Quantitative trait analysis may shed light on mechanisms regulating hematopoiesis in vivo. Strain-dependent variation existed among C57BL/6 (B6), DBA/2, and BXD recombinant inbred mice in the responsiveness of primitive progenitor cells to the early-acting cytokines kit ligand, flt3 ligand, and thrombopoietin. A significant quantitative trait locus was found on chromosome 2 that could not be confirmed in congenic mice, however, probably because of epistasis. Because it has been shown that alleles of unknown X-linked genes confer a selective advantage to hematopoietic stem cells in vivo in humans and in cats, we also analyzed reciprocal male D2B6F1 and B6D2F1 mice, revealing an X-linked locus regulating the responsiveness of progenitor and stem cells to early-acting factors. Among DBA/2, B6, and BXD recombinant inbred mice, correlating genetic variation was found in the absolute number and frequency of Lin−Sca1++kit+ cells, which are highly enriched in hematopoietic progenitor and stem cells, and in the number of Lin−Sca1++kit− cells, a population whose biologic significance is unknown, suggesting that both populations are functionally related. Suggestive quantitative trait loci (QTLs) for the number of Lin−Sca1++ cells on chromosomes 2, 4, and 7 were confirmed in successive rounds of mapping. The locus on chromosome 2 was confirmed in congenic mice. We thus demonstrated genetic variation in the response to cytokines critical for hematopoiesis in vivo and in the pool size of cells belonging to a phenotype used to isolate essentially pure primitive progenitor and stem cells, and we identified loci that may be relevant to the regulation of hematopoiesis in steady state.
Introduction
Hematopoiesis consists of an ordered series of events in which primitive hematopoietic stem cells renew or differentiate into mature blood cells of at least 8 lineages. The exact mechanisms responsible for the regulation of self-renewal versus differentiation of hematopoietic stem cells in vivo are unknown but may involve stochastic mechanisms and a balance between stimulatory and inhibitory signals regulating renewal, differentiation, and apoptosis.1
One strategy to gain insight into the regulation of hematopoiesis is to study naturally occurring genetic variation in the hematopoietic system. This may lead to the identification of regulatory mechanisms that are relevant in vivo. Pool size and cycling activity of the stem cell compartment are under complex intrinsic genetic control in inbred mouse strains.2-8 Putative stem cell pool size, as determined by the day 35 cobblestone area-forming cell assay (CAFCd35), varies widely among inbred mouse strains.2 This was not the case for earlier appearing, and therefore more mature, CAFCd7, indicating that the gene(s) involved act on the more primitive progenitor compartment.8 Using BXD recombinant inbred (RI) mouse strains, a locus involved in the regulation of CAFCd35 frequency was mapped to mouse chromosome 18, in a region syntenic with and corresponding to a critical segment of human chromosome 5q, which is frequently deleted in myelodysplasia and acute myeloblastic leukemia.2 Muller-Sieburg and Riblet, using a different assay in BXD mice, mapped 2 candidate loci regulating the number of long-term culture-initiating cells (LTC-IC) to mouse chromosome 1.3 Further evidence for the existence of intrinsic genetic control of hematopoietic stem cell kinetics comes from embryo-aggregated chimeric DBA/2↔B6 mice. In these mice, DBA/2-derived stem cells contribute significantly and stably to hematopoiesis in young adults, but with aging, B6-derived hematopoiesis becomes predominant, if not exclusive.5 Hematopoietic recovery after the administration of the myelotoxic drug 5-fluorouracil to young B6↔DBA/2 chimeras was significantly faster in the DBA/2-derived stem cell compartment than in the B6-derived compartment.6 Similarly, when chimeric bone marrow was transplanted, early engraftment of irradiated hosts was predominantly DBA/2 derived, significantly out of proportion to the DBA/2 representation in the marrow graft, further suggesting a proliferative advantage for DBA/2-derived stem cells. Because the microenvironment in these chimeric mice was identical for both DBA/2- and B6-derived stem cells, stem cell–intrinsic mechanisms must be at the basis of these differences.6 Genetic variation has also been demonstrated in the repopulation capacity of hematopoietic stem cells. One of the CXB RI strains had a significantly higher capacity for long-term competitive repopulation in CByB6F1 recipient mice than any of the other CXB strains.7 Loci affecting the efficiency of mobilization of progenitor cells to the peripheral blood have been identified on chromosomes 2 and 11.9
In humans and in cats, evidence for genetically determined regulation of hematopoiesis comes from studies addressing X inactivation in the hematopoietic system in vivo.10-13 In as many as 50% of aging women, progressive skewing of X inactivation occurs in the hematopoietic system.10,11 Similar data were obtained in cats.12 Furthermore, skewed X chromosome inactivation in all hematopoietic lineages also occurs after bone marrow transplantation12 and after repeated chemotherapy with busulfan in cats.13,14 Human female monozygotic twins that show skewed X inactivation in the hematopoietic system with aging tend to inactivate the X chromosome from the same parent.11,15Thus, alleles on the X chromosome confer a growth, survival, or reconstitution advantage to stem cells. Progressive skewing of X inactivation in the hematopoietic system is therefore likely to be caused to a large extent by hemizygous selection,12 ie, a selective advantage of one X chromosome over another, whereas stochastic mechanisms are less dominant.15
The aim of this study was to test the hypothesis that strain-dependent variation would exist in the response of primitive hematopoietic progenitor and stem cells, as defined by the Lin−Sca1++kit+phenotype,16-18 to the early-acting factors kit ligand (KL), flt3 ligand (flt3L), and thrombopoietin (TPO). These factors have been shown to be critical to the function of the hematopoietic stem cell compartment in vivo.19-21 We show here that there is indeed strain-dependent variation in the response to these early-acting factors and that at least one locus for this trait maps to the X chromosome. In addition, we found strain-dependent variation in the absolute number and frequency of Lin−Sca1++cells (c-kit+ and c-kit− subpopulations). A quantitative trait locus (QTL) for this trait on chromosome 2 was confirmed in congenic mice.
Materials and methods
Mice
Female mice of strains C57BL/6J (B6), DBA/2J, and BXD RI, aged 6 to 8 weeks, were all purchased from Jackson Laboratories (Bar Harbor, ME). The mice were maintained in a germ-free environment and fed ad libitum. Experiments and animal care were performed in accordance with the Mount Sinai Institutional Animal Care and Use Committee.
Antibodies and cytokines
Unconjugated Ter119 (erythroid), CD2 (T and NK cells), CD3, CD4, CD8 (T cells), B220 (B cells), Ly6G/Gr1 (granulocytes), Mac1 (macrophages), phycoerythrin-conjugated Sca1, biotin-conjugated anti–c-kit, Cychrome-conjugated streptavidin, and fluorescein isothiocyanate–conjugated goat antirat antibodies were purchased from PharMingen (San Diego, CA). Recombinant mouse flt3L and thrombopoietin (TPO) and anti–transforming growth factor-β (anti–TGF-β) antibodies were purchased from R&D Systems (Minneapolis, MN). Supernatants from BHK/HM-5, BHK/MKL (both a kind gift of Dr J. Matous, University of Washington), and WEHI 3B (a kind gift of Dr S Tsai, Mount Sinai School of Medicine, New York, NY) cells were used as a source of granulocyte macrophage–colony-stimulating factor (GM-CSF), KL, and interleukin-3 (IL-3), respectively.
Isolation of Lin−Sca1+/+ cells
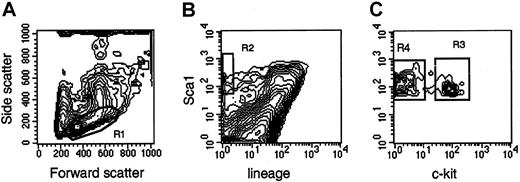
Femurs and tibias were flushed with Iscoves modified Dulbecco medium (IMDM; Gibco BRL, Grand Island, NY) supplemented with 10% fetal calf serum (FCS). Low-density bone marrow cells, obtained after density centrifugation, were stained with Ter119, CD2, CD3, CD8, CD4, B220, Mac1, and Gr1 for 20 minutes at 4°C, washed, and stained with goat antirat antibodies for 20 minutes at 4°C. After washing, the cells were stained for 20 minutes at 4°C with phycoerythrin-conjugated Sca1 and biotin-conjugated CD117 (c-kit), washed with phosphate-buffered saline, and stained with streptavidin-Cychrome. Cells were sorted on a MoFlo flow cytometer (Cytomation, Fort Collins, CO) at 30 psi sheath pressure and at a rate of 12 000 to 15 000 events per second. Cells with a low side scatter, a low-to-medium forward scatter (Figure1, R1), a green (lineage) fluorescence lower than the median fluorescence of cells stained with isotype-matched control antibodies, and an orange (Sca1) fluorescence twice the intensity (in terms of channel numbers) of the brightest cells in control samples (Figure 1, R2) were sorted as Lin−Sca1++ cells. Cells falling in R1 and R2 with positive red (c-kit) fluorescence (Figure 1, R3) were sorted as Lin−Sca1++kit+ cells, whereas cells falling in R1 and R2 with negative red fluorescence (Figure 1, R4) were sorted as Lin−Sca1++kit−cells. The absolute number of cells in each population was estimated by recording the number of sorted cells, as indicated by the number of sortable events on the counter of the cell sorter. The abort frequency was less than 5% in the sorting conditions we used. Bone marrow pooled from at least 2 mice was used in each experiment. Because the secondary goat antirat antibodies were not blocked with rat immune globulins, most Lin+ cells appear Sca1+ in Figure 1. This blocking step was omitted because only lineage-negative cells were isolated, and the blocking step did not affect the fluorescence or the number of the cells in the sort windows in preliminary experiments.
Sort windows used to isolate Lin−Sca1++, Lin−Sca1++kit+, and Lin−Sca1++kit− cells.
Cells with low to medium forward scatter (FSC) and low side scatter (SSC) (R1), negative green (lineage) fluorescence, orange (Sca1) fluorescence higher than twice the fluorescence level of cells stained with control antibodies (not shown) (R2), and positive red (c-kit) fluorescence (LSK+, R3) or negative red fluorescence (LSK−, R4) were isolated. For the isolation of Lin−Sca1++ cells, cells falling into R1 and R2 were isolated.
Sort windows used to isolate Lin−Sca1++, Lin−Sca1++kit+, and Lin−Sca1++kit− cells.
Cells with low to medium forward scatter (FSC) and low side scatter (SSC) (R1), negative green (lineage) fluorescence, orange (Sca1) fluorescence higher than twice the fluorescence level of cells stained with control antibodies (not shown) (R2), and positive red (c-kit) fluorescence (LSK+, R3) or negative red fluorescence (LSK−, R4) were isolated. For the isolation of Lin−Sca1++ cells, cells falling into R1 and R2 were isolated.
Pre–colony-forming cell assay
Lin-Sca1++ or Lin−Sca1++kit+ cells were cultured in triplicate at approximately 50 cells per well in flat-bottom, 96-well plates in serum-free media (StemPro34; Gibco BRL), 50 ng/mL flt3L, 50 ng/mL TPO, and 10% BHK/KL supernatant (containing kit ligand). Because the BHK/KL supernatant contained 10% FCS, the actual serum concentration in these cultures was still 1%. Three hours after plating, the exact number of cells per well was determined by visually counting the cells at × 40 magnification. After 5 days of liquid culture at 37°C and 5% CO2, the cells were counted, and 500 cells were plated in methylcellulose cultures containing IMDM, IL-3 (10% of WEHI 3B supernatant), GM-CSF (10% of BHK/HM-5 supernatant), KL (10% of BHK/MKL supernatant), erythropoietin (2 U/mL), FCS (20%), anti–TGF-β (10 μg/mL), and 2-mercaptoethanol (10−6M). After 8 days of incubation at 37°C in a humidified incubator with 5% CO2, the cultures were scored for colony formation. The same lot of fetal bovine serum was used in all the experiments.
Statistical analysis
Student t test for unpaired samples was used, unless indicated otherwise.
Congenic mice
Congenic mice were generated by marker-assisted back-crossing22 23 using B6 and BXD-31Ty as founders. These B6.DBA/2-(D2Mit133-D2Mit200) congenic animals have a segment of chromosome 2 between 66.7 and 102 cm, derived from DBA/2 on a B6 background, and were obtained after 5 generations of male back-crossing and genotype-based selection with genetic markers spaced approximately 20 cM apart. Analysis of polymorphic simple sequence repeat elements was made by polymerase chain reaction using primers obtained from Research Genetics (Huntsville, AL).
Quantitative trait analysis using BXD RI strains
Recombinant inbred strains were generated by repeated inbreeding of F2 mice derived from 2 parental inbred strains, C57BL/6 (B6) and DBA/2 in the case of BXD RI strains. The genome of RI strains is composed of a patchwork of homozygous chromosome segments derived from either progenitor strain, with each of the RI lines having a unique combination of patches from the progenitor. Genetic maps are available for each RI strain that allow the mapping of traits specified by loci at which the parental strains are polymorphic.24,25Linkage analysis is performed by determining the relevant trait in each of the strains of a set of RI strains, which results in a strain distribution pattern (SDP). The data are then analyzed using Map Manager QTb29ppc software developed by Manly.26 This software statistically analyzes the linkage of a given trait with previously typed polymorphic loci in the RI strains, which are inherited from either parental strain. This allows the assignment of a trait to a corresponding map position and the calculation of statistical significance.27
Statistical analysis.
Because of the limited number of RI strains available, the number of SDPs to be compared, and, consequently, the complexity of testing required to establish concordance, a close or identical match in SDPs may occur by chance.27,28 As with all statistical comparisons, it is necessary to make a calculation of the probability that the observed result was a false-positive. Therefore, the genome-wide probability of obtaining the observed linkages by random chance, corresponding to an error threshold of P = .05, is calculated using the robust nonparametric permutation method developed by Churchill and Doerge,27 which is implemented in the Map Manager QTb29ppc software developed by Manly et al.26 The peak logarithm of the likelihood of odds ratio (the LOD score) of the correctly ordered data obtained in our study is compared with the peak LOD scores computed for 5000 random permutations of the same data. As an example of this kind of analysis, if the actual data gave an LOD score of 6 and only 1 in 1000 random permutations exceeded this value, the genome-wide probability of a false-positive would be approximately 0.001. The P = .5 error threshold, a level considered suggestive of a QTL, corresponds to a LOD score that is exceeded by the highest LOD scores of half the permutations.27
Interval mapping.
A subroutine of the Map Manager QTb29ppc software, using computationally efficient regression equations, is used for mapping the QTLs. The probability of linkage between our trait under study and previously mapped genotypes was estimated at 1-cM intervals along the entire genome, except for the Y chromosome. The statistical power of linkage of the phenotype to individual genotypes (point-wise linkage statistics) should attain values of at least 0.0001 to reach a level of genome-wide statistical significance.27 28
Mapping databases.
Mapping databases were downloaded fromwww.nervenet.org/papers/bxn.html.29
Results
Responsiveness to early-acting factors and number of phenotypically defined progenitor cells in B6 and DBA/2 mice
A pre–colony-forming cell (pre-CFC) assay was used to measure proliferation and differentiation capacity of Lin−Sca1++kit+cells,16-18 isolated by flow cytometric cell sorting according to the sort windows shown in Figure 1, from B6 and DBA/2 mice. In this assay, Lin−Sca1++kit+ cells were cultured in liquid cultures supported by the early-acting factors KL, flt3L, and TPO.19-21 After 5 days of culture, the cells were counted and plated in methylcellulose assays supported by KL, IL-3, GM-CSF, erythropoietin, and neutralizing anti–TGF-β antibodies for the determination of CFC content. B6 and DBA/2 mice were chosen for these studies because of the known strain-dependent variation in the hematopoietic system between these 2 mouse strains2-6,8,9and because of the availability of a relatively large set of BXD RI strains together with a dense map of polymorphic markers.24-26 29
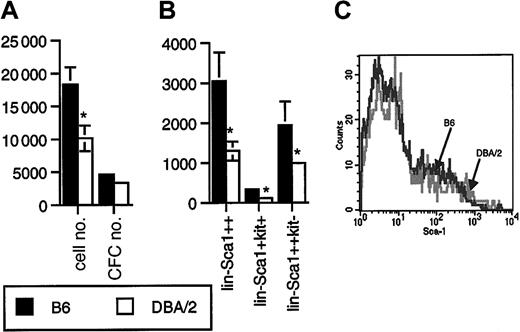
Lin−Sca1++kit+ cells from B6 mice proliferated better in response to flt3L, KL, and TPO than Lin−Sca1++kit+ cells from DBA/2 mice (P = .01, n = 12, paired t test; Figure2A). The difference in CFC generation was not statistically significant (n = 12, Figure 2A). Most colonies generated were myeloid, with 20% to 40% macroscopic colonies derived from high proliferative potential cells. During these experiments, we also recorded the number of sorted Lin−Sca1++kit+ events, as determined on the counter on the cell sorter, as well as the frequency of Lin−Sca1++kit+ cells. The frequency (not shown) and the absolute number (Figure 2B) of Lin−Sca1++kit+ cells were 2-fold higher in B6 than in DBA/2 mice (P = .04, n = 8). Long-term repopulating stem cells are enriched in the Lin−Sca1++kit+ fraction of bone marrow cells.16-18 In addition, a c-kit− subpopulation of Lin−Sca1++ cells has been identified by Randall and Weissman that does not proliferate in vitro and that has no detectable long-term repopulating capacity.30 In addition, in our hands, these cells did not respond to flt3L, KL, or TPO at all (n = 16, not shown). The biologic significance of this population is unknown. We noticed, however, that the number of Lin−Sca1++kit− cells was also 2-fold higher in B6 than in DBA/2 mice (P = .04, n = 8, Figure 2B). When, in a separate set of experiments, the total number of Lin−Sca1++ cells was quantified, the number in B6 mice was again 2-fold higher than in DBA/2 mice (P = .02, n = 6, Figure 2B). These data suggest that the size of the Lin−Sca1++ population as a whole is genetically determined. An alternative explanation might be that B6 and DBA/2 mice simply differ in the level of expression of Sca1, so that it would appear that DBA/2 mice have a lower number of Sca1++ cells. However, the range of expression levels of Sca1 on Lin− cells in B6 and DBA/2 mice (Figure 2C) was identical. It is therefore unlikely that the observed 2-fold variation in the numbers of Lin−Sca1++ cells was caused by variation in the relative level of expression of the Sca1 antigen.
Proliferative capacity and number of Lin−Sca1++kit+ and Lin−Sca1++kit− cells in B6 and DBA/2 mice.
(A) Number of cells and CFCs obtained after 5 days of liquid culture of 50 Lin−Sca1++kit+ cells from B6 and DBA/2 mice supported by KL, flt3L, and TPO. Results are given as mean ± SEM (n = 12 independent triplicate experiments; *significantly different from B6, paired t test). (B) Absolute number of Lin−Sca1++, Lin−Sca1++kit+, and Lin−Sca1++kit− cells in B6 and DBA/2 mice as determined by flow cytometric cell sorting (mean ± SEM, n = 8 independent experiments in which the bone marrow pooled from at least 2 mice was used; *significantly different from B6, paired t test). (C) Range of expression levels of Sca1 on Lin− bone marrow cells from B6 and DBA/2 mice.
Proliferative capacity and number of Lin−Sca1++kit+ and Lin−Sca1++kit− cells in B6 and DBA/2 mice.
(A) Number of cells and CFCs obtained after 5 days of liquid culture of 50 Lin−Sca1++kit+ cells from B6 and DBA/2 mice supported by KL, flt3L, and TPO. Results are given as mean ± SEM (n = 12 independent triplicate experiments; *significantly different from B6, paired t test). (B) Absolute number of Lin−Sca1++, Lin−Sca1++kit+, and Lin−Sca1++kit− cells in B6 and DBA/2 mice as determined by flow cytometric cell sorting (mean ± SEM, n = 8 independent experiments in which the bone marrow pooled from at least 2 mice was used; *significantly different from B6, paired t test). (C) Range of expression levels of Sca1 on Lin− bone marrow cells from B6 and DBA/2 mice.
Quantitative trait analysis in BXD RI strains
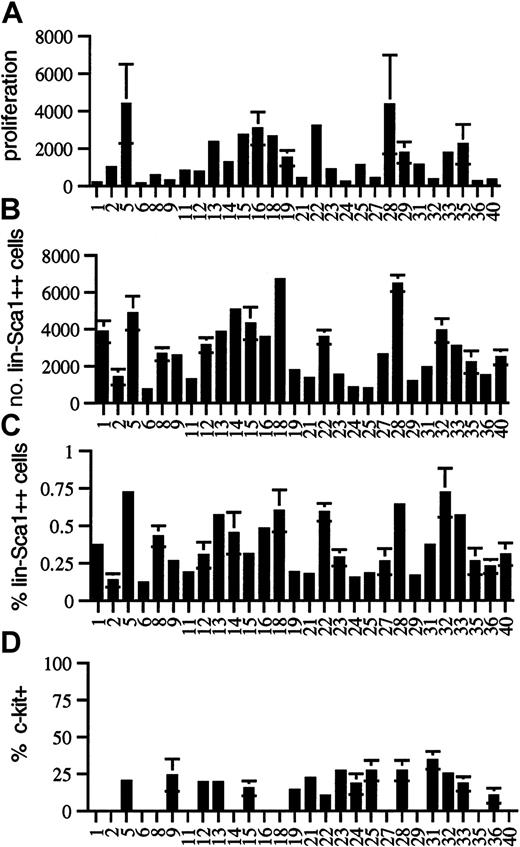
To further investigate potential genetically determined variation in the cytokine responsiveness and number of Lin−Sca1++ cells, we performed quantitative trait analysis using BXD RI strains. Twenty-eight BXD RI strains were analyzed for responsiveness to the early-acting factors flt3L, KL, and TPO, for the absolute number and frequency of Lin−Sca1++ cells, and for the fraction of c-kit+ cells among Lin−Sca1++cells. In all RI strains, at least 3 independent experiments were performed in triplicate using bone marrow pooled from at least 2 individual mice in each experiment. The strain distribution patterns are shown in Figure 3. Because Lin−Sca1++kit− cells do not respond to early-acting cytokines at all (n = 16), only Lin−Sca1++kit+ cells are responsible for cell proliferation and CFC generation from Lin−Sca1++ cells. Table1 summarizes the map positions obtained.
Strain-distribution patterns in BXD mice.
Strain-distribution pattern of (A) the proliferation of Lin−Sca1++ cells in response to KL, flt3L, and TPO (expressed as cell number after 5 days of culture per 50 input Lin−Sca1++ cells), (B) the absolute number of Lin−Sca1++ cells, (C) the relative fraction of Lin−Sca1++ cells, and (D) the fraction of c-kit+ cells among Lin−Sca1++cells in 8- to 10-week-old BXD RI mice (mean ± SEM, n = 3 independent experiments in which the bone marrow pooled from at least 2 mice was used).
Strain-distribution patterns in BXD mice.
Strain-distribution pattern of (A) the proliferation of Lin−Sca1++ cells in response to KL, flt3L, and TPO (expressed as cell number after 5 days of culture per 50 input Lin−Sca1++ cells), (B) the absolute number of Lin−Sca1++ cells, (C) the relative fraction of Lin−Sca1++ cells, and (D) the fraction of c-kit+ cells among Lin−Sca1++cells in 8- to 10-week-old BXD RI mice (mean ± SEM, n = 3 independent experiments in which the bone marrow pooled from at least 2 mice was used).
Overview of mapped QTLs
| Trait . | Chromosome . | Markers . | cM . | Maximum LRS* . | P† . | High allele . | Significance1-153 . |
|---|---|---|---|---|---|---|---|
| No. Lin−Sca1++cells | 2 | D2Mit495-D2Mit411 | 73.2-77.2 | 9.4 | .002 | B6 | Suggestive |
| 4 | D4Mit33-D4Mit343 | 77.5-79.0 | 15.9 | .0001 | DBA/2 | Suggestive | |
| 7 | D7Mit17-D7Mit238 | 51.0-53.0 | 9.2 | .002 | B6 | Suggestive | |
| flt3L/KL/TPO response | 2 | D2Mit495-D2Mit411 | 73.2-77.2 | 18.8 | .00007 | B6 | Significant |
| X | ND | ND | ND | ND | DBA/2‡ | Significant‡ |
| Trait . | Chromosome . | Markers . | cM . | Maximum LRS* . | P† . | High allele . | Significance1-153 . |
|---|---|---|---|---|---|---|---|
| No. Lin−Sca1++cells | 2 | D2Mit495-D2Mit411 | 73.2-77.2 | 9.4 | .002 | B6 | Suggestive |
| 4 | D4Mit33-D4Mit343 | 77.5-79.0 | 15.9 | .0001 | DBA/2 | Suggestive | |
| 7 | D7Mit17-D7Mit238 | 51.0-53.0 | 9.2 | .002 | B6 | Suggestive | |
| flt3L/KL/TPO response | 2 | D2Mit495-D2Mit411 | 73.2-77.2 | 18.8 | .00007 | B6 | Significant |
| X | ND | ND | ND | ND | DBA/2‡ | Significant‡ |
ND indicates not determined.
As determined by interval mapping.
P value for point-wise linkage statistics.
Determined in reciprocal male F1 hybrids.
There was wide phenotypic variation among BXD RI strains for proliferative capacity in response to flt3L, KL, and TPO. Cell number per 50 input Lin−Sca1++ cells after 5 days of culture ranged from 124 ± 72 (BXD6) to 4369 ± 2119 (BXD5) (Figure3A). CFC output completely paralleled cellular proliferation, and no strain-dependent variation was noted in the secondary cloning efficiency (not shown). It was surprising that up to 35-fold variation in the proliferative capacity was seen among BXD RI strains (Figure3A), whereas there was only a small difference in this trait among the 2 progenitor strains (Figure 2A), indicating that this is a multigenic, quantitative trait. For multigenic traits, phenotypic spread among BXD strains actually represents the phenotypic spread in the F2 generation, not of the progenitor strains. Therefore, extensive phenotypic variation can be seen within the RI strains, even when the phenotype of the 2 progenitor strains is not significantly different for that trait. The reason is that RI strains may have most of the positive or most of the negative alleles contributing to a given trait, whereas the 2 progenitor strains may have both positive and negative alleles for that trait.24 25 A major QTL for proliferative capacity was found on chromosome 2, with a likelihood ratio statistic (LRS) in the significant range as determined by permutation analysis. Interval mapping showed the highest LRS value (18.8) between D2Mit495 (73.2 cM) and D2Mit411 (77.6 cM) (Table 1). The donor of the high allele was B6.
Among the 28 BXD RI strains studied, the number of Lin−Sca1++ cells varied from 721 ± 86 (BXD6) to 6472 ± 447 (BXD28) cells per femur (Figure 3B). The percentage of Lin−Sca1++ cells in the bone marrow (Figure 3C) correlated well with the absolute number of Lin−Sca1++ cells (r = 0.870,P < .0001, not shown). This correlation indicates that the fraction of Lin−Sca1++ cells is a good measure of the absolute number of Lin−Sca1++ cells and that our data cannot be explained by genetically determined variation in bone marrow cellularity. In addition, the range of expression of Sca1 was the same in all BXD strains, indicating again that differences in the expression levels of Sca1 cannot explain the variation in the number of Lin−Sca1++ cells. Although there was some variation in the ratio of c-kit+/c-kit− cells among Lin−Sca1++ cells in BXD RI strains, no statistically significant differences were found (Figure 3D; only strains in which 3 experiments were performed are shown). Variation in the fraction of c-kit–expressing Lin−Sca1++cells thus cannot explain the 35-fold variation in proliferative capacity of Lin−Sca1++ cells. The 9-fold variation in the number of Lin−Sca1++ cells among BXD RI strains compared with the only 2-fold difference between the 2 progenitor strains, B6 and DBA/2, suggest that the number of Lin−Sca1++ cells is a complex multigenic trait.
Quantitative trait analysis was performed for the number of Lin−Sca1++ cells using the average values for the 3 independent experiments performed in each BXD strain (Table 1). This analysis revealed a highly suggestive QTL for the number of Lin−Sca1++ cells on chromosome 4 (Table 1). The highest value of the LRS (15.9) by interval mapping was found between D4Mit33 (77.5cM) and D4Mit343 (79 cM). This was just short of significant, for which an LRS of 16.6 was required according to permutation analysis.27 The frequency of Lin−Sca1++ cells mapped to the same region of chromosome 4 with a suggestive level of significance (LRS 12.7). The donor of the high allele was DBA/2. A second suggestive QTL (LRS 9.4 for absolute number and 11.4 for frequency of Lin−Sca1++ cells) was found on chromosome 2, between D2Mit495 (73.2 cM) and D2Mit411 (77.6 cM). The donor of the high allele at this locus was B6. A third, weaker QTL was found on chromosome 7 between D7Mit17 (51 cM) and D7Mit238 (53 cM); B6 was the donor of the high allele (LRS 9.2). The level of significance of the association with all 3 loci was in the suggestive range according to published criteria. This means that the association is likely but needs confirmation.25,27,28 However, the same 3 loci were also found, again with a level of statistical significance in the suggestive range, when each round of mapping was analyzed individually. Therefore, each of these QTLs has been confirmed in successive, independent rounds of linkage analysis.25
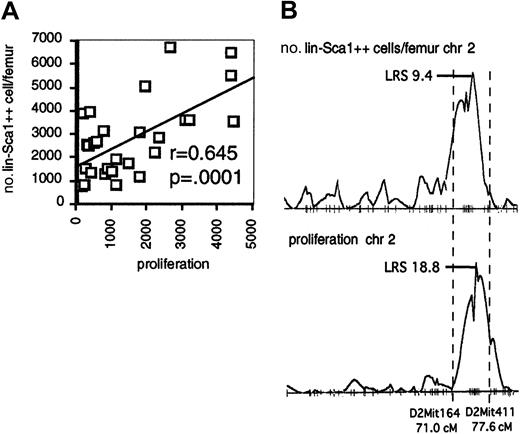
Interestingly, the QTL on chromosome 2 for the number of Lin−Sca1++ cells overlaps with the QTL on chromosome 2 for the proliferative capacity of Lin−Sca1++ cells. In addition, there was a significant correlation between proliferation and the number and frequency of Lin−Sca1++ cells among BXD RI strains (Figure 4A; data shown for absolute number of Lin−Sca1++ cells), and the pattern of LRS values along chromosome 2 was virtually identical for both traits (Figure 4B). Taken together, these data may suggest that genetic variation in the responsiveness to early-acting cytokines contributes to the regulation of the absolute number of Lin−Sca1++ cells among BXD RI strains.
Responsiveness to early-acting factors and number of Lin−Sca1++ cells in BXD RI strains.
(A) Correlation between the responsiveness to flt3, KL, and TPO and number of Lin−Sca1++ cells per femur among BXD RI strains. (B) LRS along chromosome 2 (chr 2) for the number of Lin−Sca1++ cells per femur (top) and proliferation (bottom).
Responsiveness to early-acting factors and number of Lin−Sca1++ cells in BXD RI strains.
(A) Correlation between the responsiveness to flt3, KL, and TPO and number of Lin−Sca1++ cells per femur among BXD RI strains. (B) LRS along chromosome 2 (chr 2) for the number of Lin−Sca1++ cells per femur (top) and proliferation (bottom).
Confirmation of a locus on chromosome 2 determining the number of Lin−Sca1++kit+ cells in congenic mice
Congenic mice22,23 25 were constructed in the laboratory of GvZ, where a segment from chromosome 2 between D2Mit133 (66.7 cM) and D2Mit200 (102 cM), encompassing the region containing a QTL for the number of Lin−Sca1++ cells and for their proliferative capacity (Table 1), from DBA/2 mice was crossed onto the B6 background (B6.DBA/2-[D2Mit133-D2Mit200] mice). We determined whether introgression of this DBA/2-derived segment of chromosome 2 onto the B6 background would change the number of Lin−Sca1++, Lin−Sca1++kit+, and Lin−Sca1++kit− cells and their responses to flt3L, KL, and TPO toward the values obtained for DBA/2 (Figure 2). Mice congenic for the distal region of chromosome 4 or chromosome 7 are not yet available.
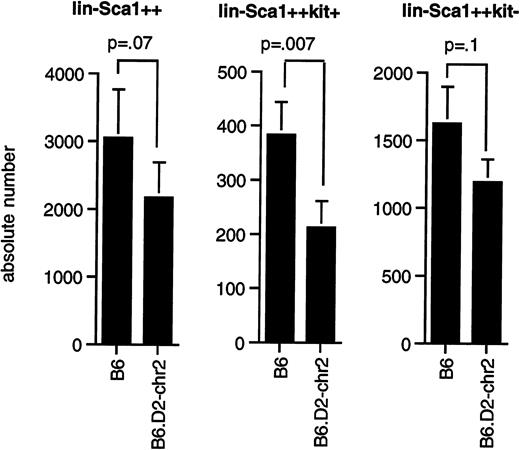
As shown in Figure 5, congenic mice had significantly fewer Lin−Sca1++kit+cells (P = .0075) than B6 mice, whereas the difference in the number of Lin−Sca1++kit−cells (P = .1) and total Lin−Sca1++ cells (P = .07) was at the limit of statistical significance by paired t test. Our data thus confirm that the segment of chromosome 2 between 66.8 and 107 cM contains at least one gene that contributes to the regulation of the pool size of Lin−Sca1++kit+ cells and possibly also of Lin−Sca1++kit− cells, that this gene shows allelic variation between B6 and DBA/2 mice, and that DBA/2 was the donor of the low allele for this trait.
Number of Lin−Sca1++, Lin−Sca1++kit+, and Lin−Sca1++kit− cells in congenic mice.
(A) Average number (mean ± SEM, n = 8) of Lin−Sca1++, Lin−Sca1++kit+, and Lin−Sca1++kit− cells per femur in B6 and B6.DBA/2-(D2Mit133-D2Mit200) congenic mice (the latter are indicated in the figure as B6.D2-chr2). Cell numbers were determined by cell sorting. P values (paired t test) are given at the tops of the figures.
Number of Lin−Sca1++, Lin−Sca1++kit+, and Lin−Sca1++kit− cells in congenic mice.
(A) Average number (mean ± SEM, n = 8) of Lin−Sca1++, Lin−Sca1++kit+, and Lin−Sca1++kit− cells per femur in B6 and B6.DBA/2-(D2Mit133-D2Mit200) congenic mice (the latter are indicated in the figure as B6.D2-chr2). Cell numbers were determined by cell sorting. P values (paired t test) are given at the tops of the figures.
We anticipated seeing lower proliferative capacity and CFC generation in Lin−Sca1++kit+ cells from congenic mice than from B6 mice. However, proliferation and CFC generation were identical in B6 and congenic mice (not shown, n = 8). The QTL on chromosome 2 for proliferative capacity could thus not be confirmed in congenic mice.
Evidence for X-linked genetic factors in the regulation of the response of primitive hematopoietic stem and progenitor cells to early-acting cytokines in mice
Alleles of unknown X-linked genes confer a selective advantage to hematopoietic stem cells in vivo in humans and in cats.10-15 No models or in vitro assays are available to investigate X-linked regulation of hematopoiesis in the mouse. Furthermore, quantitative trait analysis with the available number of BXD RI strains does not detect all QTLs involved in complex traits.24,25 We therefore tried to establish a model to detect X-linked allelic variation in the hematopoietic stem cell compartment by investigating whether reciprocal male B6D2F1 and D2B6F1 mice differed in the number of Lin−Sca1++kit+ and Lin−Sca1++kit− cells and in the responsiveness of Lin−Sca1++kit+to early-acting cytokines. Female D2B6F1 and B6D2F1 mice are heterozygous in all their loci, and X inactivation is random.31 Therefore, the phenotype of reciprocal female F1 hybrids will be the same, unless there is allelic variation in imprinted genes, so that a parent-of-origin effect is observed. Reciprocal male F1 hybrids, however, differ in the origin of the unique X chromosome (B6 in B6D2F1 mice, and DBA/2 in D2B6F1 mice). Phenotypic variation between reciprocal male F1 hybrids is thus most likely caused by allelic variation at X-linked loci. Theoretically, it is also possible that allelic variation at Y-linked loci exists.
The numbers of Lin−Sca1++kit+ and Lin−Sca1++kit− cells in female and male F1 hybrids were intermediate between those for the parental strains, indicating that it is unlikely that X-linked genes regulate the number of Lin−Sca1++kit+ or Lin−Sca1++kit− cells in vivo (not shown, n = 8). Proliferation (Figure6A) and CFC production (Figure 6B) were similar in female B6D2F1 and D2B6F1 mice (n = 4). This is in accordance with complete heterozygosity in autosomal loci and random X inactivation in female F1 hybrids.31 In contrast to female F1 hybrid mice, however, proliferation (Figure 6A) and CFC generation (Figure 6B) in Lin−Sca1++kit+cells from male D2B6F1 mice were significantly higher than in Lin−Sca1++kit+ cells from male B6D2F1 mice (n = 8). Because male D2B6F1 and B6D2F1 mice are heterozygous in all their autosomal loci but differ in the origin of the X chromosome (B6 in B6D2F1 mice and DBA/2 in D2B6F1 mice), these data suggest that at least one X-linked locus contributes to the regulation of the response of Lin−Sca1++kit+ cells to early-acting factors and that the donor of the high allele is DBA/2. As mentioned before, genetic variation in Y-linked genes could theoretically also explain our data. Although this cannot be formally excluded, the involvement of Y-linked genes is less likely given the paucity of somatically active genes on the Y chromosome.32In addition, if allelic variation between DBA/2 and B6 mice on the Y chromosome plays a role, then one would expect a differential effect of sex on this trait in B6 and DBA/2 mice, which was not the case (not shown).
Difference in proliferative capacity in reciprocal male and female F1 hybrid mice.
Cell proliferation (A) and CFC generation (B) per 50 input Lin−Sca1++kit+ cells isolated from male and female D2B6F1 and B6D2F1 hybrid mice. Results are given as mean ± SEM. P values indicated on the figure were obtained by paired t test (n = 4 for female D2B6F1 and B6D2F1 mice; n = 8 for male D2B6F1 and B6D2F1 mice).
Difference in proliferative capacity in reciprocal male and female F1 hybrid mice.
Cell proliferation (A) and CFC generation (B) per 50 input Lin−Sca1++kit+ cells isolated from male and female D2B6F1 and B6D2F1 hybrid mice. Results are given as mean ± SEM. P values indicated on the figure were obtained by paired t test (n = 4 for female D2B6F1 and B6D2F1 mice; n = 8 for male D2B6F1 and B6D2F1 mice).
Discussion
In this study, we have identified 2 significant traits involving the hematopoietic stem and progenitor cell compartment that show quantitative variation: (1) responsiveness to early-acting hematopoietic cytokines and (2) absolute number of Lin−Sca1++ cells (both c-kit+ and c-kit− subpopulations).
Variation in the fraction of Lin−Sca1+ cells among inbred mouse strains was already noted previously by Spangrude and Brooks.33 Lin−Sca1++ cells are highly enriched in stem and progenitor cells16-18 but are still heterogeneous. Long-term repopulating cells are thought to be Lin−Sca1++kithigh,1,16-18though some reports suggest that a kit− precursor of kithigh repopulating stem cells exists.34Lin−Sca1+kitlow cells have been shown to be enriched in early-lymphoid precursors,35though lymphoid-committed precursors have recently been identified in the Lin−kithigh population as well.36 A Lin−Sca1+kit− population has been described by Randall and Weissman30 in steady state bone marrow. This mystery population is quiescent, increases in size with aging, does not proliferate in vitro, and does not contain long-term repopulating activity. We did indeed reproduce these findings (J.C.L. and H.-W.S., unpublished observations, December 2001). The functional significance of this Lin−Sca1++kit− population is unknown. We show here that at least 3 loci determine the total number of Lin−Sca1++ cells: one on chromosome 2, one on chromosome 7, and one on chromosome 4. Because we found that there was little variation in the ratio of kit+/kit−cells among Lin−Sca1++ cells, despite a 10-fold variation in the total number of Lin−Sca1++ cells in BXD strains (Figure 3), we conclude that the number of Lin−Sca1++kit+ and Lin−Sca1++kit− cells are regulated by at least partially overlapping genetic mechanisms. Indeed, the number of Lin−Sca1++kit+ and Lin−Sca1++kit− cells is 2-fold higher in B6 than in DBA/2 mice. These mechanisms are not necessarily identical, nor do they necessarily affect each subpopulation to the same extent. In congenic mice, we find a significant effect of the DBA-derived segment of chromosome 2 on the number of Lin−Sca1++kit+ cells, but the effect on the number of Lin−Sca1++kit− and total Lin−Sca1++ cells numbers is at the limit of statistical significance, possibly suggesting that this locus mainly regulates the number of Lin−Sca1++kit+ cells. This will, of course, be reflected in the size of the total Lin−Sca1++ population, for which we have demonstrated linkage on chromosome 2 in BXD RI strains. Although the biologic significance of a small change in the number of Lin−Sca1++ cells caused by a single locus may be limited, the combined effect of many loci affecting pool size of Lin−Sca1++ cells, as in those BXD mice with extreme values, is likely to be important. In addition, the importance of our data lies in the fact that we show that multiple loci affecting the number of Lin−Sca1++ cells exist. This may lead to identification of the mechanisms that regulate the number of Lin−Sca1++ cells and of the kit+and kit− subpopulations thereof.
The correlation between the response to flt3L, KL, and TPO and the number of Lin−Sca1++ cells among BXD RI strains and the overlapping QTLs for both traits raise the possibility that the responsiveness to early-acting cytokines plays a role in determining the number of Lin−Sca1++ cells. However, a phenotype could be reproduced in mice congenic for the locus on chromosome 2 for the number of Lin−Sca1++kit+ and Lin−Sca1++kit− cells, but not for proliferative capacity in response to KL, flt3L, and TPO. The most likely explanation for the lack of a proliferative phenotype in the congenic mice is epistasis with other loci that show allelic variation.25,37 In addition, a locus was found on the X chromosome that clearly determines the response of Lin−Sca1++kit+ cells to early-acting factors, but it has no effect on the number of Lin−Sca1++kit+ or Lin−Sca1++kit− cells. Our data thus suggest that genetic variation in the response to early-acting cytokines does not determine the number of Lin−Sca1++kit+ or Lin-Sca1++kit− cells. This is in accordance with the data of Miller et al,38 who showed that the number of stem cells, as defined by the Lin−Sca1+WGA+ phenotype, is similar in W mutant and wild-type mice. The correlation between the response to early-acting factors and the number of Lin−Sca1++ cells is most likely explained by the fact that linked, but distinct, QTLs on chromosome 2 are major determinants for both traits.
Other QTLs related to hematopoiesis have been mapped to this segment of chromosome 2. In several studies,8,39 a suggestive QTL on chromosome 2 at approximately 50 cM, regulating the increase of the number of CAFCd35 with aging in B6 and DBA/2 mice has been reported. Hasegawa et al9 identified a significant QTL that controls the efficiency of mobilization of hematopoietic progenitor cells in response to GM-CSF on chromosome 2 between 46 and 86 cM.9It was recently shown that the mobilization of hematopoietic stem cells occurs after the M-phase of the cell cycle.40 Therefore, it is possible that an allele responsible for a higher responsiveness to early-acting factors also causes more efficient mobilization of hematopoietic stem cells. Potential candidate genes in this region of chromosome 2 known to play a role in hematopoiesis includejag1 (encoding the Notch ligand Jagged-1),41bmp2 (encoding BMP-2),42 and the il1complex (encoding IL-1α and IL-1β, respectively).43
QTL analysis, as reported here and by others,2-6,8,9demonstrates differential genetic regulation of the size of the stem and progenitor cell compartment, as defined by immunophenotype on one hand and by functional assays on the other hand. Our data show that Lin−Sca1++kit+ cells are more numerous in B6 than in DBA/2 mice, confirming an earlier report in which the WGA++Lin−rho123lowphenotype was used to isolate and identify the most primitive hematopoietic precursors.6 Yet, Lin−Sca1++kit+ cells from DBA/2 mice repopulate syngeneic recipients more efficiently than those of B6 mice and generate more CAFCd35.44 In addition, DBA/2 mice show faster recovery after the administration of 5-fluorouracil than B6 mice44 and contain more CAFCd35 and LTC-IC than B6 mice.2,3,8 Conversely, data in allophenic mice suggest that on aging, B6-derived stem cells are more efficient at sustaining hematopoiesis.4,5 In CXB mice, long-term repopulation potential does not correlate with LTC-IC and CAFC numbers either.7 QTL analysis and subsequent identification of genes responsible for quantitative variation in the stem cell compartment, as defined by function and by phenotype, may provide novel insight into the regulation of hematopoiesis by helping to resolve these discrepancies. A possible explanation may be that the number of Lin−Sca1++kit+ cells is a reflection of stem and progenitor cell pool size in steady state conditions, whereas assays such as CAFC, LTC-IC, and long-term repopulation assay45 reflect, among other things, the capacity of the stem cell compartment to respond to hematopoietic stress.
The answer to whether the putative X-linked locus we identified here is the same as the locus that causes skewed X inactivation on aging in cats and in humans will have to await gene identification of the loci involved. However, given the importance of early-acting cytokines such as KL, flt3L, and TPO in vivo,19-21 allelic variation in one or more X-linked genes affecting the responsiveness to these cytokines is an attractive candidate mechanism for the large genetic component in the skewed X inactivation in elderly females in humans11,15 and in cats.12 Our study suggests that an easy assay—measurement of the responsiveness to early-acting cytokines in vitro—may allow mapping and identification of X-linked loci involved in the regulation of hematopoiesis.
In summary, we demonstrated genetic variation in the response to cytokines critical for hematopoiesis in vivo and in the pool size of cells belonging to a phenotype used to isolate essentially pure primitive progenitor and stem cells, and we identified loci that may be relevant to the regulation of hematopoiesis in steady state.
Supported in part by National Institutes of Health grant RO1 AG16327 (H.-W.S.) and RO1 AG16653 (G.V.Z.). E.H. was supported by a fellowship from the Belgian American Educational Foundation and by the Belgian Hematological Society. H.G. was supported by a fellowship from the Deutsche Akademie der Naturforscher Leopoldina.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hans-Willem Snoeck, Carl C. Icahn Institute for Gene Therapy and Molecular Medicine, Mount Sinai School of Medicine, Box 1496, One Gustave L. Levy Pl, New York, NY 10029; e-mail:hans.snoeck@mssm.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal