Abstract
Protein 4.1R is a vital component of the red blood cell membrane cytoskeleton. Promotion of cytoskeletal junctional complex stability requires an erythroid differentiation stage–specific splicing switch promoting inclusion of exon 16 within the spectrin/actin binding domain. We showed earlier that an intricate combination of positive and negative RNA elements controls exon 16 splicing. In this report, we further identified 3 putative exonic splicing enhancers within exon 16 and investigated the function of the sequence CAGACAT in the regulation of exon 16 splicing. Mutation of these sequences leads to increased exclusion of exon 16 in both in vivo and in vitro splicing assays, indicating that CAGACAT is a functional exonic splicing enhancer. UV cross-linking further detects an approximately 33-kDa protein that specifically binds to the CAGACAT-containing transcript. An anti-SF2/ASF antibody specifically immunoprecipitates the approximately 33-kDa protein. Furthermore, SF2/ASF stimulates exon 16 inclusion in both in vitro complementation assays and minigene-transfected mouse erythroleukemia cells (MELCs). Finally, SF2/ASF expression is up-regulated and correlates with exon 16 inclusion in differentiated MELCs. These results suggest that increased splicing factor 2/alternative splicing factor (SF2/ASF) expression in differentiated mouse erythroleukemia mediates a differentiation stage–specific exon 16 splicing switch through its interaction with the exonic splicing enhancer.
Introduction
Protein 4.1R (4.1R) is a vital component of the red blood cell membrane cytoskeleton. The 4.1R stabilizes the spectrin-actin lattice while attaching to the embedded membrane proteins band 3 and glycophorin. The 80-kDa erythrocyte form is the prototype of a complex array of 4.1R isoforms whose expression is regulated by coupled transcription and splicing,1 utilization of alternative translation initiation sites,2,3 alternative pre-mRNA splicing,4,5 and posttranslational modification.6 The expression of the 80-kDa erythrocyte form during erythropoiesis is tightly regulated through 2 splicing events in a differentiation stage–specific manner.7,8 At an early stage, before erythroid colony-forming units (CFU-Es), suppression of exon 2′ inclusion causes the omission of an upstream translation start site used for the synthesis of “nonerythroid” 135-kDa forms. In erythroblasts, a later event is the induction of the inclusion of exon 16, which encodes a critical portion of the spectrin/actin binding (SAB) domain required to promote cytoskeletal junctional complex stability.9,10
Correct gene expression requires accurate and efficient removal of introns from pre-mRNAs. In addition to the information contained in canonical splice signals (5′ splice site, branch site, and 3′ splice site), precise exon definition requires additional regulatory cis elements present in the exon and its flanking introns.11 These elements become particularly important in the presence of weak splice sites or in alternative splicing.12,13 Exonic splicing enhancers (ESEs) and exonic splicing silencers (ESSs), which are ubiquitous in both constitutively and alternatively spliced exons, account for some of these signals.14,15 ESEs act as binding sites for serine/arginine-rich (SR) proteins, a family of highly conserved and structurally similar splicing factors that act at multiple steps of the pre-mRNA splicing pathway.16 SR proteins bind to ESEs through their RNA binding domain and promote splicing by recruiting spliceosomal components via protein-protein interactions mediated by the arginine serine-rich (RS) domain.17,18 ESSs, on the other hand, function as binding sites for repressor proteins.14,19,20 The members of the heterogeneous nuclear ribonucleoprotein (hnRNP) are often involved in negative regulation by binding to ESSs.14,21,22 A number of test cases have shown that SR proteins and hnRNPA1 antagonistically affect 5′ splice site recognition and that subtle changes in the relative concentrations of these 2 factors can determine 5′ splice site recognition.23,24 The balance between positive and negative regulation of splice site selection likely depends on the cis element's identity and on changes in cellular splicing factors under physiologic or pathologic conditions.25,26
Splicing of 4.1R exon 16 is highly regulated in a differentiation stage–specific manner during erythroid differentiation; exon 16 is omitted from much of the 4.1R mRNA of pre-erythroid cells but is included in most of the mRNA produced in late erythroid cells.7,8 Using a mouse erythroleukemia cell (MELC) system, we have shown (1) that exon 16 is preferentially included in mature 4.1R mRNA in induced MELCs and (2) that the protein isoform bearing exon 16–encoding peptide is rapidly recruited into the membrane.8 We have also shown that exons and abutting intronic sequences contain both positive and negative elements serving as primary targets to promote exon skipping in predifferentiated MELCs and to allow exon inclusion as the cells differentiate.27 Among these elements, the 15 nucleotides (nt's) at the 5′ end and the 42 nt's at the 3′ end of exon 16 have been identified as ESSs.27,28 Our comparison of these regions to known putative ESE sequences29 revealed 3 SR putative protein binding sites within exon 16. In this report, we showed that SR protein SF2/ASF binds to ESEs (CAGACAT) and regulates the extent of exon 16 inclusion in in vitro and in vivo assays. We also demonstrated a correlation between the differentiation-specific up-regulation of SF2/ASF expression and the increased efficiency of exon 16 splicing in maturing erythroid cells. These results suggest that SF2/ASF is important in regulated differentiation stage–specific exon 16 splicing through its interaction with the ESE (CAGATAG) located on exon 16.
Materials and methods
Plasmid constructs
The wild-type (WT), 15GE, and 42GE exon 16 minigene constructs composed of a 3-exon13,16,17 splicing cassette in pRc-cytomegalovirus (pRc-CMV) vector (Invitrogen, Carlsbad, CA) has been described.27 The 15GE and 42GE constructs were made by replacing the 15 nt's at the 5′ and the 42 nt's at the 3′ of exon 16 with the corresponding β-globin exon 2 sequences, respectively.27 The exonic mutation constructs EX-1, EX-2, and EX-3 were made by 2-step polymerase chain reaction (PCR) using the wild type as a template and amplified with the outer primer set (5′-GAAGACGAGCCACCTGAGCAA-3′, 5′-CTAGGGGCAACCCAGCAGAG-3′) and its respective inner primer set. The inner primer sets (with mutated nucleotides underlined; 5′-GAGAATTTTTTTATGGTGAAAACATTTATATCAGAC-3′, 5′-CACCATAAAAAAATTCTCTCTTTTTCTGTG-3′; 5′-TTTTTTAGACATAGCAATTTAATGTTGGAGG-3′, 5′-GCTATGTCTAAAAAAAATGTTTTCACCATCTAG-3′; and 5′-TTTTTTTAGCAATTTAATGTTGGAGGTTTG-3′, 5′-TAAATTGCTAAAAAAAATATAAATGTTTTCACCATC-3′) were used for the incorporation of EX-1, EX-2, and EX-3 mutation, respectively. For EX-2.1, EX-3.1, and EX-3.2 constructs, the inner primer sets (5′-TATTATAGACATAGCAATTTAATGTTGGAGG-3′, 5′-GCTATGTCTATAATAAATGTTTTCACCATCTAG-3′; 5′-TTATATTAGCAATTTAATGTTGGAGGTTTG-3′, 5′-TAAATTGCTAATATAAATATAAATGTTTTCACCATC-3′; and 5′-TTATTATAGCAATTTAATGTTGGAGGTTTG-3′, 5′-TAAATTGCTATAATAAATATAAATGTTTTCACCATC-3′) were used, respectively. The resultant second-step PCR products were digested with BstEII and NheI and subsequently subcloned into the same restriction sites in the wild type.
The UV cross-linking constructs wt, ex-1, ex-2, and ex-3 were prepared by PCR amplification using its respective WT, EX-1, EX-2, and EX-3 minigene construct as template and primer sets (5′-CATTAAGCTTTTTTTCCCGCAAGAACAACATTTTA-3′, 5′-ATCATCTAGAGGCAACCAACTGAAATATGCTTCAAGT-3′) with HindIII and XbaI linkers. The amplified fragments were then subcloned into pcDNA3 vector (Invitrogen) between the HindIII and XbaI sites. SF2/ASF expression construct was described.30
Cell culture, transfection, and flow cytometry analysis of MEL differentiation and cell sorting
MELCs and HeLa cells were cultured in Dulbecco modified Eagle medium (DMEM) medium with 10% fetal bovine serum (FBS). L8 cells (Rat skeletal muscle myoblast, ATCC CRL-1769; American Type Culture Collection, Manassas, VA) were cultured in 4:1 mixture of DMEM medium and M199, plus 1% chicken embryo extract and 10% horse serum. Cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's procedures. Selection of stable cell lines was performed with the addition of 800 μg/mL of G-418.
MELCs were induced to erythroid differentiation on fibronectin substratum for 2, 4, or 6 days.27 Cells were removed from fibronectin-coated plates by incubating in phosphate-buffered saline (PBS) with 10% FBS and 5 mM EDTA (ethylenediaminetetraacetic acid) at 37°C for 5 minutes. After washing with PBS containing 2% FBS, cells were immunostained with fluorescein isothiocyanate (FITC)–conjugated anti-CD71 (0.5 μg/106 cells) and phycoerythrin (PE)–conjugated anti-TER119 (0.5 μg/106 cells; BD Pharmingen, San Diego, CA) antibodies. The 7-aminoactinomycin D (7-AAD) was added to exclude dead cells from analysis. CD71 and TER119+ cells were sorted from day-4–induced MELCs by a Mo-Flo machine (DakoCytomation, Carpinteria, CA). The sorted cells were spread on Superfrost/Plus microscope slides (Fisher Scientific, Pittsburgh, PA), fixed in methanol, air dried, and stained in Giemsa Stain according to the manufacturer's procedure (Sigma). The samples were viewed with a Nikon Eclipse E800 microscope (Nikon, Inc, USA) equipped with Spot Flex camera (Diagnostic Instruments Inc, Sterling Heights, MI) under a 40 × Plan Apo objective. The images were collected using Spot software (Diagnostic Instruments, Inc) and processed using Photoshop software (Adobe Systems, Inc).
Semiquantitative RT-PCR
Total RNA was isolated using TRIZOL reagent (Invitrogen) from stable lines as well as from transient transfected cells 48 hours after transfection. Three micrograms of RNA was reverse transcribed using the SuperScript II reverse transcription (RT) kit (Invitrogen) in a 20-μL reaction containing 2 pmol SP6 primer. PCR amplifications were performed with primers (5′-AGAGCCCACAGAAGCATGGA-3′ and 5′-GTGTGTAGATAAGCCCTTGTCCCA-3′) located at exon 13 and 17, respectively. A limiting cycle amplification protocol31 was used to obtain the PCR reaction within its linear range. Aliquots of the PCR reactions were initially withdrawn for agarose gel analysis at 20 cycles. The reaction was then resumed for 2 additional cycles, and aliquots were withdrawn again. This procedure was repeated several more times to determine the optimal number of amplification cycles. Twenty-two amplification cycles, the minimal number of cycles sufficient for detecting clearly visible PCR bands, were used for semiquantitative analysis of the spliced products. The bands of interest were quantified using analysis software from ChemiImager 5500 system (Alpha Innotech, San Leandro, CA).
HeLa nuclear extract preparation and SR protein purification
HeLa S3 cells were grown as suspension culture in Joklik Minimum Essential Medium (JMEM) (Sigma, St Louis, MO) and 5% horse serum in spinner flasks at a density of 2 × 105 to 3 × 105 cells/mL. The purification of nuclear extracts from HeLa cells was as described.32 In brief, 10 liters of HeLa cells were collected and washed with PBS. The cell pellet was suspended in buffer A (10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], pH 7.6; 1.5 mM MgCl2; 10 mM KCl; 0.2 mM phenylmethylsulfonyl fluoride [PMSF]; and 0.5 mM dithiothreitol [DTT]) and Douncer homogenized. The nuclei were separated from the cytosol fraction by centrifugation. The nuclei were resuspended in buffer C (20 mM HEPES, pH 7.6; 1.5 mM MgCl2; 420 mM KCl; 0.2 mM EDTA; 25% glycerol; 0.2 mM PMSF; and 0.5 mM DTT) and rotated at 4°C for 30 minutes. The nuclear extracts were recovered by centrifugation at 30 500g, dialyzed in buffer D (20 mM HEPES, pH 7.6; 100 mM KCl; 0.2 mM EDTA; 20% glycerol; 0.2 mM PMSF; and 0.5 mM DTT), and frozen in liquid N2. The cytosol fraction was centrifuged at 100 000g for 1 hour. The S100 fraction collected from the supernatant was dialyzed in buffer D, frozen in liquid N2, and stored at -80°C. SR proteins were purified from HeLa cells as described.33
In vitro transcription and in vitro splicing
Pre-mRNA substrates for in vitro splicing and UV cross-linking were produced from plasmids linearized with XbaI and transcribed with T7 RNA polymerase (Amersham Pharmacia Biotech, Piscataway, NJ) in the presence of a Ribo m7G Cap Analog (Promega, Madison, WI) and α-32P–nucleoside triphosphate (α-32P–NTP; PerkinElmer, Shelton, CT).
In vitro splicing reactions were performed in a 25-μL splicing reaction containing 20 ng α-32P–labeled pre-mRNA substrates (∼20 000 counts per minute [cpm]), 0.5 mM adenosine triphosphate (ATP), 20 mM creatine phosphate, 3.2 mM MgCl2, 7.5 μL splicing dilution buffer (20 mM Tris-Cl, pH 7.6; 100 mM KCl), and 7.5 to 10 μL HeLa nuclear extract. The reactions were incubated at 30°C for 3 hours. The reactions were stopped by the addition of 70 μL H2O, 100 μL 2 × protein kinase (PK) buffer (100 mM Tris-Cl, pH 7.5; 20 mM EDTA; 20 mM NaCl; 4% sodium dodecyl sulfate [SDS]), and 5 μL (10 mg/mL) proteinase K (Roche Molecular Biochemicals, Indianapolis, IN) and incubated at 37°C for 10 minutes. The reactions were phenol extracted, ethanol precipitated, and analyzed on a 4% sequencing polyacrylamide gel electrophoresis (PAGE) gel. The spliced products were visualized by autoradiography. The complementation assays were performed in a splicing reaction using S100 fraction and with the addition of recombinant SF2/ASF purified from baculovirus that was purchased from ProteinOne (College Park, MD).
UV cross-linking, immunoprecipitation, and immunoblotting
The reaction conditions for UV cross-linking were identical to that of in vitro splicing. In brief, 20 ng α-32P–labeled pre-mRNA was incubated under splicing conditions using HeLa nuclear extracts or SR proteins in the presence of a 5-fold molar excess of a nonspecific tRNA or a specific wt competitor. The reactions were incubated at 30°C for 20 minutes and exposed to 254 nm UV light using an UV stratalinker 2400 (Stratagene, La Jolla, CA) with a total energy of 12 000 μJ. Samples were treated with 5 μL RNase Cocktail (Ambion, Austin, TX) for 30 minutes at 37°C and analyzed by SDS-PAGE electrophoresis. The cross-linked proteins were visualized by autoradiography.
For immunoprecipitation assays, RNase-treated samples were diluted to 500 μL with immunoprecipitated (IP) buffer (50 mM Tris-HCl, pH 8.0; 150 mM NaCl; 0.1% nonidet P–40 [NP40]; and 1 mM PMSF). The samples were precleared with 10 μL rabbit preimmune serum bound to 50 μL protein G–Sepharose. The precleared samples were then incubated with 5 μg each of anti-SF2/ASF (Zymed Laboratories, South San Francisco, CA), anti-9G8 (Santa Cruz Biotechnology, Santa Cruz, CA), or anti-SC35 (BD Pharmingen) antibodies bound to 50 μL of protein G–Sepharose at 4°C overnight. After extensive washing with IP buffer, the samples were heated at 90°C for 5 minutes in SDS sample buffer and analyzed by a 10% SDS-PAGE gel. The immunoprecipitated proteins were visualized by autoradiography.
For immunoblotting, 107 cells were lysed in 500 μL triple-lysis buffer.34 The protein contents were determined using a standard bicinchoninic acid (BCA) assay determination kit (Pierce Chemical, Rockford, IL). Proteins were transferred to nitrocellulose membranes and the detection of SF2/ASF was carried out by immunoblotting with an anti-SF2/ASF antibody using an enhanced chemiluminescence (ECL) detection kit (Amersham Pharmacia Biotech). Anti–β-actin antibody (Sigma) was used as a loading control.
Results
Identification of 3 exonic splicing enhancers
We established an exon 16 minigene system in MELCs that mimics endogenous exon 16 splicing patterns during dimethyl sulfoxide (DMSO)–induced erythroid differentiation.27 Deguillien et al27 and Hou et al28 have also demonstrated silencing activity at the 15 nt's at the 5′ and the 42 nt's at the 3′ end of exon 16. Analysis of exon 16 sequences with the ESE prediction program ESEfinder29 revealed similarity between 3 sequences within the silencer regions and the putative ESEs responsive to human SR proteins. These sequences (AGACTAG, nt's 13-19; TATATC, nt's 34-39; and CAGACAT, nt's 39-45) display sequence scores above the selected threshold values for SR proteins SRp40, SRp55, and SF2/ASF, respectively. However, SR protein binding motifs are relatively degenerate within a broader range of sequences.29,35 Thus, the putative SR binding motifs in exon 16 do not present as unique binding sites for their respective SR proteins.
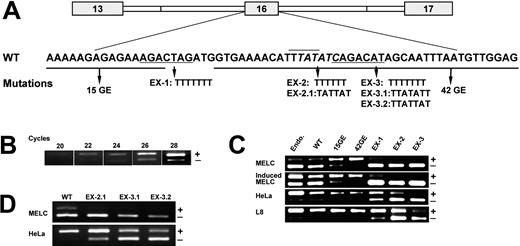
To determine whether the putative ESE elements affect exon 16 splicing, we generated mutated forms of exon 16 minigenes in which we replaced the putative ESE sequences with U residues (Figure 1A). Substituting U residues for ESEs has been used widely to analyze ESE function.36,37 This replacement also avoids creating any missense or nonsense mutation codons that could affect splicing activity.38 We standardized the semiquantitative RT-PCR analysis of the spliced products from WT-transfected HeLa using 22 cycles, the minimal number of amplification cycles that is sufficient for detecting clearly visible PCR bands (Figure 1B). This standardization was used throughout the study.
Effect of exon mutations on exon 16 splicing during MELC induction as well as in HeLa and L8 cell lines. (A) Diagram of exon 16 minigene construct, GE15, GE42, and exonic mutations (EX-1, EX-2, EX-2.1, EX-3, EX-3.1, and EX-3.2) within exon 16. The wild-type and mutated sequences are indicated. (B) Standardization of semiquantitative RT-PCR. Agarose gel electrophoretic analysis of RT-PCR performed around exon 16 from WT-transfected HeLa at an increasing number of amplification cycles. + indicates exon 16 inclusion; and -, exon 16 exclusion. The minimal number of cycles, 22 cycles, that is sufficient for detecting clearly visible bands was subsequently used to analyze the spliced products. (C) Analysis of exonic mutations on exon 16 splicing by RT-PCR. The minigene and its mutation construct stably transfected MELCs were induced to differentiate with DMSO as described in “Materials and methods.” Total RNA was collected from cells as indicated, subjected to RT-PCR, and analyzed by 2% agarose gel. + indicates exon 16 inclusion; and -, exon 16 exclusion. (D) Effect of EX-2.1, EX-3.1, and EX-3.2 on exon 16 splicing by RT-PCR. + indicates exon 16 inclusion; and -, exon 16 exclusion. Endo. indicates endogenous exon 16 splicing patterns.
Effect of exon mutations on exon 16 splicing during MELC induction as well as in HeLa and L8 cell lines. (A) Diagram of exon 16 minigene construct, GE15, GE42, and exonic mutations (EX-1, EX-2, EX-2.1, EX-3, EX-3.1, and EX-3.2) within exon 16. The wild-type and mutated sequences are indicated. (B) Standardization of semiquantitative RT-PCR. Agarose gel electrophoretic analysis of RT-PCR performed around exon 16 from WT-transfected HeLa at an increasing number of amplification cycles. + indicates exon 16 inclusion; and -, exon 16 exclusion. The minimal number of cycles, 22 cycles, that is sufficient for detecting clearly visible bands was subsequently used to analyze the spliced products. (C) Analysis of exonic mutations on exon 16 splicing by RT-PCR. The minigene and its mutation construct stably transfected MELCs were induced to differentiate with DMSO as described in “Materials and methods.” Total RNA was collected from cells as indicated, subjected to RT-PCR, and analyzed by 2% agarose gel. + indicates exon 16 inclusion; and -, exon 16 exclusion. (D) Effect of EX-2.1, EX-3.1, and EX-3.2 on exon 16 splicing by RT-PCR. + indicates exon 16 inclusion; and -, exon 16 exclusion. Endo. indicates endogenous exon 16 splicing patterns.
We stably transfected the WT and mutated minigene constructs EX-1, EX-2, and EX-3 into MELCs, induced differentiation with DMSO, and analyzed them for splicing activity. We also included both 15GE and 42GE constructs (Figure 1A) as controls since they have been shown to activate exon 16 splicing.27 Earlier, we reported exon 16 expression in selected tissues and cell lines, including HeLa and L8 (rat skeletal muscle myoblast).39,40 Thus, we also analyzed these 2 cell lines for exon 16 splicing. Consistent with our earlier report,27 we showed that endogenous exon 16 inclusion increases from approximately 15% in uninduced to approximately 50% in induced MELCs (Figure 1C lanes Endo/MELC, Endo/Induced MELC). WT minigene behavior is the same as the endogenous exon 16 splicing patterns that occur during induced differentiation of MELCs (Figure 1C lanes WT/MELC, WT/Induced MELC). The replacement of the 15 nt's (Figure 1C lanes 15GE/MELC, 15GE/Induced MELC) or 42 nt's (Figure 1C lanes 42GE/MELC, 42GE/Induced MELC) within exon 16 drastically increased exon 16 inclusion in both induced and uninduced MELCs. Approximately 70% of 15GE and approximately 100% of 42GE included exon 16 in both uninduced and induced MELCs. These results suggest that the 15 nt's and the 42 nt's as a whole act to silence and repress exon 16 splicing in both undifferentiated and differentiated MELCs. However, the effect of EX-1, EX-2, and EX-3 on exon 16 splicing was quite different when compared with that of 15GE and 42GE in MELCs. Mutation in EX-1 did not affect exon 16 inclusion in uninduced MELCs (Figure 1C lane EX-1/MELC), but it reduced exon 16 inclusion to approximately 10% in induced MELCs (Figure 1C lane EX-1/Induced MELC). Both EX-2 and EX-3 almost completely abolished exon 16 inclusion in both uninduced and induced MELCs (Figure 1C lanes EX-2/MELC, EX-2/Induced MELC; EX-3/MELC, EX-3/Induced MELC). These results strongly suggest that enhancers and silencers are likely to be adjacent in exon 16.
To examine whether the effects of the aforementioned silencers and enhancers are specific for MELCs, we tested exon 16 minigene and mutation constructs on HeLa and L8 cell lines. In endogenous 4.1R, exon 16 inclusion was approximately 80% in HeLa cells and approximately 95% in L8 cells (Figure 1C lanes Endo/HeLa, Endo/L8). WT behavior is the same as the endogenous exon 16 splicing pattern (Figure 1C lanes WT/HeLa, WT/L8). The 15GE and 42GE resulted in approximately 100% exon 16 inclusion in both cell lines (Figure 1C lanes 15GE/HeLa, 15GE/L8; 42GE/HeLa, 42GE/L8), suggesting that these sequences have silencing activity as in MELCs. EX-1, EX-2, and EX-3 repressed exon 16 inclusion in HeLa cells to approximately 60%, 30%, and 0% (Figure 1C lanes EX-1/HeLa, EX-2/HeLa, EX-3/HeLa), respectively. These mutations also repressed exon 16 inclusion in L8 cells to approximately 80%, 40%, and 10% (Figure 1C lanes EX-1/L8, EX-2/L8, EX-3/L8), respectively. The densitometric quantification of the effects of EX-1, EX-2, and EX-3 on exon 16 inclusion from 3 independent experiments is shown in Table 1. Taken together, these results suggest that sequences AGACTAG, TATATC, and CAGACAT enhance splicing, with CAGACAT being the most potent enhancer.
Data from 3 independent transfection experiments expressed as percentages of exon 16 inclusion in both uninduced and induced MELCs, HeLa, and L8 cells
. | Exon 16 inclusion, % . | . | . | . | . | ||
|---|---|---|---|---|---|---|---|
| RNA . | Experiment 1 . | Experiment 2 . | Experiment 3 . | Average, % . | SD . | ||
| MELC | |||||||
| WT | 19.4 | 20 | 16.5 | 18.6 | 1.9 | ||
| EX-1 | 10.1 | 9.8 | 9.1 | 9.7 | 0.5 | ||
| EX-2 | 3.0 | 3.0 | 2.4 | 2.8 | 0.4 | ||
| EX-3 | 0 | 0 | 0 | 0 | 0 | ||
| Induced MELC | |||||||
| WT | 55.5 | 46.6 | 45.3 | 49.1 | 5.6 | ||
| EX-1 | 7.5 | 3.4 | 8.2 | 6.4 | 2.6 | ||
| EX-2 | 2.4 | 2.6 | 3.6 | 2.9 | 0.6 | ||
| EX-3 | 0 | 0 | 0 | 0 | 0 | ||
| HeLa | |||||||
| WT | 81.6 | 73.9 | 84.2 | 79.9 | 5.4 | ||
| EX-1 | 53.9 | 50.8 | 58.1 | 54.3 | 3.7 | ||
| EX-2 | 23.1 | 27.1 | 30.7 | 26.9 | 3.8 | ||
| EX-3 | 0 | 0 | 0 | 0 | 0 | ||
| L8 | |||||||
| WT | 96.5 | 96.8 | 95.7 | 96.3 | 0.6 | ||
| EX-1 | 82.4 | 85.4 | 81.4 | 83.1 | 2.1 | ||
| EX-2 | 42.0 | 33.7 | 42.1 | 39.3 | 4.8 | ||
| EX-3 | 7.8 | 6.0 | 7.7 | 7.2 | 1.0 | ||
. | Exon 16 inclusion, % . | . | . | . | . | ||
|---|---|---|---|---|---|---|---|
| RNA . | Experiment 1 . | Experiment 2 . | Experiment 3 . | Average, % . | SD . | ||
| MELC | |||||||
| WT | 19.4 | 20 | 16.5 | 18.6 | 1.9 | ||
| EX-1 | 10.1 | 9.8 | 9.1 | 9.7 | 0.5 | ||
| EX-2 | 3.0 | 3.0 | 2.4 | 2.8 | 0.4 | ||
| EX-3 | 0 | 0 | 0 | 0 | 0 | ||
| Induced MELC | |||||||
| WT | 55.5 | 46.6 | 45.3 | 49.1 | 5.6 | ||
| EX-1 | 7.5 | 3.4 | 8.2 | 6.4 | 2.6 | ||
| EX-2 | 2.4 | 2.6 | 3.6 | 2.9 | 0.6 | ||
| EX-3 | 0 | 0 | 0 | 0 | 0 | ||
| HeLa | |||||||
| WT | 81.6 | 73.9 | 84.2 | 79.9 | 5.4 | ||
| EX-1 | 53.9 | 50.8 | 58.1 | 54.3 | 3.7 | ||
| EX-2 | 23.1 | 27.1 | 30.7 | 26.9 | 3.8 | ||
| EX-3 | 0 | 0 | 0 | 0 | 0 | ||
| L8 | |||||||
| WT | 96.5 | 96.8 | 95.7 | 96.3 | 0.6 | ||
| EX-1 | 82.4 | 85.4 | 81.4 | 83.1 | 2.1 | ||
| EX-2 | 42.0 | 33.7 | 42.1 | 39.3 | 4.8 | ||
| EX-3 | 7.8 | 6.0 | 7.7 | 7.2 | 1.0 | ||
The poly U mutation at both EX-2 and EX-3 sites drastically reduces exon 16 inclusion and raises the question of whether the poly U introduces a silencer element rather than disrupting an enhancer element. For this reason, we tested additional mutants EX-2.1, EX-3.1, and EX-3.2 (Figure 1A) in which 2 purines (A) disrupt poly U sequences. EX-2.1, EX-3.1, and EX-3.2 all resulted in almost complete exon 16 exclusion in MELCs while reducing exon 16 inclusion to approximately 60%, 40%, and 30% in HeLa cells, respectively (Figure 1D). These results lead to the conclusion that the reduction of exon 16 inclusion does not solely rely upon the poly U in EX-2 and EX-3 but also responds to the sequences disrupted by purines in EX-2.1, EX-3.1, and EX-3.2.
Although the WT sequences corresponding to EX-1, EX-2, and EX-3 could possibly serve as binding sites for SRp40, SRp55, and SF2/ASF as predicted in program ESEfinder,29 we chose to concentrate this study on the interaction of SF2/ASF with EX-3 since SF2/ASF antibody was the only antibody available at the present time.
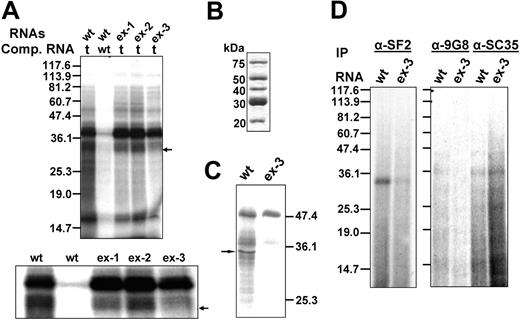
To determine whether the exonic elements affected exon 16 splicing in vitro as they did in vivo, we subjected the WT and EX-3 transcripts to an in vitro splicing assay using HeLa nuclear extracts. We incubated the 32P-labeled pre-mRNA transcripts from the WT and the EX-3 construct in HeLa nuclear extracts, performed the splicing reaction at 30°C for 3 hours, and analyzed the RNA products. The WT substrate produced several splicing intermediates as well as the final products with exon 16 inclusion and exclusion (Figure 2 lane WT). EX-3 resulted in fewer intermediates and products that completely excluded exon 16 (Figure 2 lane EX-3). These results suggest that the enhancer CAGACAT appears to activate exon 16 inclusion.
In vitro splicing of WT and EX-3 substrates. The WT and EX-3 mutation pre-mRNAs were synthesized and subjected to in vitro splicing reactions using HeLa nuclear extracts. The RNAs were resolved on a 4% polyacrylamide gel containing 8 M urea and visualized by autoradiography. The precursors, intermediates, and final products are indicated at left. The RNA molecular weight markers in nt are at right. The arrow indicates product with exon 16.
In vitro splicing of WT and EX-3 substrates. The WT and EX-3 mutation pre-mRNAs were synthesized and subjected to in vitro splicing reactions using HeLa nuclear extracts. The RNAs were resolved on a 4% polyacrylamide gel containing 8 M urea and visualized by autoradiography. The precursors, intermediates, and final products are indicated at left. The RNA molecular weight markers in nt are at right. The arrow indicates product with exon 16.
SF2/ASF binds specifically to splicing enhancer CAGACAT
Given its activity as a splicing enhancer, the exonic element CAGACAT probably binds to a splice-enhancing protein. To identify proteins binding to the enhancer sequences, we performed UV cross-linking on 32P-labeled RNA transcripts from 67 nt's upstream to 37 nt's downstream of exon 16 in the presence of nonspecific competitor tRNA or a wt-specific competitor. The exon 16 in wt pre-mRNA is unaltered; ex-1, ex-2, and ex-3 contain exon 16's respective mutated sequence. We first analyzed the binding proteins presented in HeLa nuclear extracts. Two major groups of bands corresponding to approximately 33- to 40-kDa proteins bind to wt in the presence of tRNA (Figure 3A wt/t). The binding of these bands to the wt transcript is specific because the competition with wt transcripts completely abolished the cross-linked bands (Figure 3A lane wt/wt). The same molecular mass proteins also cross-linked to ex-1 and ex-2 transcripts (Figure 3A lanes ex-1/t, ex-2/t), which suggests that these proteins bind to the sequences shared by wt, ex-1, and ex-2. Interestingly, in the ex-3 reaction, we detected a group of strong bands at about 38 to 40 kDa with a much lighter approximately 33-kDa band (Figure 3A lane ex-3/t). An enlarged figure of the region surrounding approximately 30 to 40 kDa is presented in the bottom panel in Figure 3A. These results suggest that an approximately 33-kDa protein or proteins from the HeLa nuclear extracts specifically binds to the wt exon 16 ESE element CAGACAT.
UV cross-linking analysis of proteins that bind to the exonic splicing enhancer sequences. (A) UV cross-linking of wt, ex-1, ex-2, and ex-3 to HeLa nuclear extracts. 32P-labeled pre-mRNA consisting of exon 16 and its flanking upstream (67 nt's) and downstream (36 nt's) intronic sequences were subjected to UV cross-linking using HeLa nuclear extracts. Either tRNA or WT sense transcripts were added to compete with WT substrates. tRNAs were added to ex-1, ex-2, and ex-3 substrates as competitor. t indicates tRNA; and wt, wt sense transcript. (Top) The full-length gel. (Bottom) The enlargement of the approximately 30- to 40-kDa region of the gel. (B) Purified SR proteins. (C) UV cross-linking of wt or ex-3 using purified SR proteins in the presence of tRNA. The arrow indicates approximately 33 kDa cross-linked band. (D) Identification by immunoprecipitation of SR proteins cross-linked to wt and ex-3. 32P-labeled RNA substrates from wt or ex-3 were subjected to UV cross-linking using HeLa nuclear extracts. The cross-linked protein-RNA mixtures were digested with RNAse A, precleared with protein A–Sepharose beads, and immunoprecipitated overnight at 4°C using anti-SF2, anti-9G8, or anti-SC35 antibodies. The immunoprecipitates were washed, subjected to SDS-PAGE electrophoresis on a 12% gel, and autoradiographed.
UV cross-linking analysis of proteins that bind to the exonic splicing enhancer sequences. (A) UV cross-linking of wt, ex-1, ex-2, and ex-3 to HeLa nuclear extracts. 32P-labeled pre-mRNA consisting of exon 16 and its flanking upstream (67 nt's) and downstream (36 nt's) intronic sequences were subjected to UV cross-linking using HeLa nuclear extracts. Either tRNA or WT sense transcripts were added to compete with WT substrates. tRNAs were added to ex-1, ex-2, and ex-3 substrates as competitor. t indicates tRNA; and wt, wt sense transcript. (Top) The full-length gel. (Bottom) The enlargement of the approximately 30- to 40-kDa region of the gel. (B) Purified SR proteins. (C) UV cross-linking of wt or ex-3 using purified SR proteins in the presence of tRNA. The arrow indicates approximately 33 kDa cross-linked band. (D) Identification by immunoprecipitation of SR proteins cross-linked to wt and ex-3. 32P-labeled RNA substrates from wt or ex-3 were subjected to UV cross-linking using HeLa nuclear extracts. The cross-linked protein-RNA mixtures were digested with RNAse A, precleared with protein A–Sepharose beads, and immunoprecipitated overnight at 4°C using anti-SF2, anti-9G8, or anti-SC35 antibodies. The immunoprecipitates were washed, subjected to SDS-PAGE electrophoresis on a 12% gel, and autoradiographed.
ESEs are common in both alternative and constitutive exons, where they act as binding sites for SR proteins. To determine whether CAGACAT is an SR protein–dependent ESE, we performed UV cross-linking using purified SR proteins (Figure 3B). The wt cross-linked several bands corresponding to approximately 50-, 40-, and 33-kDa SR proteins (Figure 3C lane wt). Although the ex-3 also cross-linked to a prominent approximately 50-kDa protein and less intensely to approximately 40-kDa SR proteins, ex-3 did not cross-link to approximately 33-kDa SR proteins (Figure 3C lane ex-3). These results suggest that an approximately 33-kDa SR protein or proteins bind to the CAGACAT enhancer sequences.
Among the known human SR proteins, SF2/ASF, 9G8, and SC35 are approximately 33 kDa in size. To identify the nature of proteins that bind to exon 16, we followed UV cross-linking with immunoprecipitation experiments using anti-SF2, anti-SC35, and anti-9G8 antibodies. We resolved the immunoprecipitated proteins by SDS-PAGE and subjected them to autoradiography. Antibody to SF2/ASF immunoprecipitated a strong 33-kDa band from the wt (Figure 3D α-SF2, lane wt) but a barely detectable band from the ex-3 cross-linked products (Figure 3D α-SF2, lane ex-3). The anti-9G8 antibody did not precipitate any band (Figure 3D α-9G8), whereas the anti-SC35 antibody precipitated an approximately 35-kDa band (Figure 3D α-SC35) in both wt and ex-3 transcripts. These results suggest that SR protein SF2/ASF binds to the enhancer element.
SF2/ASF enhances exon 16 splicing activity in vivo as well as in vitro
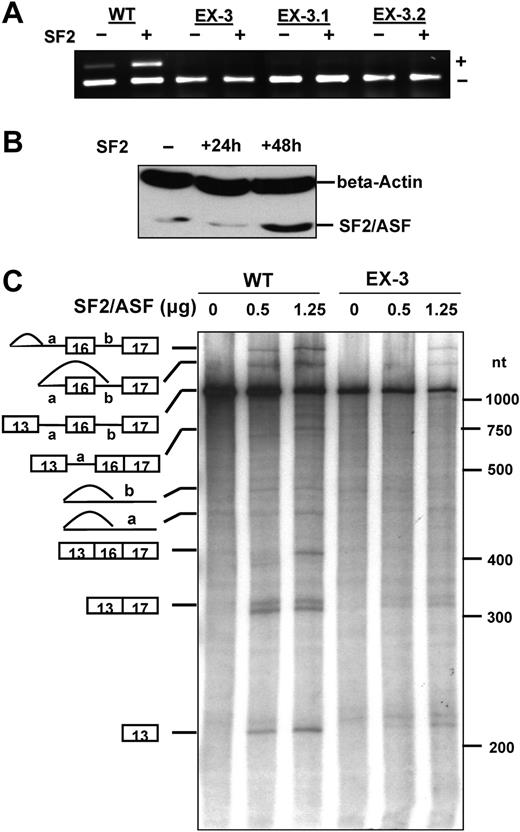
We next examined whether SF2/ASF identified in UV cross-linking experiments has an effect on exon 16 splicing by expressing SF2/ASF in MELCs that have been stably transfected with either WT or EX-3 series (EX-3, EX-3.1, and EX-3.2). Transfection of SF2/ASF into the WT stable line increased exon 16 splicing from approximately 10% to 30% 48 hours after transfection (Figure 4A lane WT). However, SF2/ASF expression did not stimulate exon 16 splicing in any of EX-3 series stable line (Figure 4A lanes EX-3, EX-3.1, and EX-3.2). To verify the expression of SF2/ASF, we analyzed SF2/ASF expression by Western blotting EX-3 stably transfected MELC lysates collected before and 24 hours or 48 hours after SF2/ASF transfection (Figure 4B). An approximately 5-fold increase in SF2/ASF expression was detected in EX-3 lines 48 hours after transfection. These results suggest that the enhancer CAGACAT responds to SF2/ASF and increases exon 16 inclusion.
Effect of SF2/ASF on exon 16 splicing in vivo and in vitro. (A) Effect of transiently expressed SF2/ASF on exon 16 splicing. The WT, EX-3, EX-3.1, and EX-3.2 stably transfected MELCs were transiently transfected with either an empty vector (lanes denoted by -) or a SF2/ASF expression construct (lanes denoted by +). Total RNA was collected 48 hours after transfection, subjected to RT-PCR, and analyzed on a 2% agarose gel. + indicates exon 16 inclusion; and -, exon 16 exclusion. (B) Overexpression of SF2/ASF in EX-3 stably transfected MELCs. The EX-3 stably transfected MELCs were transiently transfected with a SF2/ASF expression construct. Protein was collected at 0, 24, or 48 hours after transfection, fractionated on a 12% PAGE gel, and subjected to immunoblotting analysis with an anti-SF2/ASF antibody. Beta-actin served as a loading control. (C) Purified recombinant SF2/SF2 specifically activates WT exon 16 splicing in an in vitro splicing assay using S100 fraction. In vitro splicing reactions were carried out using HeLa S100 fraction and substrates pre-mRNA derived from either WT or EX-3 minigene constructs in the absence (0 μg) or presence of 0.5 or 1.25 μg of SF2/ASF. Schematic representations of precursors, intermediates, and final products are shown at left. RNA molecular markers in nt are at right.
Effect of SF2/ASF on exon 16 splicing in vivo and in vitro. (A) Effect of transiently expressed SF2/ASF on exon 16 splicing. The WT, EX-3, EX-3.1, and EX-3.2 stably transfected MELCs were transiently transfected with either an empty vector (lanes denoted by -) or a SF2/ASF expression construct (lanes denoted by +). Total RNA was collected 48 hours after transfection, subjected to RT-PCR, and analyzed on a 2% agarose gel. + indicates exon 16 inclusion; and -, exon 16 exclusion. (B) Overexpression of SF2/ASF in EX-3 stably transfected MELCs. The EX-3 stably transfected MELCs were transiently transfected with a SF2/ASF expression construct. Protein was collected at 0, 24, or 48 hours after transfection, fractionated on a 12% PAGE gel, and subjected to immunoblotting analysis with an anti-SF2/ASF antibody. Beta-actin served as a loading control. (C) Purified recombinant SF2/SF2 specifically activates WT exon 16 splicing in an in vitro splicing assay using S100 fraction. In vitro splicing reactions were carried out using HeLa S100 fraction and substrates pre-mRNA derived from either WT or EX-3 minigene constructs in the absence (0 μg) or presence of 0.5 or 1.25 μg of SF2/ASF. Schematic representations of precursors, intermediates, and final products are shown at left. RNA molecular markers in nt are at right.
We further evaluated the role of SF2/ASF in exon 16 splicing by examining whether SF2/ASF increased splicing when added in vitro. In these experiments, we used recombinant SF2/ASF purified from baculovirus to complement an in vitro splicing using HeLa S100 fraction. HeLa S100 extracts are depleted for SR proteins and can be supplemented with individual SR protein family members to activate splicing.23,24,41,42 We used pre-mRNAs derived from the WT and EX-3 constructs to follow the splicing pathways leading to either the inclusion or skipping of exon 16. The addition of SF2/ASF to WT splicing reaction mixtures activated splicing pathways leading to the inclusion of exon 16 (Figure 4C WT, lanes 0, 0.5, and 1.25). S100 alone did not support WT splicing while the addition of 0.5 μg of SF2/ASF stimulated the splicing pathway that skipped exon 16. However, the addition of 1.25 μg of SF2/ASF drastically switched the splicing pathway from exon 16 skipping to exon 16 inclusion. Mutation of the enhancer sequence in EX-3 RNA significantly reduced stimulation caused by SF2/ASF (Figure 4C EX-3, lanes 0, 0.5, and 1.25). Although a small amount of exon 16–excluded product was detected when 0.5 μg and 1.25 μg SF2/ASF was added into the reaction, no obvious product with exon 16 was detected. This suggests that SF2/ASF binding to the exon sequence can specifically enhance exon 16 splicing and supports our hypothesis that the ESE element affects splicing through the activation by SF2/ASF.
Increased SF2/ASF expression correlates with exon 16 inclusion in differentiated erythroid cells
Using the MELC culture system that supports terminal erythroblast proliferation and differentiation, we have shown that exon 16 is preferentially included in mature 4.1R mRNA in induced MELCs.8 In this study, we further enriched the undifferentiated and differentiated populations via flow cytometry in order to distinguish MELCs at different developmental stages. We collected MELCs grown on fibronectin-coated dishes at 0, 2, 4, and 6 days after DMSO-induced differentiation, double-labeled them for erythroid-specific TER119 and nonerythroid transferrin receptor (CD71), and analyzed them via flow cytometry.43 The day-0 cells contained mainly the CD71highTer119low cells (Figure 5A Day 0). Day 2, 4, and 6 cells shifted from a less mature population of CD71highTer119low to a more mature population of CD71lowTer119high (Figure 5A Day 2, Day 4, and Day 6). After we stained CD71highTer119low and CD71lowTer119high populations sorted at day 4 (Figure 5B) with May-Grünwald Giemsa (Figure 5C), their morphologies resembled erythroblasts at different development stages, with CD71highTer119low being the least differentiated with large and light-stained nuclei (Figure 5C CD71+) and CD71lowTer119high as having the most differentiated with small and dense nuclei (Figure 5C TER119+).
Analysis of SF2/ASF expression during MELC differentiation using cells sorted by flow cytometry. (A) Flow cytometry of MELCs at 0, 2, 4, and 6 days after DMSO-induced differentiation. MELCs were double-stained for a PE-conjugated anti-TER119 monoclonal antibody (mAb) and an FITC-conjugated anti-CD71 mAb and analyzed by flow cytometry. Axes indicate relative logarithmic fluorescence units for PE (x-axis) and FITC (y-axis). (B) Regions defined by characteristic staining pattern of day-4–induced MELCs, including CD71highTER119low (CD71+), CD71highTER119high (CD71+/TER119+), and TER119highCD71low (TER119+). (C) CD71+ and TER119+ cells were sorted by fluorescence-activated cell sorter (FACS) from day-4–induced MELCs and stained with May-Grünwald Giemsa. (D) Temporal relationship between 4.1R exon 16 splicing and SF2/ASF expression in differentiating MELCs. CD71+ and TER119+ cells were sorted from day-4–induced MELCs. Twenty micrograms of total proteins from CD71+ or TER119+ cells were analyzed with anti-SF2/ASF antibody. Immunoblotting with anti–β-actin antibody served as a loading control. RT-PCR was used to analyze exon 16 expression from total RNA isolated from the same samples.
Analysis of SF2/ASF expression during MELC differentiation using cells sorted by flow cytometry. (A) Flow cytometry of MELCs at 0, 2, 4, and 6 days after DMSO-induced differentiation. MELCs were double-stained for a PE-conjugated anti-TER119 monoclonal antibody (mAb) and an FITC-conjugated anti-CD71 mAb and analyzed by flow cytometry. Axes indicate relative logarithmic fluorescence units for PE (x-axis) and FITC (y-axis). (B) Regions defined by characteristic staining pattern of day-4–induced MELCs, including CD71highTER119low (CD71+), CD71highTER119high (CD71+/TER119+), and TER119highCD71low (TER119+). (C) CD71+ and TER119+ cells were sorted by fluorescence-activated cell sorter (FACS) from day-4–induced MELCs and stained with May-Grünwald Giemsa. (D) Temporal relationship between 4.1R exon 16 splicing and SF2/ASF expression in differentiating MELCs. CD71+ and TER119+ cells were sorted from day-4–induced MELCs. Twenty micrograms of total proteins from CD71+ or TER119+ cells were analyzed with anti-SF2/ASF antibody. Immunoblotting with anti–β-actin antibody served as a loading control. RT-PCR was used to analyze exon 16 expression from total RNA isolated from the same samples.
This system allowed us to analyze the timing of SF2/ASF expression and exon 16 splicing. We used Western blotting with an anti-SF2 antibody to analyze SF2/ASF expression. We observed an approximately 3- to 5-fold increase in SF2/ASF expression in the CD71lowTer119high population compared with the CD71highTer119low population (Figure 5D SF2/ASF). We also examined exon 16 splicing via RT-PCR analysis (Figure 5D Exon 16). Exon 16 expression also increased in the CD71lowTer119high population to approximately 40%; the less mature CD71highTer119low population showed only approximately 10% exon 16 inclusion. Thus, SF2 expression correlates with exon 16 inclusion in differentiated MELCs.
Discussion
Exon 16 expression in 4.1R is required during late erythroid differentiation for 4.1R to interact with spectrin and actin.7-10 Earlier, we demonstrated how the complex interplay of positive and negative regulatory signals in the pre-mRNA controls the developmental stage–specific splicing of exon 16.27 In this study, we identified 3 novel ESEs within the earlier reported ESS sequences.27,28 We showed that an ESE (CAGACAT) functions as a binding site for the SR protein SF2/ASF. We further established the physiologic importance of this interaction by demonstrating that changes in the level of endogenous SF2/ASF protein expression can mediate an important 4.1R exon 16 pre-mRNA splicing switch during erythroid differentiation.
Both our work27 and that of Hou et al28 prove that the 15 nt's and 42 nt's within exon 16 regulated exon 16 splicing. Exon 16 contains 2 copies of UAG triplets, a known core element for hnRNPA1 binding ESS.19,22 The first UAG overlaps with a putative SR protein–binding site for SRp40 (EX-1); the second UAG partially overlaps with a putative SF2/ASF binding site (EX-3). We designed our mutation constructs to allow for disruption of the first UAG but preservation of the second one. One would expect that the disruption of the UAG sequence would destroy its silencing activity and enhance exon 16 splicing. However, when the first UAG and its upstream 4 nt's were replaced with T (EX-1), exon 16 splicing was drastically reduced in all tested cell lines. Thus, the silencing effect of EX-1 must derive from the disruption of the putative enhancer sequences. The mutation in EX-2 and EX-2.1 also resulted in reduced exon 16 in tested cell types, suggesting a constitutive splicing enhancer interacting with the putative SRp55 binding sequences. Due to the lack of the SRp40 and SRp55 antibodies, we are unable to confirm whether SRp40 and SRp55 act as positive regulators through their interaction with the ESE and exert splicing effects on exon 16 inclusion. In this study, we concentrated on EX-3 because of the availability of the SF2/ASF antibody.
The mutation in EX-3 (CAGACTAG) did not alter the second UAG motif but changed its upstream 5 nt's. It resulted in complete exclusion of exon 16 in all tested cell lines except L8 (with ∼80% exon 16 exclusion), which suggests that the ESE responds to a constitutive splicing factor or factors. The reduction of exon 16 inclusion does not solely rely on the poly U in EX-3 but also responds to poly U disrupted by purines in EX-3.1 and EX-3.2, although to a lesser extent. These results rule out the possibility that the poly U in EX-3 acts as a silencer element. Using UV cross-linking and immunoprecipitation assays, we showed that SF2/ASF binds to the ESE. The interaction of SF2/ASF with this ESE correlates well with the overexpression of SF2/ASF in MELCs as well as with the addition of recombinant purified SF2/ASF protein in an in vitro splicing assay that enhances exon 16 splicing. The up-regulation of endogenous SF2/ASF expression, which led to an induction of exon 16 splicing in differentiated MELCs, further supports the physiologic importance of SF2/ASF. In this study, we used DMSO to commit MELCs to late erythroid differentiation. DMSO is a polar solvent that promotes or inhibits differentiation and apoptosis in a variety of cell lines.44,45 Also, DMSO has recently been shown to activate splice site selection and enhance SR protein activity in HeLa extracts in vitro.46 Using several cell lines and different inducers of erythroid differentiation, Theoleyre et al47 systemically analyzed the effect of chemical inducers on exon 16 splicing. They47 showed that exon 16 inclusion in mature 4.1R mRNA is not a direct effect of DMSO but rather is linked to the inducer-activated erythroid-terminal differentiation program. It is possible that increased SF2/ASF protein levels mediate a pre-mRNA splicing switch in differentiated MELCs via interaction with the ESE.
Our identification of a positive regulator suggests a mechanism for exon 16 inclusion in induced MELCs, although Hou et al28 have shown that a negative regulator is involved in exon 16 regulation. They used an in vitro splicing assay to show that the exclusion of exon 16 correlates with hnRNPA1 in a concentration-dependent manner. The hnRNPA/B binds to the 42 nt's silencer element in exon 16 to repress exon 16 inclusion. They have also shown that decreased hnRNPA/B expression during erythropoiesis mediates an exon 16 splicing switch. Thus, 2 regulation pathways, one that increases SR proteins and one that decreases hnRNP proteins, seem to simultaneously switch on exon 16 splicing in differentiated erythroid cells. It is well established that changes in the relative amounts of hnRNPA/B and SR proteins can alter either the choice of alternative splicing site or the inclusion-exclusion ratio of selected alternative exons.48-50
Tissue-specific or differentiation stage–specific splicing is regulated in a combinatorial manner, with multiple positive and negative inputs. Our current results and those of Hou et al28 show that both SF2/ASF and hnRNPA/B are involved in the regulation of exon 16 splicing. As in other systems,23,51 SF2/ASF may stimulate binding of spliceosomal components such as U2AF and U1snRNP to the splice sites. SF2/ASF may also antagonize the negative effect of inhibitory proteins such as hnRNPA/B. Some ESE elements antagonize the negative effect of a juxtaposed ESS.16,26,51 Studies of splicing of the HIV-1 tat exon 3 suggest that one function of SR proteins appears to bind to an ESE and then counteracts a nearby splicing repressor complex consisting of hnRNP proteins bound to an ESS.51 Binding of hnRNPA/B to the 42 nt silencer elements represses exon 16 inclusion; however, it is also possible that binding of SF2/ASF to a juxtaposed ESE element(s) may block hnRNPA/B binding to ESS. Splicing of exon 16 can thus reflect competition between positive and negative factors, with SF2/ASF possibly functioning through the ESE and hnRNPA1 possibly functioning through the ESS. Another possibility is that proper regulation requires concerted interactions among SR proteins, hnRNPA1, and other splicing factors to orchestrate the developmental switch in exon 16 splicing during erythropoiesis.
The precise mechanism by which SF2/ASF enhances exon 16 splicing efficiency is unknown. How SF2/ASF is regulated during erythroid differentiation also remains to be determined. Nevertheless, our data show that SF2/ASF protein is necessary in increasing the exon 16 splicing efficiency and that changes in SF2/ASF levels in differentiating erythroid cells can mediate an important switch in exon 16 pre-mRNA splicing. Elucidation of the mechanisms controlling alternative splicing of 4.1R pre-mRNA will enhance our understanding of how regulated splicing in red blood cell membranes affects biogenesis and possibly affects the expression of other key erythroid protein forms during late erythropoiesis.
Prepublished online as Blood First Edition Paper, November 2, 2004; DOI 10.1182/blood-2004-05-1757.
Supported by National Institutes of Health grant HL24385 (E.J.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Paula J. Grabowski and Mona Ashiya (University of Pittsburgh) for the HeLa cells and the instruction on nuclear extract preparation, in vitro splicing, and UV cross-linking experiments. We thank Ms Hui Cheng for the characterization of the intermediate spliced products from in vitro splicing reaction.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal