion of RBM12 in human CD34+ HSPCs via 2 independent sgRNAs (n = 3 biological replicates). (B, C, E, and F) Plotted are means ± standard deviation. (A) Western blot analysis with indicated antibodies at day 15 of differentiation. (B) Growth curve displaying total cell number from day 8 to 12 of differentiation. (C) Left: Representative contour plots of CD71 vs CD235ab at day 15 of differentiation. Right: Quantification of CD71loCD235ab+ and CD71loCD235ab+/HbF+ cells. ∗P < .05 by Student’s t test. (D) Representative May-Grünwald Giemsa images of HSPCs at day 15 of differentiation. (E) qRT-PCR analysis of globin genes at day 12 of differentiation. Results are normalized to HPRT1. ∗P < .05 by Student’s t test. (F) qRT-PCR analysis of differentiation markers GATA1, BAND3, and ALAS2 at day 12 of differentiation. Results are normalized to HPRT1.
RBM12 depletion in primary human CD34+HSPCs does not impair erythroid maturation. CRISPR/Cas9 depletion of RBM12 in human CD34+ HSPCs via 2 independent sgRNAs (n = 3 biological replicates). (B, C, E, and F) Plotted are means ± standard deviation. (A) Western blot analysis with indicated antibodies at day 15 of differentiation. (B) Growth curve displaying total cell number from day 8 to 12 of differentiation. (C) Left: Representative contour plots of CD71 vs CD235ab at day 15 of differentiation. Right: Quantification of CD71loCD235ab+ and CD71loCD235ab+/HbF+ cells. ∗P < .05 by Student’s t test. (D) Representative May-Grünwald Giemsa images of HSPCs at day 15 of differentiation. (E) qRT-PCR analysis of globin genes at day 12 of differentiation. Results are normalized to HPRT1. ∗P < .05 by Student’s t test. (F) qRT-PCR analysis of differentiation markers GATA1, BAND3, and ALAS2 at day 12 of differentiation. Results are normalized to HPRT1.
RBM12 characterization
RBM12 (also known as SH3/WW domain anchor protein in the nucleus, SWAN) contains 5 RRMs, 2 proline-rich regions, and several putative transmembrane domains.27 Alternative splicing of RBM12 results in 4 isoforms, all sharing the same coding sequence.28 Increased RBM12 expression has been correlated with poor prognosis in hepatocellular and Meibomian cell carcinoma,29,30 and various mutations in RBM12 have been found to be associated with endometrial and colorectal cancer diagnoses.31,32 Additionally, truncating mutations in RBM12 have been reported in a cohort of patients with psychosis.33 Despite the various studies citing its potential role in cancer and neurological disorders, there are no studies characterizing its molecular function to date.
The roles of RBPs can be narrowed down by assessing their nuclear localization.34 We carried out anti-RBM12 immunofluorescence staining in primary human erythroblasts and HeLa cells and found that RBM12 displayed nuclear localization (supplemental Figure 3A), suggesting that it functions at the level of pre-mRNA synthesis or processing. Available tissue-expression databases display expression across various tissue types, including the hematopoietic system (supplemental Figure 3B).35 In the erythroid compartment, RBM12 levels are initially high, but fade during terminal differentiation (supplemental Figure 3C-D).36,37 To assess whether RBM12 expression is different between fetal and adult stages, as is the case for several developmental globin regulators,18,19,38,39 we analyzed published data sets and found its expression to not be developmentally selective (supplemental Figure 3E).38,40 In sum, RBM12 is a nuclear RBP with broad tissue expression that remains unchanged between fetal and adult erythroid stages.
RBM12 influences developmental control of β-globin genes in mice
Human and mouse orthologs of RBM12 share 86.4% amino acid identity, with the highest regions of conservation at the 5 RRMs (supplemental Figure 4A). To test whether RBM12 activity is conserved across mammals, we depleted it via retroviral delivery of sgRNAs into embryonic day 14.5 murine fetal liver erythroblasts (representing adult erythroid cells) expressing Cas9 (supplemental Figure 4B). Unlike humans, mice do not express a fetal form of β-globin, but express 2 embryonic genes, Hbb-y and Hbb-bh1, both of which also undergo a developmental switch that is controlled by BCL11A and ZBTB7A/LRF, among other factors.6,41 Upon Rbm12 depletion, transcript levels of Hbb-bh1, but not Hbb-y, increased (supplemental Figure 4C-D), reminiscent of the increase of HBG1/2 mRNA but not HBE1 mRNA in primary human HSPCs upon RBM12 loss (Figure 2E). Additionally, in line with results obtained in human cells, Rbm12 depletion did not delay erythroid maturation, as shown by the normal activation of adult α- and β-globin genes Hba and Hbb-b1 and the repression of the Kit gene (supplemental Figure 4E). In sum, RBM12 modulates Hbb-bh1 levels in murine primary erythroid cells, suggesting that its regulatory influence on the β-globin genes is conserved.
RBM12 depletion increases HbF levels and reduces cell sickling in erythroid cells from patients with SCD
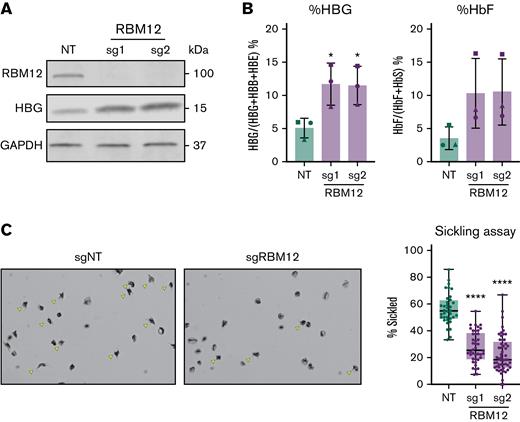
Increased levels of HbF can diminish the propensity of sickle hemoglobin to polymerize under hypoxic conditions and thus inhibit the pathognomonic changes in cell shape.1,42 To assess whether RBM12 depletion can increase HbF levels sufficient to exert antisickling effects, we measured cell sickling in RBM12-depleted erythroid cells in SCD patient–derived CD34+ HSPCs upon exposure to 2.5% O2 for 2 hours. Consistent with results from healthy donors, RBM12 loss triggered a >2-fold increase in HBG1/2 mRNA and percentage of HbF protein (Figure 3A-B). As a result, we observed a ∼50% decrease in cell sickling in the RBM12-depleted samples relative to control (Figure 3C). Together, these results suggest that RBM12 depletion induces HbF to levels sufficient to attenuate cell sickling.
RBM12 depletion in CD34+HSPCs derived from patients with SCD. CD34+ HSPCs isolated from apheresis waste product of 3 patients with SCD targeted with 2 independent RBM12 sgRNAs along with an NT control. (A) Western blot analysis with indicated antibodies at day 10 of differentiation. (B) HbF induction assayed via qRT-PCR and high-performance liquid chromatography. Plotted are means ± standard deviation (n = 3). ∗P < .05 by Student’s t test. Left: qRT-PCR of HBG1/2 mRNA as percentage of total β-like globin transcripts at day 12 of differentiation. Results are normalized to HPRT1. Right: High-performance liquid chromatography at day 15 of differentiation. (C) Left: Representative images of sickling assay results. Sickled cells are indicated by yellow arrows. Original magnification ×40 on an Olympus BX40 microscope fitted with an Infinity Lite B camera (Olympus) and the coupled image capture software. Right: Quantification of percent sickled cells. Each dot represents a distinct field of view. Each sample has approximately 1000 total cells quantified. ∗∗∗∗P < .0001 by Student’s t test.
RBM12 depletion in CD34+HSPCs derived from patients with SCD. CD34+ HSPCs isolated from apheresis waste product of 3 patients with SCD targeted with 2 independent RBM12 sgRNAs along with an NT control. (A) Western blot analysis with indicated antibodies at day 10 of differentiation. (B) HbF induction assayed via qRT-PCR and high-performance liquid chromatography. Plotted are means ± standard deviation (n = 3). ∗P < .05 by Student’s t test. Left: qRT-PCR of HBG1/2 mRNA as percentage of total β-like globin transcripts at day 12 of differentiation. Results are normalized to HPRT1. Right: High-performance liquid chromatography at day 15 of differentiation. (C) Left: Representative images of sickling assay results. Sickled cells are indicated by yellow arrows. Original magnification ×40 on an Olympus BX40 microscope fitted with an Infinity Lite B camera (Olympus) and the coupled image capture software. Right: Quantification of percent sickled cells. Each dot represents a distinct field of view. Each sample has approximately 1000 total cells quantified. ∗∗∗∗P < .0001 by Student’s t test.
Effects of RBM12 depletion on the erythroid transcriptome and proteome
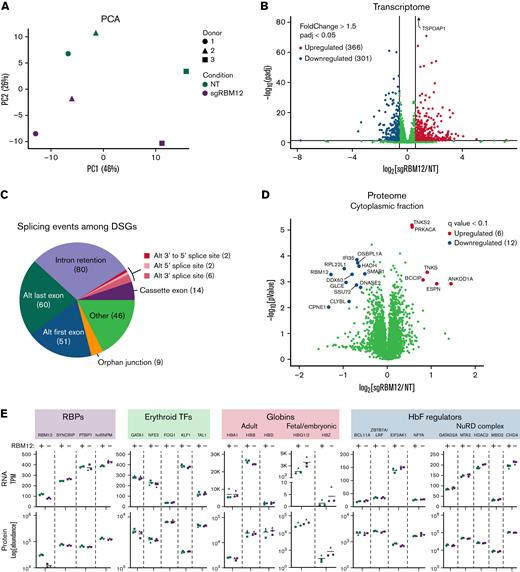
We next assessed the global impact of RBM12 depletion on the erythroid transcriptome. We performed next-generation sequencing on polyA-selected mRNA (RNA-seq) of primary human erythroblasts from 3 healthy donors following CRISPR/Cas9-mediated depletion of RBM12 (supplemental Figure 5A). Principal component analysis (PCA) revealed clustering of samples from the same donors regardless of RBM12 status, suggesting that RBM12 depletion does not cause widespread changes in the erythroid transcriptome (Figure 4A). Differential gene expression analysis identified 366 upregulated and 301 downregulated genes (fold change >1.5, adjusted P < 0.05; Figure 4B). Gene set enrichment analysis against various gene sets available on the Molecular Signatures Database43,44 found that downregulated transcripts were strongly enriched for interferon-γ and -α response signatures (supplemental Figure 5C). Gene set enrichment analysis against additional gene sets, such as those curated from a variety of sources (including online pathway databases and the literature), also showed enrichment of immune system related pathways among the downregulated genes (supplemental Table 1).
Effects of RBM12 depletion on transcriptome and proteome. (A) PCA of normalized read counts from RBM12-knockout (KO) RNA-seq in primary human erythroblasts at day 7 of differentiation. (B) Volcano plot of differentially expressed transcripts. Highlighted are transcripts found to be differentially expressed with a fold change ≥1.5 and adjusted P < 0.05. (C) Pie chart displaying abundance of alternative splicing events occurring among differentially spliced genes (DSGs). (D) Volcano plot of the cytoplasmic fraction of RBM12-KO liquid chromatography/tandem mass spectrometry in primary human erythroblasts at day 7 of differentiation. Highlighted and labeled are proteins found to be differentially expressed with a q-value <0.1. (E) Transcripts per million (TPM; top) and fold change protein abundance (bottom) of RBM12 KO vs NT control samples (n = 3 independent donors).
Effects of RBM12 depletion on transcriptome and proteome. (A) PCA of normalized read counts from RBM12-knockout (KO) RNA-seq in primary human erythroblasts at day 7 of differentiation. (B) Volcano plot of differentially expressed transcripts. Highlighted are transcripts found to be differentially expressed with a fold change ≥1.5 and adjusted P < 0.05. (C) Pie chart displaying abundance of alternative splicing events occurring among differentially spliced genes (DSGs). (D) Volcano plot of the cytoplasmic fraction of RBM12-KO liquid chromatography/tandem mass spectrometry in primary human erythroblasts at day 7 of differentiation. Highlighted and labeled are proteins found to be differentially expressed with a q-value <0.1. (E) Transcripts per million (TPM; top) and fold change protein abundance (bottom) of RBM12 KO vs NT control samples (n = 3 independent donors).
Many nuclear RRM-containing RBPs regulate alternative splicing. Therefore, we analyzed our RNA-seq data for differential splicing using Modeling by Alternative Junction Inclusion Quantification software.45 Differentially spliced genes (DSGs) were identified as transcripts that contained ≥1 local splicing variation with a >15% difference in inclusion (delta percent spliced in >0.15) of alternative junctions observed between the RBM12-knockout (KO) and NT control conditions. Upon RBM12 depletion, we detected 127 DSGs that contained 133 splicing events. Many of these events that occur upon RBM12 KO included alternative first and last exon splicing and intron retention (Figure 4C).
We next probed changes in the erythroid proteome upon RBM12 depletion. We analyzed nuclear and cytoplasmic extracts from the same set of samples used in RNA-seq by liquid chromatography/tandem mass spectrometry (LC-MS/MS). Samples showed a high degree of correlation with PCA separating samples based on the subcellular origin (supplemental Figure 5D-E). Of the 6620 proteins detected, only 18 were found to be significantly up- or downregulated in the cytoplasmic fraction (q-value <0.1; Figure 4D; supplemental Table 2). No statistically significant changes were detected in the nuclear fraction (data not shown). Several of the proteins that were differentially expressed have been linked to cancer prognosis,46,47 which is in line with previous reports of RBM12’s association with various cancers. Additionally, proteins implicated in innate immune system regulation, such as DDX60 and IFI35, were significantly downregulated,48,49 providing a link to the downregulated immune signatures identified at the transcript level.
Intersecting the RNA-seq and LC-MS/MS data to identify changes commonly found in both datasets, we specifically looked at the expression of the RBP candidates from the CRISPR/Cas9 screen, erythroid transcription factors, globin chains, and known HbF regulators (Figure 4E and supplemental Figure 5F).50 As expected, HBG1/2 was upregulated in mRNA and protein abundance. HBZ, which encodes for the embryonic α- globin chain, was also found to be upregulated, albeit at low baseline expression. We note that, even though HBG1/2 and HBZ upregulation were found to be statistically significant in transcript abundance, they did not reach the q-value cutoff in protein abundance. We found that RBM12 depletion does not alter the expression of major known HbF regulators and cofactors, suggesting that it functions via a novel pathway. This was individually confirmed via Western blotting and qRT-PCR for BCL11A and ZBTB7A/LRF (supplemental Figure 5G).
In sum, RBM12 reduction in primary human HSPCs does not cause large changes in the erythroid transcriptome or proteome, in line with the observation that RBM12 loss seems to be of little consequence to erythroid fitness and maturation. Furthermore, the lack of changes in known HbF regulators implicates a pathway that does not involve altering the expression of major known HbF repressors and their coregulators.
RBM12 preferentially binds mRNA at the 5'UTR
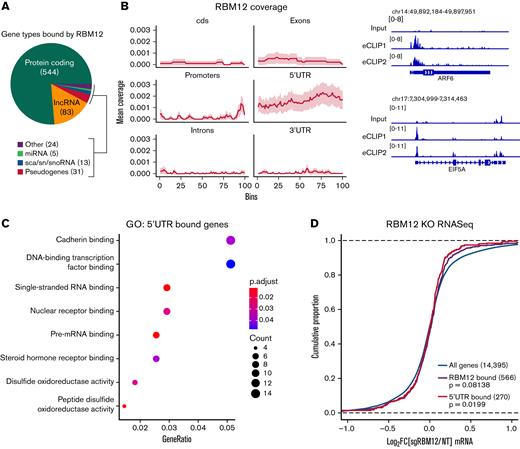
The position of RBPs on their target RNAs might inform their function.51 To probe the molecular function of RBM12, we queried its binding pattern via eCLIP-seq.22 After reproducing a published PTBP1 eCLIP-seq experiment in K562 cells as a technical pilot (supplemental Figure 6), we assayed RBM12 binding in WT primary human erythroblasts. We obtained a total of 1358 high-confidence RBM12-bound peaks at 680 transcripts. Many of the transcripts bound were protein coding genes, followed by long noncoding RNAs (Figure 5A). Interestingly, RBM12-bound peaks were enriched at the 5′UTR (Figure 5B). Gene Ontology analysis revealed that a majority of these 5′UTR-bound transcripts play a role in a variety of transcriptional and posttranscriptional functions (Figure 5C). Intersection with the RNA-seq data revealed that levels of transcripts bound by RBM12 at the 5′UTR, and in general, were unchanged following RBM12 depletion (Figure 5D). Based on these results, RBM12 displays preferential binding at the 5′UTR of its target mRNAs in primary human erythroblasts. The transcripts that are bound by RBM12 are also unchanged at the transcriptional level, indicating a mechanism that likely does not converge on mature transcript abundance.
RBM12 binding patterns. RBM12 eCLIP-seq in WT primary human erythroblasts at day 8 of differentiation. (A) Pie chart displaying the distribution of gene types bound by RBM12. (B) Left: Coverage profile of RBM12 binding. Each feature was divided into 100 bins of equal length. Right: Representative Integrative Genomics Viewer snapshots of RBM12-bound genes. Displayed are the size-matched input control and 2 replicate eCLIP tracks. (C) Gene Ontology (GO) analysis of transcripts bound at the 5′UTR by RBM12. (D) Empirical cumulative distribution function plot displaying log2 fold change in transcript abundance of RBM12 KO samples over control along with RBM12 bound peaks (in general and specifically at 5′UTR). P values calculated by Kolmogorov-Smirnoff test.
RBM12 binding patterns. RBM12 eCLIP-seq in WT primary human erythroblasts at day 8 of differentiation. (A) Pie chart displaying the distribution of gene types bound by RBM12. (B) Left: Coverage profile of RBM12 binding. Each feature was divided into 100 bins of equal length. Right: Representative Integrative Genomics Viewer snapshots of RBM12-bound genes. Displayed are the size-matched input control and 2 replicate eCLIP tracks. (C) Gene Ontology (GO) analysis of transcripts bound at the 5′UTR by RBM12. (D) Empirical cumulative distribution function plot displaying log2 fold change in transcript abundance of RBM12 KO samples over control along with RBM12 bound peaks (in general and specifically at 5′UTR). P values calculated by Kolmogorov-Smirnoff test.
Mechanistic insight into RBM12 mediated HbF regulation
Although RBM12 depletion was well tolerated in primary human erythroblasts, we were unable to derive homozygous RBM12 KO clonal HUDEP2 cell lines. Even haploinsufficient RBM12 HUDEP2 cells (RBM12+/−) grew slower than their parental counterparts (supplemental Figure 7A-B). Nevertheless, RBM12+/− cells did exhibit increased HbF levels relative to WT, and restoration via expression of full-length RBM12 (RBM12-FL) rescued its HbF-repressive function (supplemental Figure 7C-D).
We used the RBM12+/− clones to rule out a potential confounder. RBM12 shares its promoter with another gene, CPNE1. Genes that share promoters are often coregulated,28 and, in line with this observation, we found that CPNE1 expression is strongly correlated with RBM12 expression, as its expression is depleted in RBM12+/− clones and is restored upon RBM12-FL overexpression (supplemental Figure 8A-B). Therefore, we tested CPNE1’s ability as the true HbF repressor behind RBM12 depletion by overexpressing CPNE1 in RBM12+/− clones. We found that this was not the case, as CPNE1 failed to rescue HbF repression (supplemental Figure 8C-D). Thus, we can conclude that the altered HbF levels are truly RBM12-mediated.
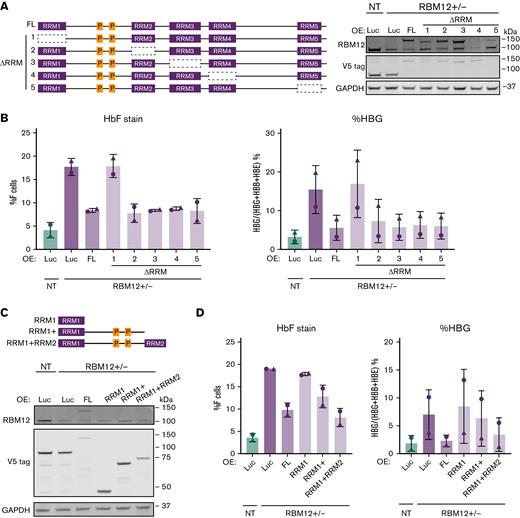
We next asked which of the 5 RRM domains are important for RBM12’s function in HbF repression. The existence of multiple RRMs in RBPs allows for increased affinity and specificity in binding mRNA targets.52 We overexpressed in 2 independent RBM12+/− clones, RBM12-FL and mutants lacking one of each RRM (ΔRRM1-5), and measured HbF induction (Figure 6A). Luciferase overexpression served as negative control. RBM12-FL and every ΔRRM mutant except ΔRRM1 were able to silence HbF expression (Figure 6B). We next tested whether this RRM1 domain is sufficient to repress HbF by overexpressing various truncated forms of RBM12 (Figure 6C). The RRM1 domain alone was insufficient to repress HbF expression; however, the addition of the linker region between RRM1 and RRM2 to RRM1 restored RBM12’s HbF-repressive activity (Figure 6D). This linker region contains 2 proline-rich regions, which have been implicated in protein-protein interactions.53,54 Taken together, the RRM1 domain of RBM12 is necessary for HbF repression and, with the addition of the proline-rich linker region, is sufficient for RBM12-mediated HbF repression. These results suggest that RBM12-mediated HbF regulation likely requires interaction with auxiliary proteins.
Structure function analysis of RBM12 RRM domains. Two independent RBM12+/− clones were infected with lentivirus containing conditional RBM12 overexpression (OE) and luciferase control constructs. (B and D) Plotted are means ± standard deviation. (A) Left: Schematic of various forms of RBM12 OE constructs. Right: Representative Western blot of mutant RBM12 OE constructs with indicated antibodies. (B) HbF flow cytometry (left) and qRT-PCR of HBG1/2 mRNA as percentage of total β-like globin transcripts (right) at day 5 of differentiation. (C) Top: Schematic of RBM12 truncated OE constructs. Bottom: Representative Western blot of RBM12 truncated OE constructs with indicated antibodies. (D) HbF flow cytometry (left) and qRT-PCR of HBG1/2 mRNA as percentage of total β-like globin transcripts (right) at day 5 of differentiation.
Structure function analysis of RBM12 RRM domains. Two independent RBM12+/− clones were infected with lentivirus containing conditional RBM12 overexpression (OE) and luciferase control constructs. (B and D) Plotted are means ± standard deviation. (A) Left: Schematic of various forms of RBM12 OE constructs. Right: Representative Western blot of mutant RBM12 OE constructs with indicated antibodies. (B) HbF flow cytometry (left) and qRT-PCR of HBG1/2 mRNA as percentage of total β-like globin transcripts (right) at day 5 of differentiation. (C) Top: Schematic of RBM12 truncated OE constructs. Bottom: Representative Western blot of RBM12 truncated OE constructs with indicated antibodies. (D) HbF flow cytometry (left) and qRT-PCR of HBG1/2 mRNA as percentage of total β-like globin transcripts (right) at day 5 of differentiation.
Discussion
We performed an RBP-targeted CRISPR/Cas9-based screen in HUDEP2 cells and identified RBM12 as a novel regulator of HbF expression. The degree of HbF induction upon RBM12 loss was sufficient to attenuate cell sickling in SCD-derived erythroid cells. Rbm12 loss in primary murine erythroid cells triggered derepression of embryonic β-type globin genes, suggesting conserved function across mammals. RBM12 depletion displayed minimal alterations to the erythroid transcriptome and proteome, consistent with an ostensible lack of detrimental effects to erythroid maturation. Additionally, differential splicing analysis yielded an abundance of intron retention and alternative first- and last-exon splicing events. Mechanistically, the HbF-silencing activity of RBM12 can be reduced to a single RRM domain plus a proline-rich linker region. eCLIP-seq demonstrates that RBM12 preferentially binds 5′UTRs of mRNA transcripts, providing a hint into its molecular function.
HUDEP2 cells present a powerful screening tool in which, historically, most hits have been validated in parallel cell systems.21,55 In this case, among 4 hits, only the loss of RBM12 produced significant HbF induction in primary cultured erythroid cells in the absence of detrimental effects to cell maturation. Although this discrepancy between systems is not sufficient to rule out the other 3 proteins as HbF regulators, it does highlight potential limitations of the in vitro cell systems used. SYNCRIP, hnRNPM, and PTBP1 belong to a group of RBPs known as heterogeneous nuclear ribonucleoproteins (hnRNPs).17 Interestingly, these hnRNPs have been reported to regulate the alternative splicing of each other’s transcripts.56 Studying the potential cooperative effects among these hnRNPs might confirm or rule out any role in HbF regulation. Because RBM12 was validated in both cell systems, follow-up studies focused on this protein.
To explore the molecular mechanisms by which RBM12 regulates HbF levels, we characterized the subcellular localization of RBM12, assessed the transcriptome and proteome of RBM12-depleted cells, and interrogated its RNA binding profiles. This led to the following insights: first, RBM12 is localized to the nucleus, making a direct role in mRNA translation or stability unlikely, favoring a role in pre-mRNA synthesis or processing. The majority of the 680 mRNAs bound by RBM12 lack corresponding changes in protein levels and are also not differentially spliced (supplemental Figure 7). In the case of mRNAs bound that do overlap with DSGs, the RBM12 eCLIP-seq peaks do not overlap with the splice junction coordinates. Hence, simple binding to mRNA by RBM12 might not be sufficient to effect measurable changes in mRNA processing. Second, eCLIP-seq analysis revealed that RBM12 preferentially binds mRNA in a specific manner, particularly at the 5′UTR. This pattern has previously been reported to occur among select nuclear RBPs such as NCBP2/CBP20, a subunit of the cap binding complex.51 In the nucleus, the cap binding complex plays a role in nuclear export, pre-mRNA processing, and transcriptional regulation.57,58 Involvement in mRNA nuclear export might be expected to ultimately impinge on mRNA translation. This remains a distinct possibility because our mass spectrometry analysis failed to capture all cellular proteins. Additionally, there remains the possibility that RBM12 plays a role in transcription initiation. Other proteins that are associated with the 5′UTR of nascent mRNA regulate transcription via RNA polymerase II (RNAPII) promoter proximal pausing, such as negative elongation factor (NELF), which contains subunits that bind RNA.59,60 It is therefore possible that, like NELF, RBM12 plays a role in regulation of RNAPII pause-release.61,62 Attempts to detect RBM12 by ChIP at 5′ ends of active genes have failed, which argues against but does not rule out a role in transcription initiation or pause-release.
Because BCL11A and ZBTB7A/LRF are the major repressors of HbF expression, there is a high chance that a putative HbF repression pathway converges on these transcription factors and their coregulators. We found that RBM12 does not measurably impact the levels of either regulator or their cofactors, suggesting that it functions in yet to be discovered pathways or that it posttranscriptionally controls subunits of these protein complexes that were not detected by mass spectrometry. Hence, the pathway by which RBM12 selectively impacts fetal genes remains elusive.
Last, our structure-function analysis of RBM12 not only validated the HbF-regulatory role of RBM12, but also identified the region necessary to carry out HbF modulatory functions, providing additional insight into the role of this undercharacterized RBP. Although the RRM1 region is necessary for HbF repression, the truncated form that includes RRM1 and 2 proline-rich regions displayed sufficiency. The fact that the HbF regulatory function can be reduced to a relatively small portion of the molecule might facilitate means to perturb this function without interfering with other RBM12 functions. It is noteworthy that targeting RBPs via small molecules is an avenue that is currently being explored via disrupting the RNA binding domain and perturbing interactions with target transcripts.63,64 In the case of RBM12, the RRM1 domain might be a candidate for small molecule inhibitor development to induce HbF expression. However, its broad expression pattern will require further investigation into its suitability as a drug target.
In summary, we identified RBM12 as a novel HbF repressor in human erythroid cells. In addition to the discovery of a potential therapeutic target, we provide the first functional characterization of RBM12. The insights into RBM12 hold promise in the field of erythroid biology by highlighting an undercharacterized branch of HbF regulation.
Acknowledgments
The authors thank the members of the laboratory of G.B.; Kristen Lynch, Zissimos Mourelatos, and Russ Carstens for helpful discussions; Stella Chou for providing the SCD patient apheresis waste samples; the Fred Hutch Cooperative Centers of Excellence in Hematology Core for providing the CD34+ enriched HSCs (National Institute of Diabetes and Digestive and Kidney Diseases [NIDDK] grant DK106829); and the Wistar Proteomics Core for performing and analyzing the LC-MS/MS experiment.
This work was supported by National Institutes of Health Hematopoiesis Training Grant 5T32DK007780-19 (A.W.); NIDDK grants NRSA F31 Fellowship 1F31DK122649 (A.W.), K08DK129716 (S.A.P.), K08DK128571 (E.K.), and R24DK106766 (R.C.H.); Doris Duke Charitable Foundation Physician Scientist Fellowship Grant 2020062 (S.A.P.); and National Heart, Lung, and Blood Institute (NHLBI) grant R01HL119479 (G.A.B.). The authors thank the DiGaetano family for their generous support.
Authorship
Contribution: A.W. and G.A.B. conceived the study, designed the experiments, and wrote the manuscript; A.W., M.Q.-V., C.A.K., B.M.G., R.C.H., and J.S. analyzed the data; A.W., M.K., M.S., A.J.T., M.S.S., O.A., S.A.P., and E.K. performed experiments; and all authors provided input on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gerd A. Blobel, Department of Hematology, The Children’s Hospital of Philadelphia, 316H Abramson Research Center, 3615 Civic Center Blvd, Philadelphia, PA 19104; e-mail: blobel@e-mail.chop.edu.
References
Author notes
RNA sequencing and enhanced cross-linking and immunoprecipitation followed by high-throughput sequencing data have been deposited into the Gene Expression Omnibus (GEO) database (accession GSE200677).