Abstract
We examined the expression of two members of theNotch family, Notch-1 and Notch-2, and one Notch ligand, Jagged-1, in hematopoietic cells. Both Notch-1 and Notch-2 were detected in murine marrow precursors (Lin−Sca-1+c-kit+). The Notch ligand, Jagged-1, was not detected in whole marrow or in precursors. However, Jagged-1 was seen in cultured primary murine fetal liver stroma, cultured primary murine bone marrow stroma, and in stromal cell lines. These results indicate a potential role for Notch-Notch ligand interactions in hematopoiesis. To further test this possibility, the effect of Jagged-1 on murine marrow precursor cells was assessed by coculturing sorted precursor cells (Lin−Sca-1+c-kit+) with a 3T3 cell layer that expressed human Jagged-1 or by incubating sorted precursors with beads coated with the purified extracellular domain of human Jagged-1 (Jagged-1ext). We found that Jagged-1, presented both on the cell surface and on beads, promoted a twofold to threefold increase in the formation of primitive precursor cell populations. These results suggest a potential use for Notch ligands in expanding precursor cell populations in vitro.
HEMATOPOIETIC STEM CELLS either self-renew, thereby maintaining stem cell properties, or alternatively, give rise to cells increasingly committed to differentiate into the various hematopoietic lineages. It is unclear if this cell fate decision is controlled by a purely stochastic mechanism1 or is the result of environmental cues mediated through specific receptor ligand interactions. In several invertebrate and vertebrate developmental systems, cell fate is influenced both by soluble molecules and by molecules acting via cell-cell interactions, including those mediated by the Notch receptor family.2 3
The Notch superfamily encodes cell-surface receptors that influence numerous cell-fate decisions in both invertebrates and vertebrates, including decisions made during central and peripheral nervous system development, wing, eye, bristle, and ovary development in Drosophilamelanogaster,4-8 the germinal vesicle and nervous system development in Caenorhabditis elegans,9 and eye development inXenopus.10 Moreover, NotchmRNA and protein have been found in hematopoietic precursors, indicating a potential role for Notch interactions in hematopoiesis11 (and Flowers et al, submitted).
Both Notch and its ligands are transmembrane proteins that consist of several epidermal growth factor (EGF) repeats in the extracellular domain. At present, four Notch homologs have been identified in vertebrates: Notch-1, -2, -3, and -4.12-16 Also, candidate ligands for Notch have been cloned from vertebrates, including two homologs for Drosophila Serrate. The first Serrate homolog has been referred to asJagged-1 in rat17 and C-Serrate-1 in chicken,18 and the second Serrate homolog asJagged-2 in rat19 and in human.20 Also, two homologs for D Delta have been cloned. The firstDelta homologue has been referred to as C-Delta-1 in chicken21 and Delta-like-1 in mouse,22and the second as Delta-like-3 in mouse.23 At the present time very little is known about the specificity of the various Notch receptors for each ligand. Notch ligands consist of a highly homologous DSL (the first letters of Delta,Serrate, and Lag-2)24 domain at the amino terminus followed by EGF repeats (see Fig 1). In Jagged-1 and -2, as in their Drosophila homolog Serrate, there is a cysteine-rich region between the EGF repeats and the transmembrane domain. Delta is missing this domain in both vertebrates and in Drosophila. The function of the cytoplasmic domain, whose structure varies among the different ligands, is unknown but deletion analyses have shown it to be essential for wild-type function.10 25-28
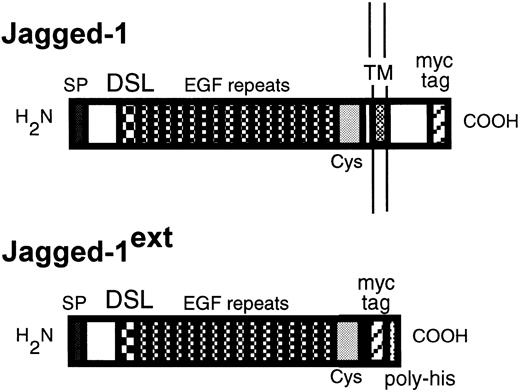
A schematic diagram of Jagged-1 and Jagged-1ext. The top diagram depicts the protein product ofJagged-1 expressed in 3T3 cells and the bottom diagram the truncated product of Jagged-1ext. Indicated are the signal peptide (SP), the DSL domain, the EGF repeats, the cysteine rich region (Cys), the transmembrane domain (TM), the myc tag epitope recognized by the 9E10 antibody (myc tag), and the poly-histidines (poly-his).
A schematic diagram of Jagged-1 and Jagged-1ext. The top diagram depicts the protein product ofJagged-1 expressed in 3T3 cells and the bottom diagram the truncated product of Jagged-1ext. Indicated are the signal peptide (SP), the DSL domain, the EGF repeats, the cysteine rich region (Cys), the transmembrane domain (TM), the myc tag epitope recognized by the 9E10 antibody (myc tag), and the poly-histidines (poly-his).
To identify a potential role for Notch in hematopoiesis, we first assessed the expression of two members of the Notch family,Notch-1 and Notch-2, in murine whole marrow and in purified precursors. We also determined the expression of the Notch ligand, Jagged-1, in murine cells including whole marrow, purified precursors, primary cultured fetal liver stroma, primary cultured bone marrow (BM) stroma, and in stromal cell lines.29 30 To assess the effect of Jagged-1 on early precursors, we cultured purified primitive mouse marrow cells (Lin−Sca-1+c-kit+) either with a 3T3 layer expressing full-length Jagged-1 or in wells containing beads coated with truncated Jagged-1, consisting of the extracellular domain of Jagged-1 (Jagged-1ext, see Fig1). The results presented in this report show the potential for Notch and Jagged-1 interactions within the marrow and further demonstrate that Jagged-1 affects the in vitro development of a primitive precursor cell population.
MATERIALS AND METHODS
Transfections, retroviral infections, and full-length and mutant Jagged-1 generation.
Full-length human Jagged-1 cDNA in pBluescript (KS+; Stratagene, San Diego, CA; Gray and Artavanis, in preparation; GenBank accession no. U61276) was subcloned into a PCS2 vector that contained sequences encoding six consecutive myc epitopes (generous gift from David Turner and Hal Weintraub, Fred Hutchinson Cancer Research Center), so that six myc-epitopes were expressed from the carboxyl terminus of Jagged-1(Fig 1). An EcoRI fragment containing Jagged-1 plus the myc tags was subcloned into LXSN, a retroviral vector.30 Viruses were made and NIH-3T3 cells were infected. Five G418-resistant clones were then assessed for Jagged-1 expression with Western blots using 9E10, a monoclonal antibody (MoAb) that recognizes the myc tag epitopes.32
To generate the extracellular Jagged-1ext, Jagged-1 sequences encoding from amino acid 1047 to the carboxy terminus including the transmembrane and cytoplasmic domain were deleted. The remaining sequence encoding the extracellular domain was then subcloned into the PCS2 vector with the myctag. A sequence encoding six consecutive histidines and a stop codon was then added in frame to the 3′ end (Fig 1). The cDNA encoding the extracellular domain of Jagged-1 plus the myc tags and the poly-histidine tail was then subcloned into the expression vector pcDNA 3.1 (Invitrogen, San Diego, CA). COS cells were transfected with Jagged-1ext using a modified method developed by Hirano et al.33 COS cells to be transfected were plated onto 150-mm dishes in 20 mL of serum-free medium (Dulbecco's modified Eagle's medium [DMEM] supplemented with Nutridoma HU; Boehringer Mannheim, Indianapolis, IN; at the recommended concentration) containing 10 mmol/L chloroquine diphosphate (Sigma, St Louis, MO). A solution of diethyl aminoethyl dextran (1 mg/mL final concentration; Pharmacia Biotech, Piscataway, NJ) and DNA (1.5 μg/mL final concentration) in phosphate-buffered saline (PBS) was added. After a 2- to 3-hour incubation at 37°C, transfected cells were washed with serum-free medium and subsequently maintained in serum-free medium. Control cells were treated in an identical manner, but DNA was not added. Conditioned medium was collected from both cells expressingJaggedext and control cells after 3 and 5 days. Conditioned medium from each cell type was then concentrated and dialyzed against PBS (10 mmol/L Na2HPO4, 100 mmol/L NaCl, pH 7.2). Jagged-1ext–containing medium and control medium were then bound to a Nickle column (Ni-NTA Agarose; Qiagen, Chatsworth, CA) using the His-Bind buffer Kit (Novagen, Madison, WI) according to manufacturers' instructions. After elution with imidizol (5 mmol/L to 1 mol/L), fractions that contained Jagged-1ext were identified with Western blots using the 9E10 antibody. Positive fractions were then pooled, concentrated, and dialyzed with PBS and analyzed by polyacrylamide gel electrophoresis (PAGE) in reducing conditions. The same fractions from the control elutions were also pooled, concentrated, and dialyzed.
To further purify Jagged-1ext, concentrated and dialyzed pooled fractions were added to 9E10 crosslinked Sepharose beads (Sigma) overnight at 4°C in a tube roller. About 10 μg of ligand was added to about 20 μL of beads in 0.5 mL of a 10 mg/mL solution of bovine serum albumin (BSA) in PBS. To make 9E10 crosslinked beads, 1 mg 9E10 antibody was crosslinked to 0.9 g cyanogen bromide activated Sepharose 4B (Sigma) according to manufacturer's instructions. Briefly, beads were swollen in 1 mmol/L HCl for 15 minutes and washed two times with coupling buffer (0.1 mol/L NaHCO3 pH 8.3, 0.5 mol/L NaCl). Antibody was then resuspended in sterile coupling buffer to a final concentration of 2 mg/mL then mixed with 0.5 mL of a 50% bead solution and incubated for 2 hours at room temperature. To block remaining active groups, 0.2 mol/L glycine, pH 8.0, was added for 2 hours at 4°C. Beads were then washed according to manufacturer's instructions. Antibody beads were blocked with 10 mg/mL BSA (Sigma) in PBS before use.
Antibodies, Western blots, immunoprecipitations.
MoAbs GR-1 (clone RB6-8C5) and Mac-1 (clone M1/70), CD2 (clone RM2.2), CD3 (clone KT3-1.1), CD5 (clone 53-7.3), CD8 (clone 53-6.7), B220 (clone RA36B2), and TER-119 were a generous gift from G. Spangrude (University of Utah, Salt Lake City). The methods for developing the rat monoclonals BHN6 and 18G are described elsewhere.34 To generate rabbit polyclonal antibody SER10, a fragment (bp 3241-3550) of the portion of C-Serrate-1 cDNA that encodes the cytoplasmic domain of C-Serrate-1 was subcloned intopet23b vector (Novagen), expressed in Escherichiacoli, and purified on a Nickel column under nondenaturing conditions according to the manufacturer's instructions (Novagen).
For Western blot analysis, total cell lysates were prepared from 1 to 5 × 106 cells (per lane) using lysis buffer (50 mmol/L Tris, pH 8.0, 0.15 mol/L NaCl, 20 mmol/L EDTA, 1.0% Triton-X-100). Triton soluble proteins from lysates were separated using either an 8% or a 6% sodium dodecyl sulfate (SDS)-PAGE using a mini-gel apparatus (Owl Scientific, Woburn, MA). Before loading gels, lysates were resuspended in reducing sample buffer (.06 mol/L Tris, pH 6.8, 1% SDS, 12.5% glycerol, 1.25% β-mercaptoethanol, 0.025% bromophenol blue). Separated proteins were transferred to nitrocellulose (Schleicher and Schull, Keene, NH) and immunoblotted with the respective MoAbs. Immunoreactivity was detected using a horseradish peroxidase (HRP)-conjugated sheep anti-mouse IgG antibody (Amersham, Arlington Heights, IL) and enhanced chemiluminescence Western blot reagents (Amersham).
To biotinylate surface proteins before immunoprecipitation, 1 × 106 cells were incubated with sulfo-NHS-biotin (Pierce, Rockford, IL) in 0.1 mol/L HEPES in PBS, pH 8.0 according to the manufacturer's instructions. Cells were removed and washed three times with TBS (25 mmol/L Tris, pH 8.0, 140 mmol/L NaCl, 2 mmol/L KCl) and lysed for 15 minutes on ice with lysis buffer plus protease inhibitors (2 μg/mL aprotinin, 1 μmol/L pepstatin A, 100 μmol/L leupeptin [all from Boehringher Mannheim, Indianapolis, IN], and 1 mmol/L phenylmethylsulfonyl fluoride [PMSF; Calbiochem, San Diego, CA]).
For immunoprecipitation of Jagged-1 with 9E10, biotinylated cell lysates were first incubated with beads crosslinked with a control antibody. Lysates were then incubated with 9E10 crosslinked beads (see below) for 2 hours at 4°C. Beads were washed once with Lysis buffer and then with RIPA buffer (50 mmol/L Tris, pH 8.3, 0.45 mol/L NaCl, 0.5 % NP-40). Beads were then resuspended in reducing sample buffer and proteins were separated using an 8% SDS-PAGE. Separated proteins were transferred and biotinylated proteins were detected with HRP streptavidin (Amersham) and enhanced chemiluminescence reagents (Amersham).
Hematopoietic cells and flow immunocytometry.
C57BL/6J (Ly5.2) female mice were obtained from The Jackson Laboratory (Bar Harbor, ME). All animals were housed in specific pathogen-free conditions and maintained on acidified drinking water and autoclaved chow ad libitum. Mice were used at 8 to 12 weeks of age. For enrichment of hematopoietic precursor cells, suspensions of BM cells were obtained by flushing femurs and passaging the marrow through a 23-gauge needle into PBS containing 2% heat-inactivated fetal calf serum (PBS/FCS). Low-density cells were enriched by equilibrium centrifugation over a cushion of Ficoll-Hypaque (Pharmacia and Nycomed-Amersham, Princeton, NJ) at a density of 1.077 g/mL. Retrieved cells were washed, then resuspended at a density of 5 × 107 cells/mL in PBS/FCS containing a predetermined saturating solution of MoAbs specific for murine T lymphocytes (CD2, CD3, CD5, CD8), B lymphocytes (B220), macrophages (Mac-1), granulocytes (Gr-1), and erythrocytes (TER-119).
After 30 minutes of incubation on ice, the cells were washed, resuspended in PBS/FCS at a concentration of 108 cells/mL, and transferred to a 50-mL conical tube. Twice-washed immunomagnetic particles (Dynabeads M-450, sheep anti-rat IgG specificity; Dynal Inc, Great Neck, NY) were slowly added to the cells to a final ratio of 4 beads/cell. The cell/bead suspension was allowed to stand for 5 minutes at room temperature, then centrifuged at 400 rpm for 3 minutes. The cells and bead mixture was resuspended in 1 mL of PBS/FCS. The magnetic-particle-free fraction was retrieved by exposure to a magnetic field, and the magnetic depletion procedure was repeated on this fraction.
The bead-free cells were then preincubated with FcγRII block (clone 2.4G2; Pharmingen, San Diego, CA) for 10 minutes on ice and then stained with biotinylated anti-Ly6A/E (Sca-1, rat IgG2a, clone E13-161.7) and fluorescein isothiocyanate (FITC)-conjugated anti-CD117 (c-kit, rat IgG2b, clone 2B8) MoAbs (Pharmingen) for 30 minutes on ice. Aliquots of the bead-free cells were also stained with isotype-matched controls. The cells were then washed with 10 mL PBS/FCS, stained with streptavidin-phycoerythrin (Pharmingen) for 30 minutes on ice, washed, and resuspended in PBS/FCS containing 1 mg/mL propidium iodide. The sample was filtered through a 70-μm pore-size nylon screen.
Fluorescence-activated cell sorting was performed on a Vantage (Becton Dickinson, Mountain View, CA), using the CloneCyt direct cloning application (Becton Dickinson). Cells sorted were propidium iodide (PI)-negative, c-kit+, Sca-1+, with intermediate forward- and right-angle scatters.
For 7-day primary culture, 50 purified precursors were added to a single well of a round-bottom 96-well plate (Corning, Corning, NY) containing beads (about 1,000 beads per well) and culture medium (Iscove's medium, 20% FCS) plus cytokines (stem cell factor [SCF], Flt3-ligand, interleukin-6 [IL-6]) (each at 100 ng/mL), and IL-11 (10 ng/mL). At 7 days of culture, cells were removed and 1/12 (approximately 10,000 cells) of the culture was replated into a 35-mm culture dish (Nunc, Naperville, IL) containing semisolid medium (40% α-MEM–based methylcellulose [Stem Cell Technologies, Vancouver, BC, Canada], 10% WEHI conditioned medium,35 30% FCS, and 1 × 10−4 mol/L 2-mercaptoethanol [2-ME], 100 ng/mL IL-6 and SCF, 50 ng/mL megakaryocyte growth and development factor [MGDF], and 3 U/mL erythropoietin).
After 10 days of culture in semisolid medium, colonies were viewed with an inverted microscope and colony types determined and counted. Colonies were designated HPP-mix if they consisted of granulocytes, macrophages, and erythroid clusters and were larger than 1.5 mm. Colonies were evaluated by preparing cytocentrifuge slides. In each colony evaluated the presence of each of these cell types as well as megakaryocytes was confirmed. A mean number of all colony types was determined for all 10 wells and then a final mean for five experiments. Statistical differences were assessed using analysis of variance.36
For time-course experiments, 200 purified precursors were added to a single well of a round-bottom 96-well plate containing beads and culture medium as described above. In experiment 1 there were 8 replicate wells and in experiment 2 there were 10. After primary culture for 5 and 8 days, cells were harvested, counted, and 25,000 cells were replated with fresh beads for further culturing. At the same time 1/12 of the culture (approximately 10,000 cells) was replated into 35-mm plates containing semisolid medium so that there were about 50 colonies in each plate. Each well was plated in this manner in quadruplicate. After 10 days in secondary culture, types and size of colonies were assessed.
For replating experiments all HPP-mix colonies from day 0, 5, and 8 primary cultures were picked and replated into fresh semisolid medium. After 5 to 7 days in the tertiary cultures, types and size of colonies were assessed.
To analyze Notch-1 or Notch-2 expression, suspensions of BM cells were enriched for low-density cells as described above. Cells were washed twice, then fixed and permeabilized by incubation with PermeaFix (Ortho, Raritan, NJ), washed twice, and then incubated at room temperature with the rat anti-human Notch-1 antibody 18G or anti-human Notch-2 antibody BHN6. Cells were washed twice and incubated at 4°C with mouse-absorbed FITC-conjugated monoclonal mouse anti-rat IgG1 antibody (Pharmingen). In addition, whole marrow was enriched for hematopoietic precursor cells by magnetic depletion using lineage-specific antibodies as described above. Enriched cells were then stained with anti-Ly6A/E (Sca-1) directly conjugated to phycoerythrin (Pharmingen) and anti-CD117 (c-kit) directly conjugated to biotin (Pharmingen). Cells were washed twice and incubated with RED613 conjugated to streptavidin (GIBCO-BRL, Grand Island, NY) and again washed twice. Cells were then fixed and permeabilized and stained with either Notch-1 or Notch-2 antibodies as previously described. Control staining was measured using isotype matched directly labeled or unlabeled control antibody. Cells were analyzed using a Vantage flow cytometer (Becton Dickinson). Cells with a surface antigen fluorescence intensity greater than 99% of control staining were considered positive while cells with a surface antigen fluorescence intensity less than the top 5% of control staining were considered negative.
Stromal cells.
Methods are previously described for culturing fetal liver stroma.28 Briefly, fetal livers at day 14 of gestation from C57Bl/6J were dissociated mechanically with repeated pipetting and passed through a 70-μm nylon filter. The retentate was cultured in 5% CO2 in modified Dexter media (DMEM, 10% FCS, 10% horse serum, 50 μmol/L 2-ME, 0.1 μmol/L hydrocortisone). Murine stromal cell lines derived from fetal liver were generated and maintained as previously described.28 29
To culture BM stroma, suspensions of BM cells were obtained by flushing femurs from two C57Bl/6J mice and two Balb-C mice. Cells were cultured at an initial density of 2 to 3 × 107 in a 25-cm culture flask in 5% CO2 in long-term culture medium (Iscove's medium, 12.5% FCS, 12.5% horse serum, 2% MEM-amino acids [GIBCO], 1% MEM-nonessential amino acids [GIBCO], 1% MEM-vitamins [GIBCO], 10−6 mol/L hydrocortisone, 10−1 mol/L 2-ME, 24 mmol/L NaHCO3, 100 mmol/L Na-Pyruvate). Cultures were harvested at 3 weeks and lysates prepared for Western blotting.
RESULTS
Expression of Notch by murine hematopoietic cells.
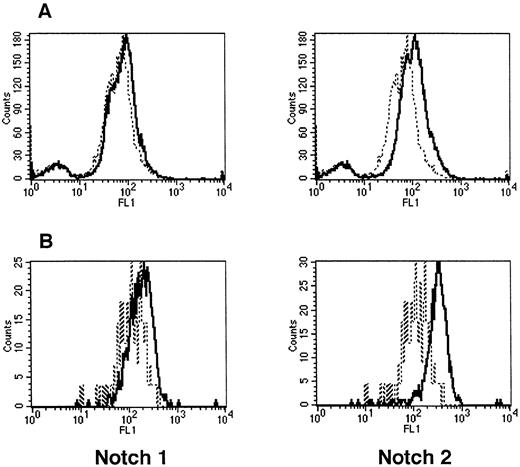
To assess Notch expression in hematopoietic cells, whole-mouse marrow and precursors (Lin−Sca-1+c-kit+) were analyzed with flow cytometry, using MoAbs that specifically recognize Notch-1 (18G) and Notch-2 (BHN6) (Fig 2). Although little or no Notch-1 was seen in whole marrow, Notch-1 was detected in precursors (Lin−Sca-1+c-kit+). Notch-2 was detected in whole marrow and, as with Notch-1, more was seen in precursors.
Expression of Notch-1 and Notch-2 by mouse BM cells. (A) Fluorescence histograms of Ficoll-Hypaque–separated marrow cells stained with MoAbs that recognize Notch-1 (18G) or Notch-2 (BHN6). (B) Fluorescence histograms of precursor cells (Lin−Sca-1+c-kit+) stained with the same MoAbs as in (A). The x-axis represents log fluorescence intensity and the y-axis represents cell number. The solid line represents staining with 18G or BHN6 antibodies and the dashed line represents staining with an isotype-matched nonspecific antibody.
Expression of Notch-1 and Notch-2 by mouse BM cells. (A) Fluorescence histograms of Ficoll-Hypaque–separated marrow cells stained with MoAbs that recognize Notch-1 (18G) or Notch-2 (BHN6). (B) Fluorescence histograms of precursor cells (Lin−Sca-1+c-kit+) stained with the same MoAbs as in (A). The x-axis represents log fluorescence intensity and the y-axis represents cell number. The solid line represents staining with 18G or BHN6 antibodies and the dashed line represents staining with an isotype-matched nonspecific antibody.
Generation of a 3T3 cell line expressing Jagged-1.
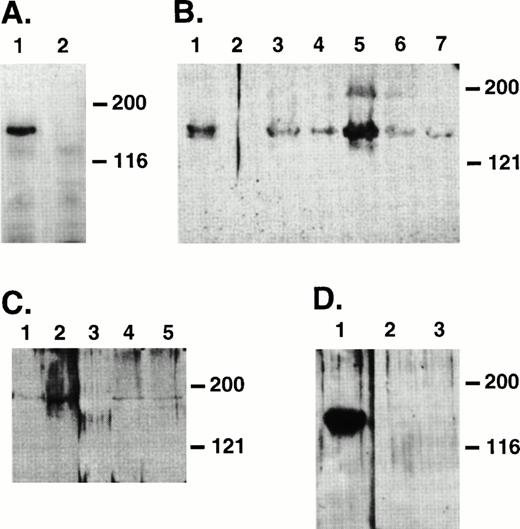
Fibroblast cell lines expressing Jagged-1 were generated by infecting a parental NIH-3T3 cell line with a retrovirus vector containing full-length human Jagged-1. Sequences encoding anmyc epitope recognized by an MoAb (9E10) were added to the 3′ end of Jagged-1 (Fig 1). Two of five G418-resistant 3T3 clones expressed ligand detectable by Western analysis using 9E10 as a probe (data not shown). To confirm cell-surface expression ofJagged-1, cells were biotinylated and lysates were prepared and immunoprecipitated with 9E10. A band of molecular weight (Mr) 150 kD, the expected Mr for Jagged-1, was seen in the expressing line but not in the parental 3T3 cell line (Fig 3A).
Immunoprecipitates of biotinylated 3T3 cells to detect surface expression of Jagged (A) and Westerns of hematopoietic cell lysates to detect expression of Jagged-1 (B, C, and D). (A) Lysates were prepared from 3T3 cells infected with a retroviral vector containing sequences encoding full-length human Jagged-1 (lane 1) or parental 3T3 cells (lane 2) that had been previously biotinylated to label surface proteins. Lysates were immunoprecipitated with 9E10 antibody and resulting proteins were separated on an 8% SDS-PAGE, transferred to nitrocellulose, and probed with HRP-streptavidin and developed using ECL. (B) Protein lysates were prepared from a 3T3 cell line expressing Jagged-1 (lane 1), a parental 3T3 cell line (lane 2 ), and murine hematopoietic stromal cell lines AFT024, CFC034, 2058, 2012, and 2018 (lanes 3 through 7, respectively). (C) Protein lysates were prepared from a 3T3 cell line expressing Jagged(lane 1), primary cultured murine fetal liver stoma (lane 2), murine BM cells (lane 3), primary cultured BM stroma from C57BL/6J (lane 4), and from Balb-C mice (lane 5). (D) Protein lysates were prepared from a 3T3 cell line expressing Jagged (lane 1), Ficoll-Hypaque–separated murine BM cells (lane 2), and sorted precursors (Lin−Sca-1+c-kit+) (lane 3). For gels in (B), (C), and (D), proteins were separated with either an 8% SDS-PAGE (B and D) or a 6% SDS-PAGE (C), transferred to nitrocellulose, and probed with SER10, a polyclonal antibody raised against C-Serrate 1.
Immunoprecipitates of biotinylated 3T3 cells to detect surface expression of Jagged (A) and Westerns of hematopoietic cell lysates to detect expression of Jagged-1 (B, C, and D). (A) Lysates were prepared from 3T3 cells infected with a retroviral vector containing sequences encoding full-length human Jagged-1 (lane 1) or parental 3T3 cells (lane 2) that had been previously biotinylated to label surface proteins. Lysates were immunoprecipitated with 9E10 antibody and resulting proteins were separated on an 8% SDS-PAGE, transferred to nitrocellulose, and probed with HRP-streptavidin and developed using ECL. (B) Protein lysates were prepared from a 3T3 cell line expressing Jagged-1 (lane 1), a parental 3T3 cell line (lane 2 ), and murine hematopoietic stromal cell lines AFT024, CFC034, 2058, 2012, and 2018 (lanes 3 through 7, respectively). (C) Protein lysates were prepared from a 3T3 cell line expressing Jagged(lane 1), primary cultured murine fetal liver stoma (lane 2), murine BM cells (lane 3), primary cultured BM stroma from C57BL/6J (lane 4), and from Balb-C mice (lane 5). (D) Protein lysates were prepared from a 3T3 cell line expressing Jagged (lane 1), Ficoll-Hypaque–separated murine BM cells (lane 2), and sorted precursors (Lin−Sca-1+c-kit+) (lane 3). For gels in (B), (C), and (D), proteins were separated with either an 8% SDS-PAGE (B and D) or a 6% SDS-PAGE (C), transferred to nitrocellulose, and probed with SER10, a polyclonal antibody raised against C-Serrate 1.
Expression of Jagged-1.
To assess expression of Jagged-1 in hematopoietic cells, proteins from marrow cells and stroma, fetal liver stroma, and hematopoietic stromal cell lines derived from fetal liver were analyzed by Western blot using a rabbit polyclonal antibody (SER10) that was raised against the cytoplasmic domain of C-Serrate-1. In 3T3 cells expressing Jagged-1, a band of Mr 150 kD, the correct molecular weight for Jagged-1, was detected (Fig 3B), whereas no bands were detected in the parental 3T3 cell line. In addition, a clear band of Mr 150 kD was seen in mouse hematopoietic stromal cell lines AFT024, CFC034, 2058, 2012, and 2018 derived from fetal liver (Fig 3B). Also, a band of Mr 150 kD was seen in lysates prepared from cultured primary fetal liver stroma and cultured primary BM stroma from two different strains of mice (Fig 3C). However, a 150-kD band was not seen in whole mouse marrow (Fig 3C, lane 3), Ficoll-Hypaque separated marrow, or purified precursors (Lin−Sca-1+c-kit+, Fig 3D).
The effect of cell-bound Jagged-1 on mouse marrow progenitor cells.
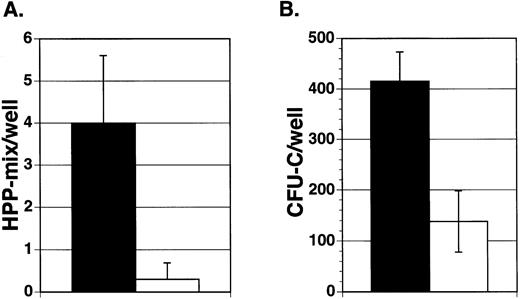
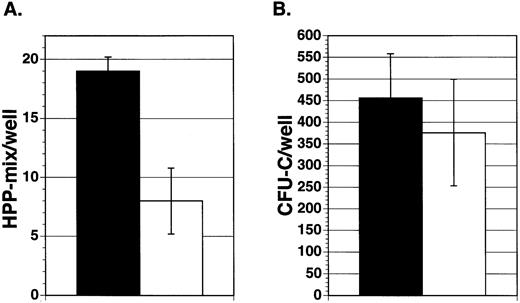
To assess the effect of Jagged-1 on marrow precursor cells, 100 enriched mouse marrow Lin−Sca-1+c-kit+cells were sorted into wells containing either mitomycin-treated 3T3 cell layers expressing Jagged-1 or mitomycin-treated parental cell layers. Cultures were supplemented with IL-11, IL-6, SCF, and Flt3-ligand. After 7 days of incubation in primary culture, hematopoietic cells were harvested from individual wells and replated in secondary culture with semisolid medium containing multiple growth factors. Similar numbers of cells were found in cultures containing cell layers expressing Jagged-1 compared to cultures with parental 3T3 cell layers (5.4 × 105 cells/well ± 0.16 × 105 compared to 3.3 × 105cells/well ± 0.6 × 105, respectively, mean for three experiments ± SEM). There was no difference in the types or numbers of mature myeloid cells found after 7 days in primary culture, as shown by flow analysis using antibodies that recognize granulocyte (Gr-1) and macrophage (Mac-1) associated antigens (data not shown). However, cell-bound Jagged-1 affected the generation or maintenance of a very early precursor cell, HPP-mix. HPP-mix colonies consisted of cells derived from multiple lineages, including granulocytes, macrophages, and erythroid clusters and were larger than 1.5 mm in diameter. There were about four times more HPP-mix colonies formed from cultures previously incubated with layers expressing Jagged-1compared to those without Jagged-1(Fig 4A). However, there were very few HPP-mix remaining in either group, suggesting that under both conditions most HPP-mix had developed into CFU-C. Indeed, cells from the 3T3-Jagged-1 cultures produced four times more colonies of all types compared to cells cultured with parental layers (Fig 4B, P < .05).
Effect of Jagged exogenously expressed from 3T3 fibroblasts on purified hematopoietic precursors. Enriched mouse marrow cells (Lin−Sca-1+c-kit+) were added to wells containing cytokines (IL-11, IL-6, SCF, and Flt3-ligand) and either mitomycin-treated Jagged expressing 3T3 cell layers (▪) or mitomycin-treated parental cell layers (□). After 7 days of incubation, hematopoietic cells were obtained from individual wells and replated in semisolid medium containing growth factors. After 10 days of incubation, the colony types were counted and the mean number of colonies per well was derived for each experiment. Each bar represents the mean number of HPP-mix (A) or CFU-C (B) colonies per well from three separate experiments ± SEM. HPP-mix refers to colonies larger than 1.5 mm in diameter and consisting of multiple lineages including at least granulocyte/macrophage and erythroid clusters.
Effect of Jagged exogenously expressed from 3T3 fibroblasts on purified hematopoietic precursors. Enriched mouse marrow cells (Lin−Sca-1+c-kit+) were added to wells containing cytokines (IL-11, IL-6, SCF, and Flt3-ligand) and either mitomycin-treated Jagged expressing 3T3 cell layers (▪) or mitomycin-treated parental cell layers (□). After 7 days of incubation, hematopoietic cells were obtained from individual wells and replated in semisolid medium containing growth factors. After 10 days of incubation, the colony types were counted and the mean number of colonies per well was derived for each experiment. Each bar represents the mean number of HPP-mix (A) or CFU-C (B) colonies per well from three separate experiments ± SEM. HPP-mix refers to colonies larger than 1.5 mm in diameter and consisting of multiple lineages including at least granulocyte/macrophage and erythroid clusters.
The effect of Jagged-1ext on mouse marrow progenitor cells.
Because the outcome of coculture experiments could be influenced by possible differences between the 3T3-Jagged-1 cell lines and the parental 3T3 cell line in the types or amounts of cytokines made, adhesion molecules expressed, or other factors unrelated toJagged-1 expression, we examined the effect of a Jagged-1ext, consisting of the extracellular domain of Jagged-1, on hematopoietic precursor cells. An myc tag epitope and six histidines were added to the carboxy terminus of the molecule to aid in purification. Jagged-1ext was purified from the supernatant of transiently transfected COS cells using a nickle column. Because several lower molecular weight contaminants were observed, further affinity purification was necessary. Hence, a fraction from the nickle column containing Jagged-1ext was added to Sepharose beads crosslinked with 9E10. To avoid possible denaturation of Jagged-1ext after dissociation from the antibody, cells were incubated directly with the Jagged-1ext-9E10-Sepharose beads. Supernatant from mock-transfected COS cells was treated with the same purification protocol and added to the 9E10 crosslinked Sepharose beads as a control.
To quantitate the effect of Jagged-1ext on hematopoietic precursor cells, 50 enriched mouse Lin−Sca-1+c-kit+marrow cells were deposited into wells containing either Jagged-1ext-9E10-beads or control-9E10-beads in the presence of IL-11, IL-6, SCF, and Flt3-ligand. After 7 days of incubation in primary culture, cells from each well were obtained and plated in semisolid medium to assess clonogenicity. In primary cultures, no difference in cell number was seen between the two culture conditions (data not shown). By 7 days there were between 1 and 5 × 105 cells/well in both culture conditions. Because there were too few cells for flow analysis, the types of mature cells found in the primary cultures were assessed by preparing cyto-centrifuge slides followed by Wright-Giemsa staining. Most mature cells were macrophages and granulocytes and no obvious differences in the distribution of these types of cells were detected between cells removed from the two conditions (data not shown). However, there were twice as many HPP-mix colonies formed from cultures incubated with Jagged-1ext than with control proteins (Fig 5A, P < .025). In addition, in four of five experiments, wells with Jagged-1ext beads contained more CFU-C than control wells (Fig 5B). The mean increase in CFU-C was 21%.
Effect of Jagged coated beads on purified hematopoietic precursors. Enriched mouse marrow cells (Lin−sca-1+c-kit+) were added to wells containing cytokines (IL-11, IL-6, SCF, and Flt3-ligand) and beads coated with Jagged-1ext (▪) or control beads (□). After 7 days of incubation cells were harvested and replated in semisolid medium containing growth factors. After 10 days of incubation the colony types were quantitated and a mean number of colonies per well was derived from each experiment. Each bar represents the mean number of HPP-mix (A) or CFU-C (B) colonies per well from five separate experiments ± SEM. HPP-mix refers to colonies larger than 1.5 mm in diameter and consisting of multiple lineages including at least granulocyte/macrophage and erythroid clusters.
Effect of Jagged coated beads on purified hematopoietic precursors. Enriched mouse marrow cells (Lin−sca-1+c-kit+) were added to wells containing cytokines (IL-11, IL-6, SCF, and Flt3-ligand) and beads coated with Jagged-1ext (▪) or control beads (□). After 7 days of incubation cells were harvested and replated in semisolid medium containing growth factors. After 10 days of incubation the colony types were quantitated and a mean number of colonies per well was derived from each experiment. Each bar represents the mean number of HPP-mix (A) or CFU-C (B) colonies per well from five separate experiments ± SEM. HPP-mix refers to colonies larger than 1.5 mm in diameter and consisting of multiple lineages including at least granulocyte/macrophage and erythroid clusters.
Time course of precursor cell generation.
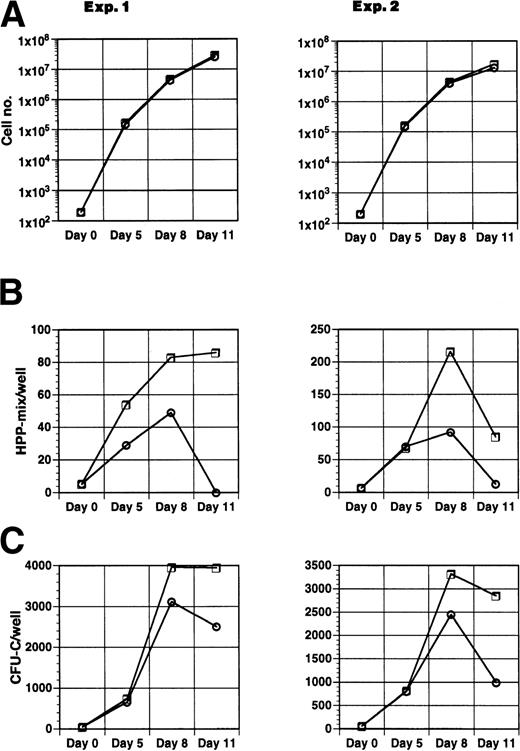
To assess whether Jagged-1ext altered the day of maximal expression of CFU-C or HPP-mix, 200 precursor cells (Lin−Sca-1+c-kit+) were plated in suspension culture in the presence of IL-11, IL-6, SCF, and Flt3-ligand either with Jagged-1ext-9E10-beads or with control-9E10-beads. To maintain density in the primary culture at below 1 × 106 cells/mL during the course of the experiment, cell numbers were reduced on days 5 and 8 by replating 25,000 cells to a new well with fresh medium and fresh Jagged-1ext or control beads. To measure clonogenicity, an aliquot of cells was replated in semisolid medium on 0, 5, 8, and 11 days of culture. Jagged-1ext had no effect on the number of cells generated (Fig 6A). However, cultures with Jagged-1ext produced more HPP-mix than did cultures without Jagged-1ext throughout the course of the experiment (Fig6B). The largest differences were observed at days 8 and 11. In addition, Jagged-1ext affected CFU-C. In both conditions, the number of CFU-C increased until about day 8, when their numbers stopped increasing and started to decrease (Fig 6C). At day 5 there were similar numbers of CFU-C in both conditions, whereas by day 8 there were about 25% more CFU-C in wells containing Jagged-1ext and at day 11 there was 1.5- to 3-fold more CFU-C in the cultures containing Jagged-1ext.
Effect of Jagged-1ext coated beads on the time course of formation of clonogenic cells. Two hundred enriched mouse marrow cells (Lin−sca-1+c-kit+) were added to wells containing cytokines (IL-11, IL-6, SCF, and Flt3-ligand) and beads coated with Jagged-1ext (□) or control beads (○). Cells were removed at time intervals and replated in semi-solid medium. After 10 days the total number of cells per 200 starting cells (A), HPP-mix (B), and CFU-C (C) were determined for each well. Each point represents the mean from 8 wells in experiment 1 and 10 wells in experiment 2.
Effect of Jagged-1ext coated beads on the time course of formation of clonogenic cells. Two hundred enriched mouse marrow cells (Lin−sca-1+c-kit+) were added to wells containing cytokines (IL-11, IL-6, SCF, and Flt3-ligand) and beads coated with Jagged-1ext (□) or control beads (○). Cells were removed at time intervals and replated in semi-solid medium. After 10 days the total number of cells per 200 starting cells (A), HPP-mix (B), and CFU-C (C) were determined for each well. Each point represents the mean from 8 wells in experiment 1 and 10 wells in experiment 2.
Secondary colony formation.
We replated HPP-mix colonies derived from the primary cultures incubated with Jagged-1ext or control beads. Because all colonies from the two conditions were replated, there were more HPP-mix replated from wells where Jagged-1ext was in the primary culture and hence there were more HPP-mix that were replated at the two time points that were evaluated. However, there was very little difference in the characteristics of the HPP-mix colonies derived from the two types of cultures. The percentage of colonies which replated into granulocyte/macrophage or erythroid colonies was the same from the two conditions and the number of granulocyte/macrophage or erythrocyte colonies formed in the tertiary culture was also roughly the same (Table 1).
Replating of Day 5 and Day 8 HPP-Mix Colonies
| . | No. of HPP-Mix in Primary Culture . | No. and Percent That Replate Gran/Macro . | No. and Percent That Replate Gran/ Macro and Eryth . | Mean No. of Colonies in Secondary Cultures . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gran/Macro . | Eryth . | |||||||||
| Jagged . | No Jagged . | Jagged . | No Jagged . | Jagged . | No Jagged . | Jagged . | No Jagged . | Jagged . | No Jagged . | |
| A. Day 5 | ||||||||||
| Exp 1 | 146 | 73 | 91 | 39 | 16 | 9 | 69 | 42 | 13 | 2 |
| 62% | 53% | 11% | 12% | |||||||
| Exp 2 | 112 | 130 | 68 | 91 | 24 | 17 | 60 | 31 | 11 | 12 |
| 61% | 70% | 21% | 13% | |||||||
| Exp 3 | 85 | 32 | 29 | 13 | 23 | 8 | 63 | 75 | 4 | 15 |
| 34% | 41% | 27% | 25% | |||||||
| Mean ± SD | 62 ± 31 | 47 ± 39 | 20 ± 4 | 13 ± 4 | 64 ± 5 | 49 ± 22 | 9 ± 5 | 9 ± 6 | ||
| 52% ± 15 | 54% ± 14 | 16% ± 5 | 13% ± 0.5 | |||||||
| B. Day 8 | ||||||||||
| Exp 1 | 33 | 21 | 18 | 15 | 4 | 1 | 48 | 63 | 15 | 2 |
| 55% | 71% | 12% | 5% | |||||||
| Exp 2 | 97 | 51 | 75 | 34 | 17 | 12 | 47 | 36 | 6 | 11 |
| 77% | 67% | 18% | 24% | |||||||
| Mean ± range | 47 ± 29 | 25 ± 10 | 11 ± 7 | 7 ± 6 | 48 ± 0.5 | 50 ± 14 | 11 ± 4 | 7 ± 6 | ||
| 66% ± 11 | 69% ± 2 | 10% ± 4 | 15% ± 10 | |||||||
| . | No. of HPP-Mix in Primary Culture . | No. and Percent That Replate Gran/Macro . | No. and Percent That Replate Gran/ Macro and Eryth . | Mean No. of Colonies in Secondary Cultures . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gran/Macro . | Eryth . | |||||||||
| Jagged . | No Jagged . | Jagged . | No Jagged . | Jagged . | No Jagged . | Jagged . | No Jagged . | Jagged . | No Jagged . | |
| A. Day 5 | ||||||||||
| Exp 1 | 146 | 73 | 91 | 39 | 16 | 9 | 69 | 42 | 13 | 2 |
| 62% | 53% | 11% | 12% | |||||||
| Exp 2 | 112 | 130 | 68 | 91 | 24 | 17 | 60 | 31 | 11 | 12 |
| 61% | 70% | 21% | 13% | |||||||
| Exp 3 | 85 | 32 | 29 | 13 | 23 | 8 | 63 | 75 | 4 | 15 |
| 34% | 41% | 27% | 25% | |||||||
| Mean ± SD | 62 ± 31 | 47 ± 39 | 20 ± 4 | 13 ± 4 | 64 ± 5 | 49 ± 22 | 9 ± 5 | 9 ± 6 | ||
| 52% ± 15 | 54% ± 14 | 16% ± 5 | 13% ± 0.5 | |||||||
| B. Day 8 | ||||||||||
| Exp 1 | 33 | 21 | 18 | 15 | 4 | 1 | 48 | 63 | 15 | 2 |
| 55% | 71% | 12% | 5% | |||||||
| Exp 2 | 97 | 51 | 75 | 34 | 17 | 12 | 47 | 36 | 6 | 11 |
| 77% | 67% | 18% | 24% | |||||||
| Mean ± range | 47 ± 29 | 25 ± 10 | 11 ± 7 | 7 ± 6 | 48 ± 0.5 | 50 ± 14 | 11 ± 4 | 7 ± 6 | ||
| 66% ± 11 | 69% ± 2 | 10% ± 4 | 15% ± 10 | |||||||
Abbreviations: HPP-Mix, colonies consisting of granulocytes, macrophages, and erythroid clusters and larger than 1.5 mm; Gran/Macro, granulocyte/macrophage colonies; Eryth, erythroid colonies.
DISCUSSION
Previously it was shown that Notch-1 mRNA was detected in human precursors (CD34+ Lin−), indicating a potential role for Notch interactions in hematopoiesis.11In the present study we further assess protein expression of bothNotch-1 and Notch-2 in murine whole marrow and in precursors. Although little or no Notch-1 was seen in whole marrow, it was detected in precursors. Notch-2 was detected in whole marrow and, again, more was seen in precursors. Interestingly, more Notch-2 was detected in murine marrow than Notch-1. Until other antibodies are used to compare Notch-1 and Notch-2 expression, it is unclear if this result is caused by differences between the two antibodies used rather than differences in Notch-1 and Notch-2 protein levels. However, these data indicate clearly that Notch protein is expressed by murine hematopoietic precursors, supporting previous suggestions of a potential role for Notch in hematopoiesis.
In this report we also examine expression of the Notch ligandJagged-1 in mouse hematopoietic cells. Interestingly, no detectible Jagged-1 was seen in whole marrow, Ficoll-Hypaque–separated marrow, or in sorted precursor cells (Lin−Sca-1+c-kit+). However, Jagged-1 was detected in murine stroma and in stromal cell lines, again supporting suggestions for a potential role for Notch-Notch-ligand interactions in hematopoiesis. Additionally, a recent paper reported Jagged-1 expression in human stromal cell lines that supported hematopoiesis.37
To address whether Notch has a role in hematopoiesis, we determined the effect of Notch-ligand interactions on primitive hematopoietic precursors using transfected fibroblasts expressing the full-length human Jagged-1. Previously it was shown that L-cell fibroblasts expressing full-length rat Jagged-1 activated full-lengthNotch-1 expressed in cells of a C2 myoblast cell line and inhibited differentiation of those cells.17 In this report we have transfected a similar fibroblast cell line with a cDNA encoding full-length human Jagged-1. Murine Jagged-1 has not yet been cloned. However, rat Jagged-1 is very similar to human Jagged-1 (95% of the amino acids are identical). In fact, the DSL domain, a region deemed critical for Notch function, is 98% identical between rat and human, differing by only one amino acid. Therefore, it is likely that human Jagged-1 would produce comparable effects to murine Jagged-1.
When we cocultured sorted murine precursor cells (Lin−Sca-1+c-kit+) that endogenously express Notch-1 and Notch-2 with fibroblasts expressing human Jagged-1, we found increased numbers of CFU-C and of a primitive precursor, HPP-mix. The primitive nature of these cells was shown by their formation of colonies of macroscopic size indicative of high proliferative potential, their multi-lineage potential, and their high replating efficiency. These results suggest that Notch ligand promotes the maintenance or expansion of precursor cells in culture presumably by activating the receptor in these cells.
Because the outcome of coculture experiments could be influenced by possible differences between the 3T3-Jagged-1 cell lines and the parental 3T3 cell line in the types or amounts of cytokines made, adhesion molecules expressed, or other factors unrelated toJagged-1 expression, we further examined the effect of the extracellular domain of the Jagged-1 molecule on hematopoietic precursors. In this report we have used Jagged-1ext bound to a substrate, consisting of beads covalently linked to an antibody that recognizes the ligand. We found the same effect on hematopoietic precursors with ligand bound beads as 3T3 cells expressing Jagged-1. In both cases, we saw an increase in the numbers of precursors including total CFU-C and HPP-mix. These results would suggest that in this case, Jagged-1ext bound to beads-activated Notch.
Previous analyses involving truncated forms of Notch ligand would not necessarily predict such an outcome. In C elegans it was shown that soluble ligand (Apx 1) exogenously expressed in vivo rescued a ligand null mutant, indicating that the soluble ligand is able to activate the Celegans Notch homologs Lin-12and Glp-1.38 It was also possible to activate theNotch homologs in some but not all developmental processes inC elegans using a construct that expressed only the DSL domain. In other studies it was shown that a soluble form of human Jagged-1 inhibited the differentiation of 32D cells that were exogenously expressing murine Notch-1.37 However, the opposite result was found in Drosophila, where it was shown that the soluble ligand (Serrate or Delta) or membrane-bound truncated ligand exogenously expressed in vivo inhibits Notchactivity.25,26 In addition, exogenous expression of the extracellular domain plus the transmembrane domain of Xenopusor chicken Delta also inhibits Notch activity duringXenopus eye development or chicken retinal development, respectively.27 28 In our situation, it is conceivable that the binding of Jagged-1ext to bivalent antibody on the beads may affect the structure in such a way that it can mimic the wild-type ligand and thereby activate Notch.
We saw no effect on the types of mature cells formed in our cultures, although we have detected Notch expression in developing myeloid cells and mature monocytes (Flowers et al, submitted). It is possible that Jagged-1 is unable to activate Notch on these cell types whereas Jagged-2, Delta-1, or Delta-3 may be able to activate Notch. It has been shown that in wing development in Drosophila,Notchactivation by Serrate is inhibited by the presence of Fringe, a secreted protein, whereas activation by Delta is not inhibited.39 Consistent with this is a recent report showing the expression of Fringe in hematopoietic cells.40
Overall, our studies show that interactions of Notch expressed on the surface of isolated hematopoietic precursors with its ligand results in increased numbers of a primitive precursor, HPP-mix. These results may indicate that Notch interactions may promote self-renewal of HPP-mix or it may promote self-renewal of an even earlier precursor cell, the transplantable stem cell. Experiments are now in progress to test these possibilities.
ACKNOWLEDGMENT
We thank John E.J. Rasko and Robert G. Andrews for reading the manuscript and for helpful suggestions. We thank David Ish-Horowitz for helpful discussions and help in preparing SER10.
Supported in part by National Institutes of Health (NIH) Grants No. HL54881, NS26084, and the Howard Hughes Medical Institute. I.L.R. is supported by the Human Capital Mobility Program No. CHBICT930657. K.A.M. is supported by NIH Grant No. HL58739-01 and National Cancer Institute Grant No. DHP-144/01. I.D.B. is also supported as a Clinical Research Professor by the American Cancer Society.
Address reprint requests to Irwin D. Bernstein, MD, Pediatric Oncology, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, C1-169, Seattle, WA 98109-1024.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal