Abstract
The 7.2 kilobase (kb) Corfu δβ thalassemia mutation is the smallest known deletion encompassing a region upstream of the human δ gene that has been suggested to account for the vastly different phenotypes in hereditary persistence of fetal hemoglobin (HPFH) versus β thalassemia. Fetal hemoglobin (HbF) expression in Corfu heterozygotes and homozygotes is paradoxically dissimilar, suggesting conflicting theories as to the function of the region on globin gene regulation. Here, we measure γ- and β-globin gene transcription, steady-state mRNA, and hemoglobin expression levels in primary erythroid cells cultured from several patients with Corfu δβ thalassemia. We show through RNA fluorescence in situ hybridization that the Corfu deletion results in high-level transcription of the fetal γ genes in cis with a concomitant reduction in transcription of the downstream β gene. Surprisingly, we find that elevated γ gene transcription does not always result in a corresponding accumulation of γ mRNA or fetal hemoglobin, indicating a post-transcriptional regulation of γ gene expression. The data suggest that efficient γ mRNA accumulation and HbF expression are blocked until β mRNA levels fall below a critical threshold. These results explain the Corfu paradox and show that the deleted region harbors a critical element that functions in the developmentally regulated transcription of the β-globin genes.
Introduction
The Corfu δ0β+ thalassemia mutation is a naturally occurring 7.2 kilobase (kb) deletion removing part of the δ-globin gene and approximately 6 kb of upstream flanking sequence1 (Figure 1). Molecular and hematologic analysis of the Corfu mutation was originally described in a Greek family of heterozygous parents and a homozygous child. The child was reported to have total hemoglobin levels of 93 g/L (9.3 g/dL; normal values, 120-150 g/L [12-15 g/dL]), which surprisingly was comprised almost entirely of fetal hemoglobin (HbF). The heterozygous parents, however, had normal levels of HbF (1%-2%) and only slightly decreased levels of adult hemoglobin (HbA).1 This paradox between extremely high levels of HbF in the homozygote and very low HbF levels with nearly normal HbA in the heterozygous parents was difficult to explain in terms of a molecular mechanism of globin gene regulation. Subsequent work showed that the β gene on the Corfu chromosome contains a splice site mutation in IVS-I position 5 (IVS-I-5) of the β-globin gene.2 Transfection experiments suggest this splice site mutation reduces the amount of properly spliced mRNA from this allele to 20%, indicating it is a β+ mutation. This mutation may explain the modest reductions in HbA in Corfu heterozygotes, but the overwhelming expression of HbF in Corfu homozygotes remains a paradox.
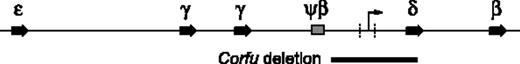
Human β-globin locus. Positions of the embryonic ϵ gene, fetal γ genes, Ψβ pseudogene, and adult δ and β genes are indicated. The Corfu deletion is represented as a black bar below the map. The minimal region of difference resulting in either HPFH or thalassemia phenotype is marked with vertical dashed lines. The arrow in this region represents the fetal-to-adult chromatin boundary/intergenic transcription start site.
Human β-globin locus. Positions of the embryonic ϵ gene, fetal γ genes, Ψβ pseudogene, and adult δ and β genes are indicated. The Corfu deletion is represented as a black bar below the map. The minimal region of difference resulting in either HPFH or thalassemia phenotype is marked with vertical dashed lines. The arrow in this region represents the fetal-to-adult chromatin boundary/intergenic transcription start site.
It has been observed previously that individuals with δβ0 thalassemia (δβ0 thal) mutations, with severely decreased HbA expression, exhibit moderately increased HbF expression. In these cases HbF expression is heterogeneously distributed in red cells and is thought to be the result of increased erythropoiesis and preferential survival of normally rare cells with demonstrable levels of HbF (F cells). However, hereditary persistence of fetal hemoglobin (HPFH) deletion mutations result in high-level, pancellular HbF expression in adult cells. Two hypotheses, which are not necessarily exclusive, have been proposed to explain the differences between the 2 phenotypes. The most widely accepted hypothesis proposes that the juxtaposition of enhancer elements, normally located downstream of the globin locus and the HPFH breakpoints, exerts a positive effect on γ-globin transcription.3 This idea is supported by transgenic mouse studies.4,5 The other hypothesis to explain the phenotypic differences between the similar δβ0 and HPFH deletions proposes that a regulatory sequence necessary for γ gene silencing is deleted in the HPFH but not the δβ0 thal alleles.6 The putative 1-kb region that represents the minimum region of difference between HPFH and thal deletions is located approximately 3 to 4 kb upstream of the δ gene and is contained within the Corfu deletion (Figure 1). However, the Corfu deletion is often cited as evidence against the regulatory element hypothesis since Corfu heterozygotes do not show significant elevations of HbF.1,7
The function of the region upstream of the δ gene has been assessed in a number of studies using transgenic mice. Detailed analysis of this region has shown that a chromatin boundary is located approximately 3 to 4 kb upstream of the δ gene, defining 2 developmentally regulated chromatin subdomains, fetal and adult.8 An intergenic transcription start site has also been mapped in close proximity to the chromatin boundary 3.3 kb upstream of the δ gene. The intergenic δβ promoter is active only in stages where adult globin gene transcription occurs, and concurrent with chromatin remodeling of the adult subdomain.8 A 2.5-kb deletion encompassing the δβ promoter in YAC-transgenic mice9 results in a failure to open the adult subdomain and a drastic reduction in adult gene expression due in part to variegated expression of the β gene8 ; however, silencing of the γ genes was unperturbed. These data suggest that the region upstream of the δ gene plays a role in controlling chromatin structure of the adult subdomain.8 A more recent transgenic study confirms that deletion of this region has no effect on γ gene silencing or the timing of β-globin switching; however, chromatin structure, absolute transcription levels, and variegation were not analyzed.10 These data show that this region is not required for γ gene silencing in transgenic mice.
Here, we analyze adult primary culture erythroid cells from healthy individuals and patients with Corfu IVS-I-5 (hereafter referred to as Corfu) heterozygous, homozygous, and compound heterozygous thalassemia (Table 1). We show that in all cases the fetal γ genes are transcribed at abnormally high levels on the Corfu chromosome, demonstrating that γ-gene silencing is indeed impaired by the deletion. In addition, we show through quantification of RNA steady-state levels and Hb polypeptides that high-level transcription of the γ genes does not necessarily lead to a corresponding accumulation of γ mRNA and increased HbF expression, indicating that γ gene expression is post-transcriptionally regulated.
Hematology
Patient . | Genotype . | Hb g/dL . | HbA% . | HbF% . | Transfusions (wk)* . |
|---|---|---|---|---|---|
| A01 | Corfu/CD39 | 9.2† | 45 | 53 | Yes (5) |
| 8.6‡ | 2 | 86 | No | ||
| A02 | Corfu/Corfu | 8.8† | 10.2 | 88 | No |
| A06 | Corfu/Corfu | 9.2† | 9.7 | 90 | No |
| A07 | Corfu/N | 11.4† | 93 | 2.1 | No |
| A08 | Corfu/IVS-I-110 | 9.2† | — | — | Yes (2) |
| 6.2‡ | 23 | 75 | No | ||
| A09 | Corfu/IVS-I-110 | 9.5† | — | — | Yes (2) |
| A10 | CD39/IVS-I-5 | 10.6† | — | 1.6 | Yes (2) |
| 6.2‡ | 13 | 84 | No | ||
| A11 | Corfu/IVS-II-1 | 9.1† | — | 95 | No |
| 8.8‡ | — | 97 | No | ||
| A12 | Corfu/IVS-I-1 | 11.1† | — | 1.5 | Yes (1) |
| 7.2‡ | — | 96 | No | ||
| A13 | Corfu/N | 11.2† | — | 0.9 | No |
| Control | N/N | 12-15 | 95 | 1-2 | No |
Patient . | Genotype . | Hb g/dL . | HbA% . | HbF% . | Transfusions (wk)* . |
|---|---|---|---|---|---|
| A01 | Corfu/CD39 | 9.2† | 45 | 53 | Yes (5) |
| 8.6‡ | 2 | 86 | No | ||
| A02 | Corfu/Corfu | 8.8† | 10.2 | 88 | No |
| A06 | Corfu/Corfu | 9.2† | 9.7 | 90 | No |
| A07 | Corfu/N | 11.4† | 93 | 2.1 | No |
| A08 | Corfu/IVS-I-110 | 9.2† | — | — | Yes (2) |
| 6.2‡ | 23 | 75 | No | ||
| A09 | Corfu/IVS-I-110 | 9.5† | — | — | Yes (2) |
| A10 | CD39/IVS-I-5 | 10.6† | — | 1.6 | Yes (2) |
| 6.2‡ | 13 | 84 | No | ||
| A11 | Corfu/IVS-II-1 | 9.1† | — | 95 | No |
| 8.8‡ | — | 97 | No | ||
| A12 | Corfu/IVS-I-1 | 11.1† | — | 1.5 | Yes (1) |
| 7.2‡ | — | 96 | No | ||
| A13 | Corfu/N | 11.2† | — | 0.9 | No |
| Control | N/N | 12-15 | 95 | 1-2 | No |
Values in parentheses indicate time since last transfusion at the day of sampling
On the day of sampling
On diagnosis, prior to transfusion treatment
Patients, materials, and methods
Patients
Buffy coat cells from healthy individuals were obtained from a local blood bank. The Corfu patients (all have the 7.2-kb deletion and linked IVS-I-5 splice consensus mutation in the β gene) and their genotypes are listed, compound heterozygotes: A01, Corfu/CD39, Codon 39 nonsense mutation in the β gene; A08, Corfu/IVS-I-110, splicing mutation in intron I of the β gene; A09, Corfu/IVS-I-110, splicing mutation in intron I of the β gene; A11, Corfu/IVS-II-1, splicing mutation in intron II of the β gene; A12, Corfu/IVS-I-1, splicing mutation in intron I of the β gene; homozygotes: A02, Corfu/Corfu; A06, Corfu/Corfu; simple heterozygotes: A07, Corfu/healthy; A13, Corfu/healthy; non-Corfu patient: A10, IVS-I-5/CD39, splicing mutation in intron I of the β gene identical to the 1 in cis with the Corfu deletion, codon 39 nonsense mutation in the β gene.
Primary cell cultures
Peripheral blood collected from healthy individuals and patients with Corfu δβ thalassemia (50 mL) was diluted 1:1 with phosphate-buffered saline (PBS) and loaded onto Ficoll-Hypaque gradients (Sigma, St Louis, MO) and centrifuged at 1200g for 15 minutes. After centrifugation, the nucleated cells were washed twice with PBS and resuspended in alpha minimal essential medium supplemented with 1 μg/mL cyclosporin A, 10% fetal calf serum, and 10% 5637 bladder carcinoma cell line–conditioned medium as described.11 After 6 days in phase 1 culture, the nonadherent cells were harvested and cultured in phase 2 medium11 with 30% fetal calf serum, 1% deionized bovine serum albumin (BSA), 10 μM 2-mercaptoethanol, 1.5 mM glutamine, 1 μM dexamethasone, 1 U/mL recombinant erythropoietin, 1 ng/mL stem cell factor, and 0.3 mg/mL human transferrin. The phase 2–cultured cells were monitored on a daily basis for hemoglobinization. Cells were harvested at 2, 4, 6, and 8 days after the first appearance of hemoglobin (after hemoglobinization).
RNA FISH
RNA fluorescence in situ hybridization (FISH) was performed as previously described12 with the modifications listed in Gribnau et al.8 In all cases, a minimum of 200 cells were counted for each data point. A mixture of oligonucleotide probes was used for specific detection of each gene. Each oligonucleotide was triple-labeled with either digoxigenin (γ) or dinitrophenol (β). The following probes were used: γ gene, CGACCTGGACTTTTGCCAGGCACAGGGTCC, TCACTCCCAACCCCAGTATCTTCAAACAGC, GCATCTTTTTAACGACCATACTTGTCCTGC, ACAGAGCTGACTTTCAAAATCTACTCCAGC; β gene, TTCCACACTGATGCAATCATTCGTCTGTTT, TGTGTACACATATTAAAACATTACACTTTA, ATTAGCAATATGAAACCTCTTACATCAGTT, AGTAATGTACTAGGCAGACTGTGTAAAGTT.
RT-PCR
Total RNA was isolated from frozen pellets from cultured erythroid cells using the commercially available RNA-Bee reagent (Ambion, Austin, TX) following the manufacturer's instructions. RNA was reverse transcribed (RT) with Superscript II (Invitrogen, Carlsbad, CA). Steady-state primary transcript levels were quantitated by SYBR Green real-time polymerase chain reaction (PCR), performed on an ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA) with combinations of the following intron primer pairs: α-globin IVS-I primers, CACCCCTCACTCTGCTTCTC and CTTGCCGTGGCCCTTAAC; α-globin IVS-II primers, CTCTCAGGGCAGAGGATCAC and GCTGTGCAGAGAAGAGGGTC; γ-globin IVS-I primers, ACAAGGGAGGGAAGGAAGG and ACCAGGAGCCTGTGAGATTG; γ-globin IVS-II primers, TGGTGGCCAAACATACATTG and ACATACTTTGCCCCCATCTG.
Relative primary transcript levels were determined against genomic DNA standards. DNA contamination on the RT-PCR reactions was monitored by real-time PCR quantification of (-)RT control samples. PCR product formation in the (-)RT samples was 60- to 250-fold lower than the (+)RT samples, showing that DNA contamination was negligible.
RT-PCR analysis of steady-state mRNA levels was carried out by TaqMan real-time PCR as described in Fibach et al.13 Real-time PCR was performed on an ABI Prism 7000 Sequence Detection System (Applied Biosystems), and relative transcript levels were determined against genomic DNA standards.
Hemoglobin HPLC
Hemolysates were prepared from approximately 107 cells by resuspending in 200 μL 0.5 × reticulocyte standard buffer (RSB) with 1% NP40. The supernatant was collected and applied to a cation-exchange high-performance liquid chromatography column. Output was detected at 415 nm.
Results
Transcription of the γ- and β-globin genes during erythroid differentiation
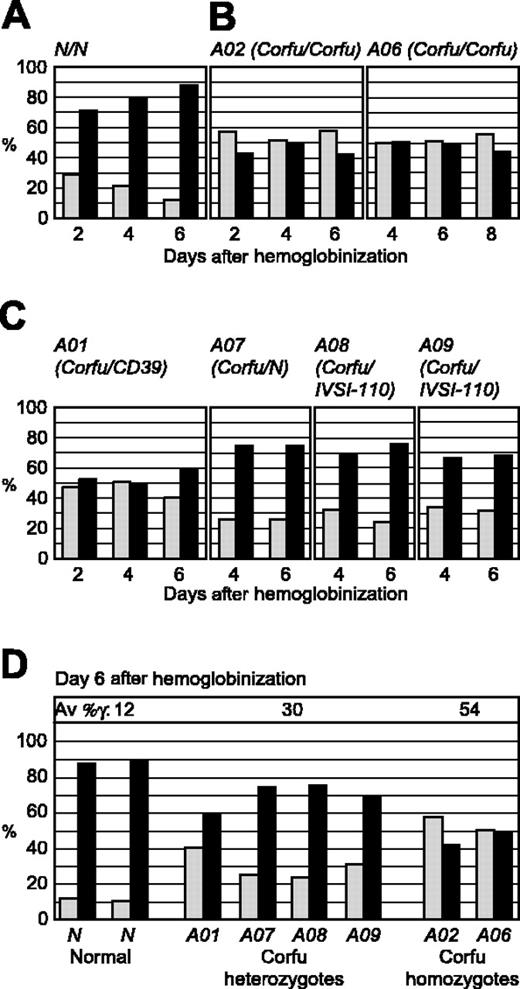
We first studied transcriptional regulation of the γ- and β-globin genes during differentiation of committed erythroid progenitors derived from peripheral blood from healthy individuals. In this procedure nucleated cells are isolated from peripheral blood and cultured according to the 2-phase liquid culture method described by Fibach and Rachmilewitz.14 Aliquots of cells were taken 2, 4, and 6 days after hemoglobin begins to accumulate (days 2, 4, 6 after hemoglobinization). The cells were fixed onto microscope slides for RNA FISH with gene-specific intron probes, and the relative amounts of γ- versus β-globin transcription foci were quantitated as previously described.12,15 From the onset of globin gene transcription the cells transcribe predominantly the β-globin gene with a small amount of γ gene transcription as expected16 (Figure 2A). As differentiation proceeds, globin loci with γ gene transcription foci decrease, while β gene transcription signals steadily increase in number. These results are consistent with previous observations, indicating that adult human erythroid cells express appreciable levels of both HbF and HbA early in erythroid differentiation16,17 prior to predominant expression of HbA late in the process, presumably reflecting γ gene silencing. These differentiation trends are highly reproducible in culture, although differentiation rates and absolute values for γ and β can vary between different batches of media.
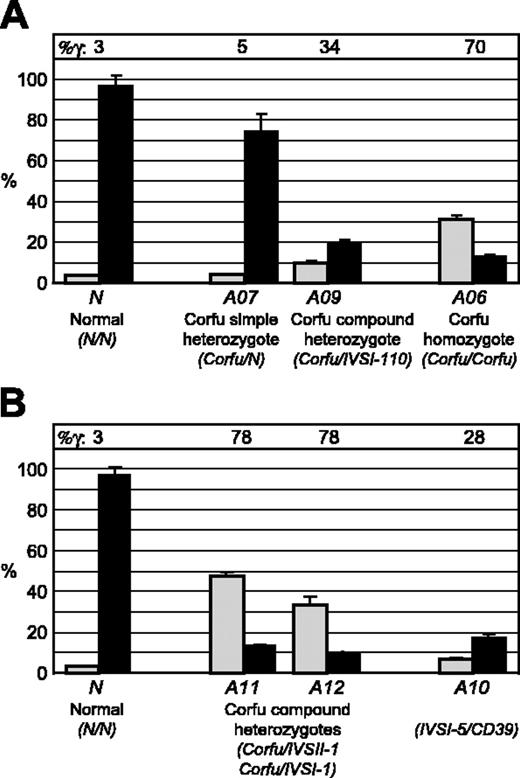
FISH analysis of γ and β gene transcription during erythroid differentiation in culture. Double-label RNA FISH was performed on cultured erythroid cells from healthy individuals and patients with Corfu deletion with gene-specific, intron probes for γ and β primary transcripts. Percentage of total γ and β gene signals are presented for 2, 4, and 6 days after hemoglobinization. ▦ indicates γ transcription; ▪, β transcription. (A) Healthy individual. Note, as cells mature, β transcription progressively increases while γ transcription is reciprocally silenced. (B) Corfu homozygotes A02 and A06. Note, γ versus β levels are stable over the culture period. (C) Corfu heterozygotes A01, A07, A08, A09. (D) γ Versus β transcription for all individuals assayed: 2 healthy subjects N; Corfu heterozygotes A01, A07, A08, A09; Corfu homozygotes A02 and A06, at day 6 after hemoglobinization when β transcription reaches 90% of total in normal cells. Average percentage of γ is indicated for each group of individuals as Av %γ, ie, 100% γ/(γ + β). Note, γ transcription increases with Corfu chromosome copy number.
FISH analysis of γ and β gene transcription during erythroid differentiation in culture. Double-label RNA FISH was performed on cultured erythroid cells from healthy individuals and patients with Corfu deletion with gene-specific, intron probes for γ and β primary transcripts. Percentage of total γ and β gene signals are presented for 2, 4, and 6 days after hemoglobinization. ▦ indicates γ transcription; ▪, β transcription. (A) Healthy individual. Note, as cells mature, β transcription progressively increases while γ transcription is reciprocally silenced. (B) Corfu homozygotes A02 and A06. Note, γ versus β levels are stable over the culture period. (C) Corfu heterozygotes A01, A07, A08, A09. (D) γ Versus β transcription for all individuals assayed: 2 healthy subjects N; Corfu heterozygotes A01, A07, A08, A09; Corfu homozygotes A02 and A06, at day 6 after hemoglobinization when β transcription reaches 90% of total in normal cells. Average percentage of γ is indicated for each group of individuals as Av %γ, ie, 100% γ/(γ + β). Note, γ transcription increases with Corfu chromosome copy number.
Transcriptional silencing of the γ genes is impaired in all Corfu individuals
To determine the effect of the Corfu mutation on globin gene transcription we performed RNA FISH on erythroid cells cultured from healthy individuals and 6 different Corfu patients (Table 1). We assayed cells from 3 compound heterozygotes (genotypes: A01, Corfu/CD39; A08, Corfu/IVS-I-110; A09, Corfu/IVS-I-110), 2 Corfu homozygotes (A02 and A06), and 1 simple Corfu heterozygote A07 (mother of A06). Patients A01, A02, A06, A08, and A09 all display greatly elevated levels of HbF, whereas A07 has normal, low-level HbF expression (Table 1). Analysis of the Corfu homozygotes A02 and A06 showed that γ transcription levels are significantly increased and β levels significantly decreased in comparison to normal cells (compare Figure 2B with 2A). Moreover, elevated γ transcription was remarkably stable throughout differentiation in A02 and A06. These results show that γ gene transcriptional silencing is impaired in the Corfu homozygotes. This was as expected, since homozygous patients are reported to have significantly elevated HbF. Elevated γ transcription was also observed in cells from compound heterozygotes A01, A08, and A09, consistent with the patients' hematology profiles; however, the amount of γ transcription was approximately half that of homozygotes (Figure 2C-D; Table 1). γ Levels in Corfu compound heterozygous cells drop only marginally with time in culture consistent with γ silencing of the allele in trans (Figure 2C).
Surprisingly, the simple Corfu heterozygote A07 also showed significantly elevated levels of γ gene transcription (Figure 2C-D). This result was unexpected since HbF is not elevated in patient A07 (Table 1), and previously reported Corfu heterozygotes do not have high HbF.1,18 In fact, the level of γ versus β transcription in A07 was very similar to the compound heterozygotes (A01, A08, A09), averaging 30% γ and 70% β on day 6 after hemoglobinization. γ Transcription levels in all Corfu heterozygous cells are nearly 2-fold lower than Corfu homozygous levels (average 30% versus 54%), indicating a clear link between Corfu chromosome dosage and the total level of γ transcription. An important conclusion from all these data is that elevated γ transcription in Corfu heterozygotes occurs regardless of whether the β locus in trans is normal or contains a β mutation.
Our RNA FISH analyses indicate that γ gene transcription is elevated in all carriers of the Corfu mutation. In view of the importance of this observation, we next sought to verify it by a completely independent method. We designed a real-time quantitative RT-PCR assay to measure γ globin primary transcript levels. We measured the abundance of γ primary transcripts using α-globin as an endogenous reference. Because of clinical limitations in obtaining patient material, the availability of RNA samples from primary erythroid cultures was severely restricted. We were only able to analyze RNA samples from 2 individuals of the original group, A07 and A09. In excellent agreement with our FISH results, γ primary transcript levels in A07 and A09 are approximately 2-fold higher than normal levels (Figure 3A), confirming that γ transcription is increased in Corfu individuals. To further investigate this, we obtained new samples from additional patients carrying the Corfu deletion: A11, Corfu/IVS-II-1; A12, Corfu/IVS-I-1; A13, simple Corfu heterozygote (Figure 3B). Although we noted some variation from the original cultures, the results clearly show that γ transcription is elevated in all Corfu subjects tested. Taken together, our data from 2 independent experimental approaches clearly demonstrate that γ transcription is abnormally high in adult individuals carrying the Corfu deletion, including simple heterozygotes.
RT-PCR analysis of γ gene transcription. Quantitative RT-PCR analysis of γ primary transcript levels in erythroid cell cultures from healthy individuals and patients with thalassemia. γ Primary transcript levels in patient versus normal cells were measured by quantitative RT-PCR with α levels as an endogenous reference. Results from 2 different sets of primary erythroid cultures are shown in panels A and B. The normal γ value is set to 1 in each experiment. (A) Relative γ transcript levels for a healthy individual N, simple Corfu heterozygote A07, and compound Corfu heterozygote A09. (B) Relative γ transcript levels for a healthy individual N, compound Corfu heterozygotes A11 and A12, simple Corfu heterozygote A13, and compound heterozygote A10 (IVS-I-5/CD39). Error bars indicate standard deviation between replicates.
RT-PCR analysis of γ gene transcription. Quantitative RT-PCR analysis of γ primary transcript levels in erythroid cell cultures from healthy individuals and patients with thalassemia. γ Primary transcript levels in patient versus normal cells were measured by quantitative RT-PCR with α levels as an endogenous reference. Results from 2 different sets of primary erythroid cultures are shown in panels A and B. The normal γ value is set to 1 in each experiment. (A) Relative γ transcript levels for a healthy individual N, simple Corfu heterozygote A07, and compound Corfu heterozygote A09. (B) Relative γ transcript levels for a healthy individual N, compound Corfu heterozygotes A11 and A12, simple Corfu heterozygote A13, and compound heterozygote A10 (IVS-I-5/CD39). Error bars indicate standard deviation between replicates.
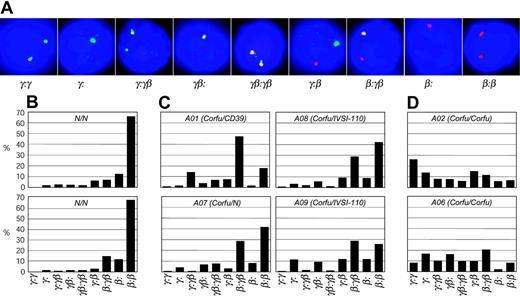
γ Gene transcription is specifically elevated in cis to the Corfu deletion
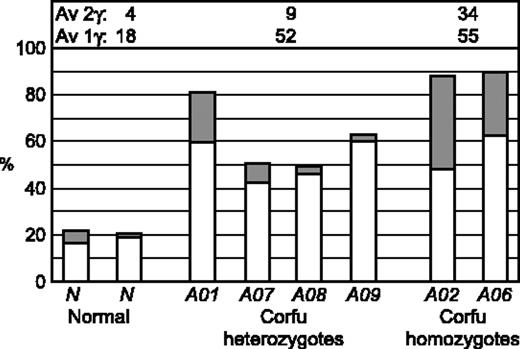
There are 9 possible transcriptional cell types resulting from the various transcriptional states of each locus: γ signal, β signal, double γβ signal, or no signal (Figure 4A). To see more clearly the pattern of γ and β transcription in the patients studied, we scored the transcriptional cell types for individuals A01, A02, A06, A07, A08, and A09 on day 6 after hemoglobinization (Figure 4B-D). Cells from healthy individuals display an overwhelming majority of β gene transcription signals with more than 65% of the cells having only β signals on both chromosomes (β:β cells, Figure 4B). All of the Corfu heterozygotes, including the simple heterozygote, show remarkably similar patterns (Figure 4C). In each case the γβ:β, γ:β, γβ, and γ cell types account for nearly all of the increase in γ transcription. These are all cell types in which only 1 chromosome has a γ signal or combination of γ and β. In the healthy individual, the “1γ cells” (γβ:β, γ:β, γβ, γ) account for about 18% of the total, while 4% are 2γ cells (γ signals on both chromosomes, ie, γβ:γβ, γ:γβ, and γ:γ) (Figure 5). In Corfu heterozygotes, however, more than half of all cells are 1γ cells (an increase of 34%), whereas 2γ cells increase only 5%. This result is wholly consistent with impairment of γ gene silencing on 1 chromosome in the Corfu heterozygotes. In homozygotes, the further rise in γ levels is almost completely accounted for by a 3-fold increase in 2γ cells to 34% (Figures 4D and 5). The percentage of 1γ cells increases only marginally compared with heterozygotes. These data show that increased γ transcription in Corfu heterozygotes is due to the propensity of 1 chromosome to transcribe the γ genes, whereas in homozygotes both chromosomes are capable of increased γ transcription. We conclude that γ gene silencing is impaired, and γ gene transcription is specifically elevated in cis to the Corfu mutation. Our data also show that γ silencing in cis to the CD39 β0 and IVS-I-110 β+ thal mutations proceeds normally, as expected.
Transcriptional cell types in human erythroid cells from healthy subjects and Corfu deletion patients. (A) Double-label RNA FISH on cells from day 6 after hemoglobinization, probed with γ (green) and β (red) primary transcript, intron probes. Nine different transcriptional cell types are observed, representing every possible combination of γ and β signals. (B) Distribution of cell types from 2 healthy individuals. (C) Corfu heterozygous individuals: compound Corfu heterozygotes A01, A08, A09; simple heterozygote A07. (D) Corfu homozygous patients A02 and A06.
Transcriptional cell types in human erythroid cells from healthy subjects and Corfu deletion patients. (A) Double-label RNA FISH on cells from day 6 after hemoglobinization, probed with γ (green) and β (red) primary transcript, intron probes. Nine different transcriptional cell types are observed, representing every possible combination of γ and β signals. (B) Distribution of cell types from 2 healthy individuals. (C) Corfu heterozygous individuals: compound Corfu heterozygotes A01, A08, A09; simple heterozygote A07. (D) Corfu homozygous patients A02 and A06.
Allele-specific transcription of the γ genes. Double-label RNA FISH was performed on erythroid cells from healthy individuals and patients with Corfu deletion. γ Primary transcript signals were quantified to highlight the distribution of transcriptional cell types with only 1 γ signal, 1γ cells (ie, γ + γβ:β + γ:β + γβ), or 2 γ signals, 1 on each allele, 2γ cells (ie, γ:γ + γβ:γβ + γ:γβ), and expressed as a percentage of the whole population of transcribing cells. □ indicates 1γ cells; ▦, 2γ cells. Average (Av) 1γ cell and 2γ cell values are indicated for normal, Corfu heterozygotes, and Corfu homozygotes. Note, increased numbers of 1γ cell numbers in heterozygote individuals A01, A07, A08, A09; increased 2γ cell numbers in homozygote individuals A02, A06.
Allele-specific transcription of the γ genes. Double-label RNA FISH was performed on erythroid cells from healthy individuals and patients with Corfu deletion. γ Primary transcript signals were quantified to highlight the distribution of transcriptional cell types with only 1 γ signal, 1γ cells (ie, γ + γβ:β + γ:β + γβ), or 2 γ signals, 1 on each allele, 2γ cells (ie, γ:γ + γβ:γβ + γ:γβ), and expressed as a percentage of the whole population of transcribing cells. □ indicates 1γ cells; ▦, 2γ cells. Average (Av) 1γ cell and 2γ cell values are indicated for normal, Corfu heterozygotes, and Corfu homozygotes. Note, increased numbers of 1γ cell numbers in heterozygote individuals A01, A07, A08, A09; increased 2γ cell numbers in homozygote individuals A02, A06.
We next wanted to determine whether the Corfu deletion or the IVS-I-5 splicing mutation was responsible for the elevated γ transcription phenotype associated with the Corfu genotype. To this end, we identified an individual (A10) who is a compound heterozygote carrying the βIVS-I-5 mutation without the Corfu deletion on 1 chromosome and a nonsense β CD39 mutation in trans. RT-PCR analysis of γ primary transcripts reveals that A10 has normal γ primary transcript levels (Figure 3B). These data show that the IVS-I-5 mutation alone does not give rise to increased γ transcription. We conclude that persistent γ transcription in Corfu individuals is dependent on the 7.2-kb deletion.
Post-transcriptional regulation of γ and β mRNA levels
We were surprised by the fact that the simple heterozygote A07 displayed increased γ transcription in the absence of any increase in HbF expression. We considered the possibility that HbF expression in adult cells is regulated at a post-transcriptional stage, such as γ mRNA processing, transport, or stability. Regulation at 1 of these stages might be expected to block accumulation of γ mRNA, hence restricting HbF expression. To determine whether elevated γ gene transcription resulted in a corresponding increase in γ mRNA level, we analyzed total RNA from the 2 groups of patient cultures. We quantitated the steady-state levels of γ-, and β-globin mRNA via real-time RT-PCR with TaqMan probes using α-globin mRNA levels as an endogenous reference. We assayed RNA from N, A06, A07, and A09 from the first batch of cultures (Figure 6A) and N, A10, A11, and A12 from the second batch (Figure 6B). Our results reveal 2 important points. First, the level of β mRNA is much lower in the Corfu patients (simple and compound heterozygotes and homozygotes) than would be predicted by the RNA FISH transcription analysis. This is undoubtedly because the Corfu β gene mutation (IVS-I-5) and the IVS-I-110, IVS-I-1, and IVS-II-1 mutations in trans would all be expected to reduce the steady-state level of β mRNA through nonsense-mediated decay.19 In each case β mRNA levels are reduced in accordance with mutant β gene dosage, with the trans β0 mutations resulting in lower β mRNA levels than β+ mutations, as expected. Second, γ mRNA levels do not correspond to transcription levels but instead vary from patient to patient, seemingly dependent on the levels of β mRNA. For example, A07 and A09 have nearly identical transcription patterns (Figure 2D), yet the level of γ mRNA in A09 is more than 2-fold higher than A07 (Figure 6A). This discrepancy becomes even more apparent when A09 is compared with A06. A06 should have twice as much γ mRNA as A09 because of the Corfu chromosome dosage is doubled in A06 and both are highly transcribing the γ gene. However, A06 actually has 3-fold higher γ mRNA levels compared with A09, and more than 6-fold higher levels than A07. We noticed in the patients with β0 mutations in trans (A11 and A12) that γ mRNA levels appeared to be even higher. This is most likely due in part to variations between cultures; however, the trans β0 mutations may also be a contributing factor (see “Discussion”). These results suggest a post-transcriptional regulatory mechanism, whereby γ mRNA steady-state levels are suppressed or specifically reduced in the presence of high steady-state β mRNA levels. This effect is even apparent in the normal blood samples (12% γ transcription; 3% γ mRNA).
γ And β mRNA steady-state levels. Total RNA was isolated from 2 batches of erythroid cultures; α, γ, and β mRNA levels were quantitated via real-time PCR. γ And β mRNA levels are presented as percentage normalized to α-globin mRNA level for each individual. ▦ indicates γ mRNA; ▪, β mRNA. Percentage of γ mRNA from total β-like mRNA is also indicated as %γ, ie, 100% γ/(γ + β). (A) Normal individual N, simple Corfu heterozygote A07, compound Corfu heterozygote A09, and Corfu homozygote A06. (B) Normal individual N, compound Corfu heterozygotes A11 and A12, simple Corfu heterozygote A13, and compound heterozygote A10 (IVS-I-5/CD39).
γ And β mRNA steady-state levels. Total RNA was isolated from 2 batches of erythroid cultures; α, γ, and β mRNA levels were quantitated via real-time PCR. γ And β mRNA levels are presented as percentage normalized to α-globin mRNA level for each individual. ▦ indicates γ mRNA; ▪, β mRNA. Percentage of γ mRNA from total β-like mRNA is also indicated as %γ, ie, 100% γ/(γ + β). (A) Normal individual N, simple Corfu heterozygote A07, compound Corfu heterozygote A09, and Corfu homozygote A06. (B) Normal individual N, compound Corfu heterozygotes A11 and A12, simple Corfu heterozygote A13, and compound heterozygote A10 (IVS-I-5/CD39).
γ mRNA level is the major determinant of HbF expression
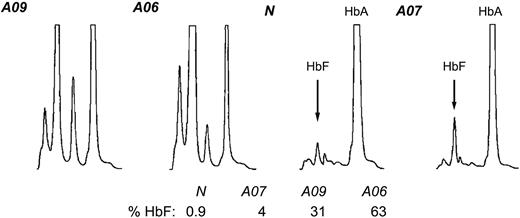
To complete our analysis of globin expression in the context of the Corfu deletion we quantitated HbF and HbA levels in the same primary erythroid cultures. We carried out HPLC analysis20 of cell lysates from A06, A07, A09, and healthy individuals (Figure 7). Normal cells contain 1% HbF and A07 has 4% HbF, whereas A09 and A06 have 31% and 63% HbF, respectively. These percentages are equivalent to the respective values for the relative abundance of γ mRNA in these cells measured by RT-PCR (compare Figure 6A with Figure 7). Moreover, they are in very good agreement with the hematology profiles (Table 1). These data suggest that HbF expression is strictly dependent on γ mRNA abundance. These findings do not rule out the possibility that the final HbF/HbA output is adjusted by additional downstream mechanisms, but their contribution is (likely to be) less significant.
Hemoglobin HPLC. Lysates from erythroid cell cultures from healthy individual N, simple Corfu heterozygote A07, compound Corfu heterozygote A09, and Corfu homozygote A06 were quantitated via cation-exchange HPLC. The peaks for HbA and HbF are indicated. The percentage of HbF from total hemoglobin, % HbF, is also indicated for each individual.
Hemoglobin HPLC. Lysates from erythroid cell cultures from healthy individual N, simple Corfu heterozygote A07, compound Corfu heterozygote A09, and Corfu homozygote A06 were quantitated via cation-exchange HPLC. The peaks for HbA and HbF are indicated. The percentage of HbF from total hemoglobin, % HbF, is also indicated for each individual.
Of particular interest is the case of the simple Corfu heterozygote A07, who has equivalent high levels of γ gene transcription to A09, but much lower levels of γ mRNA and HbF. High levels of HbF are specifically associated with pathologically low β-globin production, as in Corfu compound heterozygotes and homozygotes.
Discussion
The Corfu paradox
To gain insight into β-like gene regulation in the context of the Corfu deletion genotype we studied erythroid cultures from Corfu individuals, focusing on several key stages of the hemoglobin production pathway: primary transcription, mRNA steady-state levels, and Hb tetramer accumulation. Our results illuminate the paradox surrounding the effects of the Corfu deletion on globin gene regulation. We show that in all cases, the Corfu mutation leads to greatly increased γ gene transcription in cis. However, γ mRNA levels do not directly follow γ gene transcription levels. Instead, it appears that γ mRNA levels and hence HbF expression are dependent on the level of β mRNA. This is particularly evident through comparison of the simple heterozygote A07 with the compound heterozygotes A08 and A09. These individuals differ by the IVS-I-110 splicing mutation carried by A08 and A09 in trans to the Corfu deletion. This mutation, which creates an alternative splice site in the β transcript, would be expected to reduce the amount of mature β message without necessarily affecting the transcription rate (however, it may have a slight effect on the number of transcription foci seen by RNA FISH15,21,22 ). Indeed, A07, A08, and A09 exhibit very similar transcription profiles as assayed by RNA FISH: elevated γ transcription from the Corfu chromosome and high β transcription from the normal and IVS-I-110 chromosomes, respectively. However, the differing fate of the γ primary transcripts imposes significant differences at the mRNA and Hb levels. For example, the β mRNA level in A07 is slightly lower than normal (70%) most likely because of the IVS-I-5 mutation on the β gene on the Corfu chromosome and competition from the γ genes. This results in a corresponding decrease in HbA level. However, although simple heterozygotes have equivalent high levels of γ transcription compared with compound heterozygotes, γ mRNA does not accumulate, and they produce only marginally more HbF than healthy individuals. This suggests that HbF expression is blocked in simple Corfu heterozygotes at a post-transcriptional stage. Most likely, the same mechanism limits HbF expression in healthy individuals as well (12% γ gene transcription but only 1% HbF). Compound heterozygotes A08 and A09 carry an additional β splicing mutation in trans to the Corfu locus. Consequently, they suffer from a severe reduction of β mRNA and HbA (20% of normal level). However, they have higher absolute levels of γ mRNA and HbF than simple heterozygotes even though γ transcription is equivalent in all of them. Consistent with this trend, erythroid cells from compound heterozygotes with trans β0 mutations (A11 and A12) have even higher levels of γ mRNA.
The Corfu homozygote A06 exhibits lower β mRNA and HbA levels than the Corfu β+ compound heterozygotes since β transcription is reduced partially through both the IVS-I-5 splice mutations and competition from the activated γ genes on both chromosomes. An apparent consequence of the severely depleted β mRNA level is the accumulation of γ mRNA and HbF to very high values.
The fact that β gene mutations that greatly decrease β mRNA levels lead to increased γ mRNA levels suggests that γ mRNA accumulation is indirectly dependent on the total amount of viable β mRNA. These results provide a plausible explanation for the Corfu paradox, showing that HbF levels do not necessarily reflect what is happening at the transcriptional level, and that increased γ gene transcription per se is not sufficient for high HbF expression. We propose that there is a preference for β over γ RNA accumulation in adult erythroid cells. Previous studies have shown preferential stabilization of α- over embryonic ζ-globin mRNA, through binding of a ribonucleoprotein complex to the 3′ untranslated region (UTR) of the mRNA.23 α mRNA has a higher affinity for the messenger ribonucleoprotein (mRNP) complex than ζ mRNA. A specific yet related β mRNA-stabilizing mRNP complex has been identified.24 The affinity of this complex for γ mRNA has not been described; however, one could imagine a parallel preferential stabilization of β mRNA over γ, similar to that proposed for α-globin mRNA. The possibility of post-transcriptional regulation of HbF expression has been suggested before.25,26 However, it has not been supported by comprehensive experimental data and, as a consequence, has remained largely overlooked. The Corfu case is not the only example of a mutation displaying low HbF expression in the heterozygote state and very high HbF expression in a compound heterozygote background. A study by Weinberg et al27 showed a similar phenomenon in a simple HPFH heterozygote compared with her β0/HPFH compound heterozygous child. The overall production of HbF always seems to be modulated in the context of other mutations in the β-globin alleles.
These findings are of fundamental significance for the large number of studies that attempt pharmacologic reactivation of γ gene transcription with the aim of increased HbF production in patients with β gene hemoglobinopathies. High HbF levels are known to ameliorate the clinical symptoms of sickle cell anemia and β thalassemia. Several clinical studies have been carried out with compounds that are thought to function through HDAC (histone deacetylase) inhibition, aimed at γ gene transcriptional de-repression, with the outcome most often measured in terms of HbF levels. The results presented here show that HbF levels are not a reliable indicator of γ transcriptional status; therefore, measuring HbF to assess the effect of these compounds on γ gene activation is dubious. Our data suggest that combinatorial therapies may need to be administered to increase both γ transcription and mRNA maturation or stabilization.
γ Silencing is impaired in cis of the Corfu deletion
The Corfu case accentuates the key question about the molecular mechanisms involved in activation of the fetal genes in the adult. The fact that in heterozygous cells γ transcription is specifically elevated on the Corfu chromosome, while the γ genes in trans are not active, shows that the mechanism does not involve increased numbers of F cells. Furthermore, selective pressures that give rise to high numbers of F cells in thal patients are unlikely to be reproduced in the liquid cultures used in this study. A central question, therefore, is what maintains the γ genes active in the Corfu locus. Our data show that the adult transcription factor environment can neither turn on the silenced γ genes on the nondeletion alleles nor silence the γ genes of the Corfu locus. Therefore, the effect of putative silencing factors binding to the γ genes can be overridden in the absence of the deleted region. This suggests that if the trans-acting factor environment is playing a role in γ gene silencing, it must be doing so, at least in part, through elements located in the deleted region. However, there is no specific requirement for this region in γ silencing since other deletions that remove these sequences and the δ and β genes result in silenced γ genes.28 An integrated model of the molecular nature of globin gene switching needs to incorporate the role of chromatin domain structure and regulation. Our data in transgenic mice have shown that the fetal subdomain acquires an accessible chromatin conformation concomitant with intergenic transcription early in development as part of the normal erythroid differentiation program. Later in development intergenic transcription in the fetal domain is extinguished with simultaneous silencing of the γ genes and chromatin condensation across the subdomain, while the adult subdomain opens to allow β gene transcription.8 We have demonstrated that in transgenic mice the adult subdomain cannot acquire an open chromatin structure in the absence of a 2.5-kb fragment containing the δβ intergenic promoter.8
However, the analysis presented here of the larger Corfu deletion is phenotypically distinct from the transgenic mice. This difference could be due to the differing sizes of the deletion. It is possible that the 2.5-kb transgenic deletion that removed the intergenic promoter that is necessary for activation of the adult subdomain left behind a boundary element that acts as a chromatin barrier to shield the fetal and adult domains. Another possibility is the presence of a pyrimidine-rich (pyr) sequence located just upstream of the δ gene that is removed on the Corfu deletion but not in the transgenics. Deletion of this element resulted in a delayed γ to β switching in transgenic mice.29 Subsequent work has shown that the pyr element recruits an Ikaros-containing complex with potential chromatin remodeling activity.30 The final possibility is that mice do not regulate the human locus in exactly the same way as in human erythroid cells. For example, γ gene transcription does not occur during early differentiation of bone marrow–derived cells in transgenic mice, as it does in humans.
In conclusion, we have shown that the Corfu deletion leads to disruption of fetal gene silencing resulting in prolonged transcription of the fetal genes during adult life. However, fetal gene expression is subject to post-transcriptional regulation; fetal hemoglobin reaches high levels only in combination with additional mutations that reduce the amount of adult β-globin message.
Prepublished online as Blood First Edition Paper, November 9, 2004; DOI 10.1182/blood-2003-11-4069.
Supported in part by the Medical Research Council and the Biotechnology and Biological Sciences Research Council, United Kingdom. P.F. is a Senior Fellow of the Medical Research Council.
L.C. and C.S.O. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Wolf Reik, Gavin Kelsey, and Emmanuel Debrand for critically reading the manuscript; George Stamatoyannopoulos and Bill Wood for helpful discussions; and Rob Macleash for HPLC analysis.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal